Abstract
In many clinical trials, studying neurodegenerative diseases including Parkinson’s disease (PD), multiple longitudinal outcomes are collected in order to fully explore the multidimensional impairment caused by these diseases. The follow-up of some patients can be stopped by some outcome-dependent terminal event, e.g. death and dropout. In this article, we develop a joint model that consists of a multilevel item response theory (MLIRT) model for the multiple longitudinal outcomes, and a Cox’s proportional hazard model with piecewise constant baseline hazards for the event time data. Shared random effects are used to link together two models. The model inference is conducted using a Bayesian framework via Markov Chain Monte Carlo simulation implemented in BUGS language. Our proposed model is evaluated by simulation studies and is applied to the DATATOP study, a motivating clinical trial assessing the effect of tocopherol on PD among patients with early PD.
Keywords: joint model, item-response theory, latent variable, Markov Chain Monte Carlo, mixed model
1 Introduction
In many longitudinal studies and clinical trials, researchers often collect some longitudinal outcomes y. The follow-up may be stopped by a dependent terminal event (e.g. death and dropout) whose probability of occurrence is non-ignorable, i.e. dependent on unobserved values of outcomes or latent variables related to outcomes. The scientific focus is often to study changes in outcomes over time and/or to analyze the relationship between y and time to the terminal event. It has been shown that the methods analyzing y alone are biased while a properly specified joint model provide consistent estimates.1 The approach of joint modeling constructs two sub-models for the longitudinal data and the event time data, linked by a set of subject-specific random effects.2 Many joint models involve a mixed effects model for the longitudinal data and a semiparametric Cox proportional hazard model for the event time.3 Many extensions have been proposed in the joint model literature such as using both random effects and a latent stochastic process to link two sub-models 1; using a spline-based approach to capture the non-linear shapes of subject-specific changes for longitudinal outcomes4; relaxation of the normality assumption on the random effects5; the incorporation of a cured fraction6; and multiple event times.7
However, in many clinical trials studying neurodegenerative diseases such as Parkinson’s disease (PD), Huntington disease, and Alzheimer’s disease, multiple longitudinal outcomes are collected to fully explore the multidimensional impairment caused by these diseases. To properly analyze these longitudinal data, one has to account for three sources of correlation, i.e. inter-source (different measures at the same visit time), longitudinal (same measure at different visit times), and cross correlation (different measures at different visits).8 Multivariate generalized linear mixed effects models have been applied to analyze the multiple longitudinal outcomes in the joint model.4 But the computation associated with the high-dimensional integration is complicated and time-consuming. An alternative approach is the latent variable model.9 Specifically, a continuous latent variable is introduced to represent patients’ underlying disease severity and the observed longitudinal data can be viewed as measurements of the latent variable. Because all outcomes share the same latent variable, the dimensionality of the data can be reduced and fewer parameters are needed. To this end, multilevel item response theory (MLIRT) models have been widely used to analyze longitudinal data in social, behavioral, and health sciences.10–15 Within the MLIRT modeling framework, the observed measurements are viewed as imperfect manifestations of the interaction between subject-specific latent traits and measurement-specific parameters. The latent traits are regressed on covariates of interest (e.g. treatment and disease duration) as well as the confounding variables. All three sources of correlation are accounted for via either random effects or covariance matrix. Advantages of the MLIRT models include better reflection of multilevel data structure, simultaneous estimation of measurement-specific parameters and covariate effects, and accurate inference about high-level measures.16,17 Marginal maximum-likelihood method18 and Bayesian method19 have been used for the MLIRT model inference. Skrondal and Rabe-Hesketh20,21 have provided detailed description and summary of the IRT models.
In this article, we propose a joint model with a MLIRT sub-model for the multiple longitudinal data and a Cox proportional hazard sub-model for time to the dependent terminal event. Two sub-models are linked by random effects denoting the subject-specific disease characteristics. We develop a Bayesian approach via Markov Chain Monte Carlo (MCMC) method for parameter estimation. To the best of our knowledge, there has been no previous work on the joint analysis based on the MLIRT modeling framework. The rest of the article is organized as follows. In Section 2, we describe the joint model, Bayesian inference, and model selection criterion. In Section 3, we apply the joint model to a motivating study. In Section 4, simulation studies are conducted to assess the performance of the proposed method. Section 5 provides a summary and discussion.
2 Model
2.1 Model Formulation and Likelihood
Let yijk be the observed outcome k from patient i at time point j, where i = 1, …, N, j = 1, …, J, and k = 1, …, K. We have coded all outcomes such that larger values are worse clinical conditions. Let yij = (yij1, …, yijk, …, yijK)′ be the vector of observation for patient i at visit j and let yi = (yi1, …, yiK)′ be the outcome vector across visits. Let ti be the observed event time for patient i, and δi (1 if the event is observed and 0 otherwise) be the event indicator. We use a MLIRT sub-model for the multiple longitudinal outcomes and a Cox proportional hazard sub-model for the event time. In level 1 measurement model within the MLIRT framework, we model the binary outcome, the cumulative probabilities of ordinal outcome, and the continuous outcome by a two-parameter model,19 graded response model,19 and common factor model,22 respectively.
| (1) |
| (2) |
| (3) |
where random error for continuous outcomes , ak and bk (positive) are the outcome-specific “difficulty” parameter and “discrimination” parameter, respectively. For the ordinal outcome with nk categories, the order constraint ak1 < · · · < akl < · · · <aknk−1 must be satisfied, and the probability of being in a particular category is p(Yijk = l) = p(Yijk ≤ l |θij) − p(Yijk ≤ l − 1|θij). The continuous latent variable θij represents disease severity for patient i at time j, with higher value denoting more severe status. In the second level latent trait regression model, we postulate
| (4) |
where Xi0 and Xi1 are the covariates of interest associated with the disease severity, Xi0 and Xi1 can share part of or all the covariates. The variable tj is the visit time with t1 = 0 for baseline. The random effects ui0 and ui1 represent the subject-specific baseline disease severity and disease progression rate, respectively, and they follow normal distribution with mean 0 and variances 1 and , respectively, and correlation coefficient ρ. The regression parameter vectors β0 and β1 represent the covariate effects on the baseline disease severity and disease progression rate, respectively. For example, if θij = β01xi + ui0 + [β10 + β11xi + ui1]tj, where xi is an indicator variable of treatment (1 if treatment, 0 otherwise), then β01 is the baseline group difference, and β10 and β10 + β11 are the disease progression rates for the placebo and treatment patients, respectively. The negative significant variable β11 indicates that the treatment is efficacious in slowing down the disease progression. Note that IRT models are over-parameterized because they have more parameters than can be estimated from the data.19 Additional constraints are usually required to make models identifiable. In the aforementioned models, we set Var[ui0] = 1 to obtain Var[θij] = 1 at t = 0 (baseline) to make the discrimination parameter bk identifiable.
One key assumption in the MLIRT model is that all measurements from each patient are independent conditioning on the random effect vector ui = (ui0, ui1)′.19 The conditional likelihood of the multiple longitudinal outcomes for patient i is
| (5) |
where p(yijk|ui) is the conditional density function of yijk obtained from Models (1)–(4). Under the Cox proportional hazard sub-model, the hazard of having a terminal event at time ti is
| (6) |
where ν0 and ν1 measure the association between the two sub-models. Two sub-models are linked together via the shared random effects ui0 and ui1, which is a popular approach in joint modeling.1,3 The covariate vector Xi can be the same or different from Xi0 and Xi1. We have selected piecewise constant function to approximate the baseline hazard function h0(t) because models using a piecewise constant baseline hazard yield good estimators for both fixed effects and frailty,23,24 although fixed cut points need to specified a priori. Given a set of fixed time points 0 = τ0 < τ1 < · · · < τm, and the baseline hazard vector g = (g0, g1, …, gm−1), we define the piecewise constant hazard function as , with indicator function Il (t) = 1 if τl ≤ t < τl+1 and 0 otherwise. The likelihood of event outcome ti and δi for patient i is
| (7) |
where the survival function . Conditional on the random effect vector ui, yi is assumed to be independent of ti. The full likelihood of the joint model for patient i is
| (8) |
where p(ui) is the density function of ui. For notation convenience, we let the difficulty parameter vector be , with ak being numeric for binary and continuous outcomes and ak = (ak1, …, aknk−1)′ for ordinal outcomes. Let the discrimination vector be b = (b1, …, bK)′ and . The unknown parameter vector Φ = (a′, b′, β′, γ′, σu, ρ, σk, ν0, ν1, g′)′. We refer to the proposed joint modeling framework (8) as joint model. We refer to as reduced model, the model assuming the occurrence of the terminal event is independent to the longitudinal outcomes (i.e. ν0 = ν1 = 0).
2.2 Bayesian Estimation and Model Selection
We develop a fully Bayesian approach via the MCMC method to estimate the unknown parameters. The model fitting is implemented using the BUGS language. Vague prior distributions are imposed on all parameters. Specifically, a normal distribution N(0, 100) is used for all components in a, β, and γ and for ν0 and ν1. We let all components in b and g have Uniform[0, 20] as prior distribution to ensure non-negativity. To satisfy the order constraint of ak for the ordinal outcome with nk categories, we let ak1 ~ N(0, 100), and akl = ak,l−1 + ωl for l = 2, …, nk − 1, with ωl ~ N(0, 100)I(0, ), i.e. normal distribution left truncated at 0. We use the prior distributions σk ~ Gamma(0.01, 0.01) and ρ ~ Uniform[−1, 1]. Multiple chains with over-dispersed initial values are run to analyze data and the Gelman–Rubin diagnostic25 is used to ensure the scale reduction R̂ of all parameters are smaller than 1.1. Moreover, we use the trace plots and autocorrelation functions25 to ensure the chain convergence.
We have adopted two model selection criteria, i.e. Deviance Information Criterion (DIC)26 and Bayes factor (BF).27 The deviance statistics is defined as D(θ) = −2 log f(y|θ) + 2 log h(y), where f(y|θ) is the likelihood function for the observed data y given the parameter vector θ, and h(y) is some standardizing function of the data alone. The DIC is defined as DIC = D̄ + pD, where D̄ = Eθ|y[D(θ)] is the posterior expectation of the deviance, D(θ̄) = D(Eθ|y[θ]) is the deviance evaluated at the posterior mean of parameters, and pD = D̄ − D(θ̂) is the effective number of parameters, which captures model complexity. A smaller DIC indicates a better fit when comparing models.
BFs is a Bayesian alternative to p values for testing hypotheses and for quantifying the degree to which observed data support or conflict with a hypothesis. Let two competing models be M1 and M2. The BF in favor of model M1 over M2 is defined as:
| (9) |
where p(Mi) is the prior probability of model Mi, where i = 1, 2, p(Mi|y) is the posterior probability of model Mi, and p(y|Mi) is the predictive probability of observing y under model Mi, and p(y|Mi) = ∫ f(y|θi, Mi) p(θi|Mi)dθi, where p(θi|Mi) is the prior distribution for parameter vector θi under model Mi. When the BF is greater than 100, decisive evidence is shown in favor of model M1. To avoid the integral involved in computation of BF, the Laplace–Metropolis estimator based on the normal distribution28 is adopted to approximate the predictive probability. Specifically, p(y|Mi) ≈ (2π)di/2|Σi|1/2f(y|θ̄i, Mi) p(θ̄i|Mi), where di is the number of the parameters in θi, Σi is the posterior covariance matrix of θi, θ̄i is the posterior mean of parameters, p(θ̄i|Mi) is the prior probability of parameters evaluated at θ̄i, and f(y|θ̄i, Mi) is the likelihood when parameters are at the posterior mean values.
3 Application
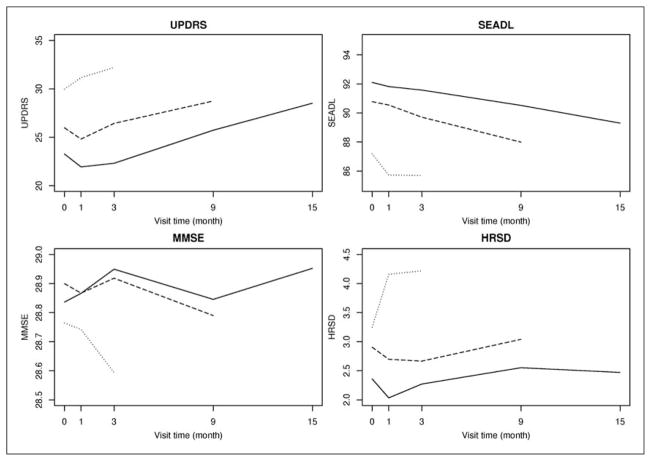
Our work is motivated by the Deprenyl And Tocopherol Antioxidative Therapy of Parkinsonism (DATATOP) study. DATATOP was a double-blind, placebo-controlled multicenter clinical trial to determine whether deprenyl or tocopherol, alone or in combination, administered to patients with early PD will prolong the time until dopaminergic therapy to treat emerging disability.29 Totally 800 patients were randomly assigned in a 2 × 2 factorial design to receive double-placebo, active tocopherol alone, active deprenyl alone, and both active tocopherol and deprenyl. In this article, we investigate the effect of tocopherol and we define the placebo group as patients who did not receive tocopherol (double-placebo and active deprenyl alone groups, 401 patients), and the treatment group as patients who received tocopherol (active tocopherol alone and both active tocopherol and deprenyl groups, 399 patients). The longitudinal outcomes are Unified Parkinson’s Disease Rating Scale (UPDRS) total score, Schwab and England activities of daily living (SEADL), Mini-Mental State Exam (MMSE), and Hamilton rating scale for depression (HRSD) collected at baseline, months 1, 3, 9, and 15. UPDRS total score evaluates patients’ mentation, behavior, activities of daily living, and motor function. It is an approximate continuous variable with integer value from 0 (normal) to 176 (severe).30 SEADL is a measurement of activities of daily living and it is an ordinal variable with integer value from 0 (severe) to 100 (normal) incrementing by 5.31 MMSE measures patients’ cognitive impairment and it is an ordinal variable with integer value from 0 (severe) to 30 (normal). HRSD, a depression test measuring the severity of clinical depression symptoms, is an ordinal variable with integer value from 0 (normal) to 52 (severe). During the course of the study, 192 and 184 patients in the placebo and treatment groups, respectively, reached a level of functional disability sufficient to warrant the initiation of dopaminergic therapy, which is a symptomatic therapy to provide temporary relief of PD symptoms. In this case, only the observed outcomes before the initiation of dopaminergic therapy can be used in the assessment of treatment efficacy because dopaminergic therapy can significantly change the values of the outcomes for a short period. Therefore, these individuals would have missing data after the initiation of dopaminergic therapy. Figure 1 displays the mean UPDRS, SEADL, MMSE, and HRSD measurements over time for DATATOP patients with follow-up time less than 6 months (dotted line), 6–12 months (dashed line), and more than 12 months (solid line). Patients with shorter follow-up time tend to have higher UPDRS and HRSD values and lower SEADL and MMSE values, indicating worse clinical outcomes. This phenomenon suggests the existence of association between the longitudinal outcomes and the time to dopaminergic therapy.
Figure 1.
Mean longitudinal measures over time. Follow-up time: less than 6 months (dotted line), 6–12 months (dashed line), and more than 12 months (solid line).
To analyze the DATATOP dataset, we have recoded the outcomes SEADL and MMSE so that higher values in all outcomes are worse clinical conditions. Moreover, we combine some categories in the outcomes SEADL, MMSE, and HRSD with zero or small number of individuals so that they have 7, 7, and 10 categories, respectively. The median follow-up time is 14 months (range: 0–25 months). We first perform the Schoenfeld residual test, the non-significant result (p = 0.43) indicates the validity of the proportionality assumption. To use the MLIRT sub-model, we let Xi0 = 0 and consider the treatment variable xi (1 treatment, and 0 if placebo) as the only covariate in Xi1. Hence, the level 2 model (4) is θij = ui0 + (β10 + β11xi + ui1)tij, with visit time being tij = (0, 1, 3, 9, 15) and the random effects ui0 and ui1 representing the subject-specific baseline disease severity and disease progression rate, respectively. The survival time is time to the initiation of dopaminergic therapy. The treatment variable is the single covariate in the Cox sub-model so that h(ti) = h0(ti) exp(γxi + ν0ui0 + ν1ui1) in Model (6).
For model selection and comparison, we compute the DIC and BF illustrated in Section 2.2. The joint model has smaller DIC (53,168), comparing with 53,502 from the reduced model. The BF in favor of the joint model over the reduced model is much larger than 100, indicating decisive evidence in favor of the joint model according to the interpretation proposed by Kass and Raftery.27 Table 1 compares the posterior mean, standard deviation (SD), and 95% equal-tail credible intervals from the reduced and the best fit joint models. The results from the joint model indicate that the placebo patients have significant disease progression at the rate of 0.392 units per month (β̂10, 95% CI: [0.343, 0.446]). In comparison, the treatment patients have disease progression rate of 0.345 units per month (β̂10 + β̂11, 95% CI: [0.237, 0.461]) with insignificant tocopherol treatment effect of slowing down the disease progression rate by −0.047 per month (β̂11, 95% CI: [−0.106, 0.015]). Moreover, tocopherol decreases the hazard of the initiation of dopaminergic therapy by 5% (γ̂ = −0.054, 1 − exp(−0.054) = 0.05, 95% CI: [−0.27,0.24]). The insignificant tocopherol effect is consistent with Shoulson.29 We observe that ν̂0 and ν̂1 are positive and significantly different from zero, (ν̂0 = 0.348, 95% CI: [0.144, 0.511], and ν̂2 = 3.854, 95% CI: [2.497, 5.642]), suggesting that the patients with worse baseline disease severity (larger ui0) and faster disease progression rate (larger ui1) tend to have higher hazard of need for dopaminergic therapy and vice versa. Both the reduced and joint models give similar estimates to the outcome-specific parameters (a and b
Table 1.
Parameter estimations from joint and reduced modeling and model comparison based on DATATOP trial.
| Parameters | Reduced model
|
Joint model
|
||||
|---|---|---|---|---|---|---|
| Mean | SD | 95% CI | Mean | SD | 95% CI | |
| For longitudinal outcomes | ||||||
| β10 | 0.307 | 0.022 | 0.264, 0.351 | 0.392 | 0.026 | 0.343, 0.446 |
| β11 | −0.051 | 0.030 | −0.108, 0.008 | −0.047 | 0.031 | −0.106, 0.015 |
| P | 0.297 | 0.070 | 0.162, 0.439 | 0.415 | 0.062 | 0.294, 0.535 |
| σu | 0.241 | 0.017 | 0.209, 0.275 | 0.287 | 0.023 | 0.244, 0.334 |
| For survival | ||||||
| Γ | −0.036 | 0.100 | −0.237, 0.154 | −0.054 | 0.138 | −0.321, 0.216 |
| ν0 | 0.348 | 0.093 | 0.144, 0.511 | |||
| ν1 | 3.854 | 0.804 | 2.497, 5.642 | |||
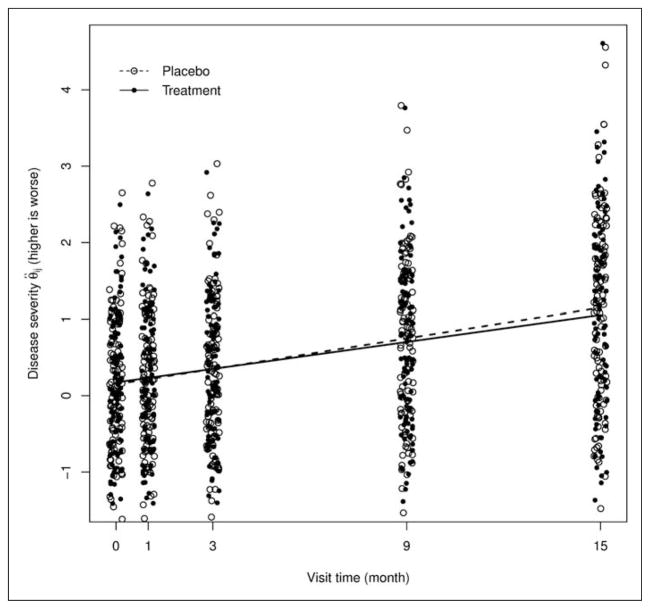
To visualize the difference in the disease progression rates in two groups, Figure 2 displays the estimates of the latent disease severity θij of 100 randomly selected patient at each visit, together with the lowess smooth curves (based on all patients) denoted by the dashed (placebo group) and solid (treatment group) lines, respectively. Figure 2 suggests that two groups have similar disease progression rate before month 9 and the placebo patients deteriorate at a slightly faster rate starting from month 9, as manifested by the departure of two curves.
Figure 2.
Estimates of the subject-specific disease severity 1 θij at each visit and the lowess curve for two groups.
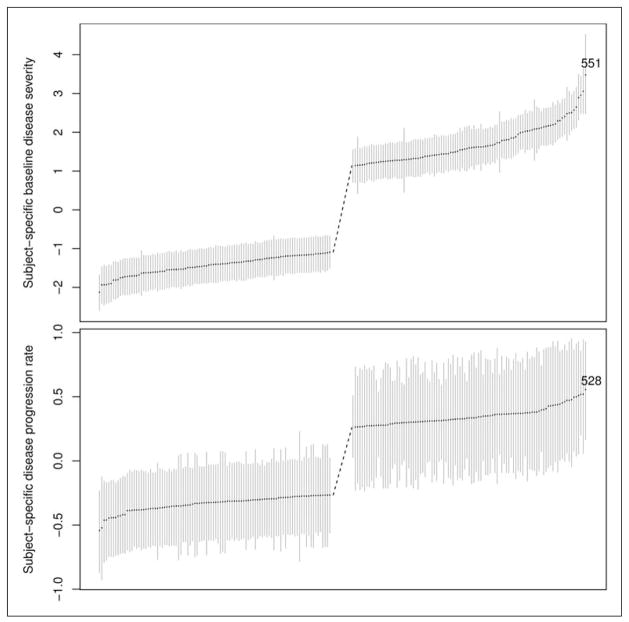
Table 1 also shows positive correlation coefficient ρ between ui0 and ui1 (0.415, 95% CI: [0.294,0.535]), suggesting that the patient with worse baseline disease severity tend to have faster disease deterioration and vice versa. To obtain more insight into ui0, ui1, and ρ, we plot in Figure 3 ui0 (upper panel) and ui1 (lower panel) with their 95% credible intervals. Patients are sorted so that patients at the left have milder disease at baseline and slower disease progression rate (larger ranks), while patients at the right have more severe disease at baseline and faster disease progression rate (smaller ranks). For clarity purpose, only patients with the smallest 100 and the largest 100 ranks are displayed in the figure. We use two patients as an example to illustrate the effect of ρ. Patient 551 has the worst baseline disease severity and he/she ranks No. 8 in the disease progression rate. Patient 528 has the fastest disease progression rate and he/she ranks No. 5 in the baseline disease severity.
Figure 3.
The ranking of subject-specific baseline disease severity (upper panel) and disease progression rate (lower panel) with point estimates and 95% CI. The numbers in the figures are patient numbers.
4 Simulation
In this section, we conduct two simulation studies to compare the performance of the proposed joint model and the reduced model. In the first simulation study, there is a strong correlation between the survival time and the longitudinal outcome (i.e. ν0 = 0.4, ν1 = 1), whereas in the second simulation study, there is no correlation (i.e. ν0 = ν1 = 0). The simulated datasets have a data structure and parameters similar to the DATATOP study. In each simulation study, we simulate 500 datasets with sample size N = 800 (400 in both treatment and placebo groups).
We simulate one continuous (yij1) and three ordinal (denoted by yij2, yij3, and yij4 with 7, 7, and 10 categories, respectively) outcomes at five visits (e.g. baseline, months 1, 3, 9, 15). Treatment variable (xi = 1 if treatment, and 0 if placebo) is the only covariate under consideration and we assume that the treatment is effective. The level 2 model (4) is θij = ui0 + (β10 + β11xi + ui1)tij, with visit time being tij = (0, 1, 3, 9, 15), and the Cox sub-model (6) is h(ti) = h0(ti) exp(γxi + ν0ui0 + ν1ui1). We set β10 = 0.4, β11 = −0.5, and γ = −0.7. Note that β11 is negative so that we expect the treated patients to have smaller θij and better clinical status. Similarly, γ is negative so that the treated patients are expected to have smaller event hazard at any specific time. We simulate random effects ui = (ui0, ui1)′ ~ N2(0, Σ), where and ρ = 0.4, σu = 1.3. For continuous outcome yij1, we set a1 = 25, b1 = 10 and σ1 = 5, and simulate from . For ordinal outcomes, we let a2 = (−2.7, − 0.6, 2, 2.8, 5, 6), b2 = 2, a3 = (−0.1, 1, 1.8, 2.6, 3.3, 4), b3 = 0.4, a4 = (−1, − 0.1, 0.5, 1, 1.5, 2, 2.4, 2.8, 3.3), b4 = 0.7, and use Model (2) to obtain the probability of being in each category for each ordinal outcome at every visit. Then, three ordinal outcomes are simulated from multinomial distributions.
The time to terminal event is simulated from the Cox sub-model with a piecewise constant baseline hazard function. Given a set of fixed time points 0 = τ0 < τ1 < · · · < τm and the baseline hazard vector g = (g0, g1, …, gm−1), we define the piecewise constant baseline hazard function as , with Il (t) = 1 if τl ≤ t ≤ τl+1. For a given interval τa ≤ ti< τa+1 with a = 0, …, m − 1, the survival function is . To solve this equation for ti, we have
| (10) |
The condition τa ≤ ti< τa+1 imposes the following constraint: . To generate the event time ti, we set the piecewise baseline hazard vector g = (0.01, 0.05, 0.13) at the fixed time points τ = (0, 8, 13, 30). We generate the censoring time from Uniform[10, 20] and δi = 1 if the event time generated from equation (10) is not larger than the censoring time.
In each simulation study, we run two parallel MCMC chains with over-dispersed initial values. Each chain is run for 10,000 iterations. The first 5000 iterations are discarded as burn-in, and the remaining 5000 samples are used to obtain the posterior distribution of the parameters. We have computed the bias (the average of the posterior means minus the true values), standard error (SE, the square root of the average of the posterior variance), SD (the standard deviation of the posterior means), and coverage probabilities (CPs) of 95% equal-tail credible intervals from the reduced and joint models.
Table 2 displays the results from the first simulation study in which the occurrence of the terminal event is strongly correlated with the longitudinal outcomes. The joint model generally provides estimates with negligible bias, SE close to SD, and the CPs reasonably close to 0.95. We notice that the CP of ν1 is slightly off from 0.95, indicating some difficulty in distinguishing the random effects as reported in Henderson et al.1 These results suggest that the joint model can generally recover the true values in the presence of dependent terminal event. In contrast, the reduced model gives severely biased estimates and the CPs are far away from the nominal value. Specifically, the treatment effect parameter β11 is biased toward zero and it is thus less likely to detect the treatment effect if the treatment effect is present. Because the parameters ν0 and ν1 are set to be positive, the patients with worse baseline disease severity (larger ui0) and faster disease progression rate (larger ui1) tend to have a terminal event earlier. By ignoring this phenomenon and treating the missing data after the terminal event as missing at random, the reduced model tends to reduce the difference between two groups and therefore underestimate the treatment effect. This finding is consistent with the literature of the univariate longitudinal data analysis with dependent dropout.32 In addition, both models provide reasonable estimates to the difficulty and discriminating parameter vectors a and b
Table 2.
Simulation results from the reduced and joint models when the terminal event is dependent on the longitudinal outcomes.
| Parameters | Reduced model
|
Joint model
|
||||||
|---|---|---|---|---|---|---|---|---|
| Bias | SE | SD | CP | Bias | SE | SD | CP | |
| For longitudinal outcomes | ||||||||
| β10 = 0.4 | −0.193 | 0.065 | 0.065 | 0.178 | 0.027 | 0.073 | 0.079 | 0.906 |
| β11 = −0.5 | 0.068 | 0.089 | 0.089 | 0.862 | −0.011 | 0.095 | 0.096 | 0.944 |
| ρ = 0.4 | −0.033 | 0.037 | 0.036 | 0.860 | 0.006 | 0.036 | 0.038 | 0.902 |
| σu = 1.3 | −0.099 | 0.047 | 0.045 | 0.466 | 0.022 | 0.054 | 0.053 | 0.930 |
| For survival | ||||||||
| γ = −0.7 | 0.209 | 0.112 | 0.115 | 0.536 | −0.072 | 0.151 | 0.152 | 0.910 |
| ν0 = 0.4 | −0.001 | 0.075 | 0.075 | 0.934 | ||||
| ν1 = 1.0 | 0.070 | 0.090 | 0.096 | 0.896 | ||||
Table 3 displays the results from the second simulation study in which the reduced model is the correct model. The reduced model provides estimates with small bias and CPs reasonably close to the nominal level. The results indicate that the reduced model can successfully recover the true values under independent terminal event. In comparison, under model overparameterization, the results from the joint model still have reasonably small bias, CPs close to nominal level, and it does not inflate the SEs. The estimates of the parameters ν0 and ν1 are correctly close to zero, suggesting that the joint model is still a reasonable model even when it is overparameterized. Moreover, both models provide reasonable estimates to the difficulty and discriminating parameter vectors a and b
Table 3.
Simulation results from the reduced and joint models when the terminal event is independent on the longitudinal outcomes.
| Parameters | Reduced model
|
Joint model
|
||||||
|---|---|---|---|---|---|---|---|---|
| Bias | SE | SD | CP | Bias | SE | SD | CP | |
| For longitudinal outcomes | ||||||||
| β10 = 0.4 | 0.002 | 0.068 | 0.070 | 0.932 | 0.001 | 0.068 | 0.069 | 0.930 |
| β11 = −0.5 | −0.003 | 0.092 | 0.095 | 0.944 | −0.001 | 0.093 | 0.096 | 0.946 |
| ρ = 0.4 | 0.004 | 0.034 | 0.035 | 0.934 | 0.005 | 0.034 | 0.035 | 0.934 |
| σu = 1.3 | 0.003 | 0.048 | 0.049 | 0.946 | 0.001 | 0.048 | 0.048 | 0.954 |
| For survival | ||||||||
| γ =−0.7 | −0.028 | 0.129 | 0.128 | 0.944 | −0.009 | 0.141 | 0.138 | 0.950 |
| ν0 = 0 | −0.009 | 0.075 | 0.078 | 0.940 | ||||
| ν1 = 0 | 0.006 | 0.061 | 0.062 | 0.926 | ||||
In conclusion, the simulation results suggest that in the presence of dependent terminal event, the joint model provide more accurate estimates for the MLIRT and Cox regression parameters and the random effects parameters. Under independent terminal event, the joint model provides results comparable with the reduced model.
5 Discussion
In clinical trials, it is quite common to have longitudinal outcomes subject to dependent terminal event. Previous work of joint modeling for this type of data has been mainly focused on a single longitudinal outcome accounting for the dependent censoring. In this article, we have proposed a joint modeling framework to jointly analyze the multiple longitudinal data subject to dependent terminal event using the MLIRT sub-model and the Cox proportional hazard sub-model. Two sub-models are linked together via shared random effects representing the subject-specific baseline disease severity and disease progression rate, respectively. The proposed joint model has a better fit than the reduced model in the analysis of the DATATOP dataset. We have found that the treatment tocopherol is insignificant in slowing the PD disease progression. Moreover, we have identified a significant positive correlation between the multiple longitudinal outcomes and the terminal event, in addition to the positive significant correlation between the baseline disease severity and disease progression rate. The simulation studies have shown that in the presence of dependent terminal event, the joint model successfully recovers the true parameters whereas the reduced model underestimates the treatment effect and has large bias in the regression and random effects parameters. Under the scenario of independent terminal event, the joint model provides results comparable with the reduced model.
Our method can be extended to robust inference to handle outlying observations in the longitudinal outcomes. One direction is to relax the normality assumption for the random errors of the continuous outcome to some long-tailed or heavy-tailed distributions, e.g. normal/ independent distributions,33 skew-normal independent distributions,34 and generalized skew-elliptical distributions.35 Another issue is about the assumption of homogeneous random covariance matrix (the matrix is the same for all subjects). Accounting for heterogeneity in random covariance matrix has been investigated in generalized linear models,36 non-linear mixed models,37 and linear mixed models.38 The use of the heterogenous random covariance matrix in the joint modeling framework of the MLIRT models warrants further investigation.
Acknowledgments
This work was supported by two NIH/NINDS grants U01NS043127 and U01NS43128. Computations were performed on the high-performance computational capabilities of the Linux cluster system at University of Texas School of Public Health (UTSPH). The authors express appreciation to UTSPH information technology staff for their technical support of the cluster.
Footnotes
Reprints and permissions: sagepub.co.uk/journalsPermissions.nav
References
- 1.Henderson R, Diggle P, Dobson A. Joint modelling of measurements and event time data. Biostatitics. 2000;1:465–480. doi: 10.1093/biostatistics/1.4.465. [DOI] [PubMed] [Google Scholar]
- 2.Tsiatis A, Davidian M. Joint modelling of longitudinal and time-to-event data: an overview. Stat Sin. 2004;14:809–834. [Google Scholar]
- 3.Wulfsohn M, Tsiatis A. A joint model for survival and longitudinal data measured with error. Biometrics. 1997;53:330–339. [PubMed] [Google Scholar]
- 4.Brown E, Ibrahim J, DeGruttola V. A flexible B-Spline model for multiple longitudinal biomarkers and survival. Biometrics. 2005;61:64–73. doi: 10.1111/j.0006-341X.2005.030929.x. [DOI] [PubMed] [Google Scholar]
- 5.Rizopoulos D, Ghosh P. A Bayesian semiparametric multivariate joint model for multiple longitudinal outcomes and a time-to-event. Stat Med. 2011;30:1366–1380. doi: 10.1002/sim.4205. [DOI] [PubMed] [Google Scholar]
- 6.Brown E, Ibrahim J. Bayesian approaches to joint cure-rate and longitudinal models with applications to cancer vaccine trials. Biometrics. 2003;59:686–693. doi: 10.1111/1541-0420.00079. [DOI] [PubMed] [Google Scholar]
- 7.Elashoff R, Li G, Li N. An approach to joint analysis of longitudinal measurements and competing risks failure time data. Stat Med. 2007;26:2813–2835. doi: 10.1002/sim.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Brien L, Fitzmaurice G. Analysis of longitudinal multiple-source binary data using generalized estimating equations. J R Stat Soc Ser C Appl Stat. 2004;53:177–193. [Google Scholar]
- 9.Wang C, Douglas J, Anderson S. Item response models for joint analysis of quality of life and survival. Stat Med. 2002;21:129–142. doi: 10.1002/sim.989. [DOI] [PubMed] [Google Scholar]
- 10.Glas C, Geerlings H, Van de laar M, et al. Analysis of longitudinal randomized clinical trials using item response models. Contemp Clin Trials. 2008;30:158–170. doi: 10.1016/j.cct.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Huang L, Wang W. The generalized multilevel facets model for longitudinal data. J Educ Behav Stat. 2012;37:231–255. [Google Scholar]
- 12.Wang W, Liu C. Formulation and application of the generalized multilevel facets model. Educ Psychol Meas. 2007;67:583–605. [Google Scholar]
- 13.Bacci S, Caviezel V. Multilevel IRT models for the university teaching evaluation. J Appl Stat. 2011;38:2775–2791. [Google Scholar]
- 14.Douglas J. Item response models for longitudinal quality of life data in clinical trials. Stat Med. 1999;18:2917–2931. doi: 10.1002/(sici)1097-0258(19991115)18:21<2917::aid-sim204>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 15.Andrade D, Tavares H. Item response theory for longitudinal data: population parameter estimation. J Multivariate Anal. 2005;95:1–22. [Google Scholar]
- 16.Maier K. A Rasch hierarchical measurement model. J Educ Behav Stat. 2001;26:307–330. [Google Scholar]
- 17.Kamata A. Item analysis by the hierarchical generalized linear model. J Educ Meas. 2001;38:79–93. [Google Scholar]
- 18.Mislevy R. Estimation of latent group effects. J Am Stat Assoc. 1985;80:993–997. [Google Scholar]
- 19.Fox J. Bayesian item response modeling: theory and applications. New York, USA: Springer-Verlag; 2010. [Google Scholar]
- 20.Skrondal A, Rabe-Hesketh S. Generalized latent variable modeling: multilevel, longitudinal, and structural equation models. Boca Raton, FL: CRC Press; 2004. [Google Scholar]
- 21.Skrondal A, Rabe-Hesketh S. Latent variable modelling: a survey. Scand J Stat. 2007;34:712–745. [Google Scholar]
- 22.Lord F, Novick M, Birnbaum A. Statistical theories of mental test scores. Boston, MA: Addison-Wesley; 1968. [Google Scholar]
- 23.Lawless J, Zhan M. Analysis of interval-grouped recurrent-event data using piecewise constant rate functions. Canad J Stat. 1998;26:549–565. [Google Scholar]
- 24.Feng S, Wolfe R, Port F. Frailty survival model analysis of the national deceased donor kidney transplant dataset using Poisson variance structures. J Am Stat Assoc. 2005;100:728–735. [Google Scholar]
- 25.Gelman A, Carlin J, Stern H, et al. Bayesian data analysis. Boca Raton, FL: CRC Press; 2004. [Google Scholar]
- 26.Spiegelhalter D, Best N, Carlin B, et al. Bayesian measures of model complexity and fit. J R Stat Soc Ser B Stat Methodol. 2002;64:583–639. [Google Scholar]
- 27.Kass R, Raftery A. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- 28.Lewis S, Raftery A. Estimating Bayes factors via posterior simulation with the Laplace–Metropolis estimator. J Am Stat Assoc. 1997;92:648–655. [Google Scholar]
- 29.Shoulson I. DATATOP: a decade of neuroprotective inquiry. Parkinson Study Group. Deprenyl and Tocopherol Antioxidative Therapy of Parkinsonism. Ann Neurol. 1998;44:S160–S166. [PubMed] [Google Scholar]
- 30.Bushnell D, Martin M. Quality of life and Parkinson’s disease: translation and validation of the US Parkinson’s disease questionnaire (PDQ-39) Qual Life Res. 1999;8:345–350. doi: 10.1023/a:1008979705027. [DOI] [PubMed] [Google Scholar]
- 31.McRae C, Diem G, Vo A, et al. Schwab & England: standardization of administration. Movement Disord. 2000;15:335–336. doi: 10.1002/1531-8257(200003)15:2<335::aid-mds1022>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 32.Touloumi G, Babiker AG, Pocock SJ, et al. Impact of missing data due to drop-outs on estimators for rates of change in longitudinal studies: a simulation study. Stat Med. 2001;20:3715–3728. doi: 10.1002/sim.1114. [DOI] [PubMed] [Google Scholar]
- 33.Lange K, Sinsheimer J. Normal/independent distributions and their applications in robust regression. J Comput Graph Stat. 1993;2:175–198. [Google Scholar]
- 34.Lachos V, Ghosh P, Arellano-Valle R. Likelihood based inference for skew-normal independent linear mixed models. Stat Sin. 2010;20:303. [Google Scholar]
- 35.Genton M, Loperfido N. Generalized skew-elliptical distributions and their quadratic forms. Ann Inst Stat Math. 2005;57:389–401. [Google Scholar]
- 36.Chiu T, Leonard T, Tsui K. The matrixlogarithmic covariance model. J Am Stat Assoc. 1996;91:198–210. [Google Scholar]
- 37.Davidian M, Giltinan D. Nonlinear models for repeated measurement data. Vol. 62. Boca Raton, FL: Chapman & Hall/CRC; 1995. [Google Scholar]
- 38.Pourahmadi M, Daniels M. Dynamic conditionally linear mixed models for longitudinal data. Biometrics. 2002;58:225–231. doi: 10.1111/j.0006-341x.2002.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]