Abstract
Variants of the contactin associated protein-like 2 (Cntnap2) gene are risk factors for language-related disorders including autism spectrum disorder, specific language impairment, and stuttering. Songbirds are useful models for study of human speech disorders due to their shared capacity for vocal learning, which relies on similar cortico-basal ganglia circuitry and genetic factors. Here, we investigate Cntnap2 protein expression in the brain of the zebra finch, a songbird species in which males, but not females, learn their courtship songs. We hypothesize that Cntnap2 has overlapping functions in vocal learning species, and expect to find protein expression in song-related areas of the zebra finch brain. We further expect that the distribution of this membrane-bound protein may not completely mirror its mRNA distribution due to the distinct subcellular localization of the two molecular species. We find that Cntnap2 protein is enriched in several song control regions relative to surrounding tissues, particularly within the adult male, but not female, robust nucleus of the arcopallium (RA), a cortical song control region analogous to human layer 5 primary motor cortex. The onset of this sexually dimorphic expression coincides with the onset of sensorimotor learning in developing males. Enrichment in male RA appears due to expression in projection neurons within the nucleus, as well as to additional expression in nerve terminals of cortical projections to RA from the lateral magnocellular nucleus of the nidopallium. Cntnap2 protein expression in zebra finch brain supports the hypothesis that this molecule affects neural connectivity critical for vocal learning across taxonomic classes.
Keywords: autism, birdsong, Caspr2, speech, zebra finch
INTRODUCTION
Language is a complex phenotype unique to humans, though facets of the behavior are shared with other species. Vocal learning, the ability to imitate or to produce novel sounds, is rare in the animal kingdom; so far found only in bats, cetaceans, elephants, pinnipeds, and songbirds. Humans are the only living primate species with this ability (Knornschild et al., 2010; Fitch, 2012; Stoeger et al., 2012). Genes underlying vocal learning and language are beginning to emerge with a major breakthrough being the identification of Forkhead Box P2 (FOXP2) as the monogenetic locus for a human speech disorder. (Abbreviations in all capitals denote the human form of the molecule, lowercase is used for animal homologs, and italics denote nucleic acids). FOXP2 is a transcription factor, and a mutation in its DNA binding domain leads to orofacial dyspraxia in a multigenerational pedigree known as the KE family (Lai et al., 2001). Additional FOXP2 mutations are associated with specific language impairment (SLI) and developmental verbal dyspraxia, further strengthening the link between the gene and language ability (Graham and Fisher, 2012). As a transcription factor, FOXP2’s effects on language must be mediated through its gene targets. Chromatin immunoprecipitation has revealed that contactin associated protein-like 2 (CNTNAP2) is a direct transcriptional target of FOXP2 (Vernes et al., 2008). CNTNAP2 is a particularly interesting target because it has independently been associated with a language-related disorder. Specifically, Old Order Amish children afflicted with cortical dysplasia-focal epilepsy (CDFE) harbor a deletion in CNTNAP2. CDFE is characterized by epilepsy, mental retardation, hyperactivity, impaired social behaviors, and language regression. A majority of affected children meet criteria for autism spectrum disorder (ASD), of which language impairment is a core deficit (Strauss et al., 2006). Within the general population, CNTNAP2 polymorphisms are associated with language-related disorders, including increased risk for ASD (Arking et al., 2008; Li et al., 2010), delayed age of first word (Alarcón et al., 2008), SLI (Newbury et al., 2011; Peter et al., 2011; Whitehouse et al., 2011), and decreased long-range connectivity of the medial prefrontal cortex (Scott-Van Zeeland et al., 2010).
The mechanistic basis of these disorders is still unclear. The best characterized function of Cntnap2 is to cluster voltage-gated potassium channels (VGKCs) to the juxtaparanodes of nerves (Poliak et al., 2003; Horresh et al., 2008). In the central nervous system, Cntnap2 may also affect synaptic development (Anderson et al., 2012). Transgenic mice lacking Cntnap2 exhibit behavioral abnormalities reminiscent of patients with CDFE, namely epilepsy, hyperactivity, diminished social activity, repetitive behaviors, and reduced frequency of ultrasonic vocalizations when pups are separated from their dams (Peñagarikano et al., 2011). This diminished vocal behavior could be due to vocal impairment or lack of motivation as a form of reduced social activity. In either case, this aspect of the model is limited because pup isolation calls are innate. Songbirds, including zebra finches, offer an advantageous model for studying the impact of Cntnap2 given that they are vocal learners with a well-characterized neural circuitry that underlies this ability.
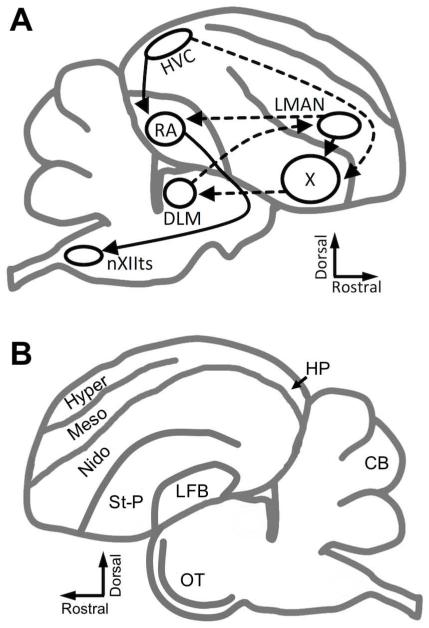
Like other songbirds, zebra finches possess a distinct set of interconnected brain nuclei dedicated to vocal learning and production termed the ‘song circuit’ (Fig. 1). The circuit consists of two pathways: the posterior vocal pathway, required for vocal production, includes a projection from the cortical nucleus HVC (proper name; Reiner et al., 2004) to the robust nucleus of the arcopallium (RA), which in turn projects to the hypoglossal nucleus (nXIIts) that controls the avian vocal organ, the syrinx (Nottebohm et al., 1976). The anterior forebrain pathway (AFP), required for song modification (Brainard and Doupe, 2000), begins with a separate subset of HVC projections to the striatopallidal nucleus area X, which projects to the medial portion of the dorsolateral nucleus of the anterior thalamus (DLM), which then projects to the lateral magnocellular nucleus of the nidopallium (LMAN), which sends nerves terminals to RA as well as back to area X. This latter pathway is a cortical-basal ganglia-thalamo-cortical loop similar to the circuitry thought to underlie vocal learning in humans (Simonyan et al., 2012). An advantage of the zebra finch model is that vocal learning behavior and anatomy is sexually dimorphic. Females have an incomplete song circuit in which area X is not fully developed (Nottebohm et al., 1976), and RA is not innervated by HVC, causing the nucleus to shrink through apoptosis (Konishi and Akutagawa, 1985; Nixdorf-Bergweiler, 1996). Consequently, males begin to sing around 35d (Immelmann, 1969; Price, 1979), whereas females have never been observed to sing in nature. The sexually dimorphic singing behavior and the underlying song circuit anatomy make zebra finches an advantageous model for studying genes related to vocal learning including human speech.
Figure 1. Diagram of the songbird brain.
A) Schematic sagittal drawing depicts simplified song control circuitry. Solid lines indicate the posterior motor pathway, beginning with HVC, which projects to RA. RA directly projects to nXIIts, which controls the motor neurons of the syrinx. Dashed lines indicate connections of the AFP, in which HVC, X, DLM, and LMAN comprise a cortical-basal ganglia-thalamo-cortical loop like those underlying procedural learning in mammalian brains. LMAN completes the song circuit by projecting to RA, as well as back to X. B) Schematic sagittal drawing depicts non-song brain regions in which Cntnap2 immunostaining was analyzed in this study.
As an initial step toward using songbirds as a model for vocal deficits associated with Cntnap2, Panaitof et al. (2010) described endogenous mRNA expression in the zebra finch. Remarkably, Cntnap2 punctuates the song circuit with differential expression in song nuclei relative to their surrounding tissues. In juvenile and adult males, Cntnap2 is enriched in two cortical song nuclei, RA and LMAN, but diminished in area X. In females, Cntnap2 levels in RA and LMAN are equivalent to or lower than those of the surrounding arco- and nidopallium, respectively (Panaitof et al., 2010). Differential Cntnap2 expression in the song circuit suggests that it serves a purpose in vocal learning (White, 2010; Hilliard et al., 2012). If so, translation is required for any effects on anatomy or physiology. Protein expression does not always follow that of the encoding mRNA, with a precedent in songbirds for socially regulated translation (Whitney and Johnson, 2005). We hypothesized that protein expression patterns would be largely similar to those for the mRNA, but with some differences due to post-transcriptional changes and to localization of the protein not only to cell bodies, but also along axons.
Here we validate an antibody against Cntnap2 for use in zebra finch tissue and describe the Cntnap2 protein distribution in the zebra finch brain at time points during male song development. We find that expression in song circuit neuronal cell bodies largely follows the mRNA but also highlights axonal connections critical for the vocal learning capacity. In line with this idea, within the sexually dimorphic nucleus RA, we identify projection neurons as the cell type that expresses Cntnap2 protein.
MATERIALS AND METHODS
Animals and tissue preparation
All animal use and experimental procedures were in accordance with NIH guidelines for experiments involving vertebrate animals and approved by the UCLA Chancellor’s Animal Care and Use Committee. Zebra finches (n=32 male and 21 female) between 25 and 500 days of age (d) used in this study were obtained from our breeding colony. Sex was determined based on sexually dimorphic plumage, or by postmortem identification of gonads at ages prior to the emergence of dimorphic plumage.
Antibody characterization
Cntnap2
In order to assess endogenous zebra finch Cntnap2 protein levels and distribution, a commercially available anti-Cntnap2 primary polyclonal antibody (Table 2) was selected for testing based on the perfect homology of the antigenic site (amino acids 1315-1331 in the C terminus of NCBI accession number NP_054860) between humans, rats, mice, and zebra finches. A translated nucleotide BLAST (National Center for Biotechnology Information, U.S. National Library of Medicine, Bethesda, MD) search revealed no other plausible targets in the zebra finch genome. The ability of this antibody to detect zebra finch Cntnap2 was vetted as described below (Fig. 2).
Table 2.
Primary Antibodies.
| Primary Antibody |
Immunogen | Manufacturer | Catalog Number |
Species |
|---|---|---|---|---|
| Cntnap2 (Caspr2) |
Synthetic peptide corresponding to amino acids 1315-1331 of rat Caspr2, accession number NP_054860) |
Millipore (Temecula, CA) |
AB5886 | Rabbit polyclonal |
| Gapdh | Purified GAPDH from rabbit muscle |
Millipore | MAB374 | Mouse monoclonal |
| Kvβ2 | Amino acids 17-22 of rat Kvβ2 (accession number NP_034728), conserved in zebra finch |
Neuromab (Davis, CA) |
K17/70 | Mouse monoclonal |
| NeuN | Purified cell culture nuclei from mouse brain |
Millipore | MAB377 | Mouse monoclonal |
| Parvalbumin | Parvalbumin purified from carp muscles |
Swant (CH) | 235 | Mouse monoclonal |
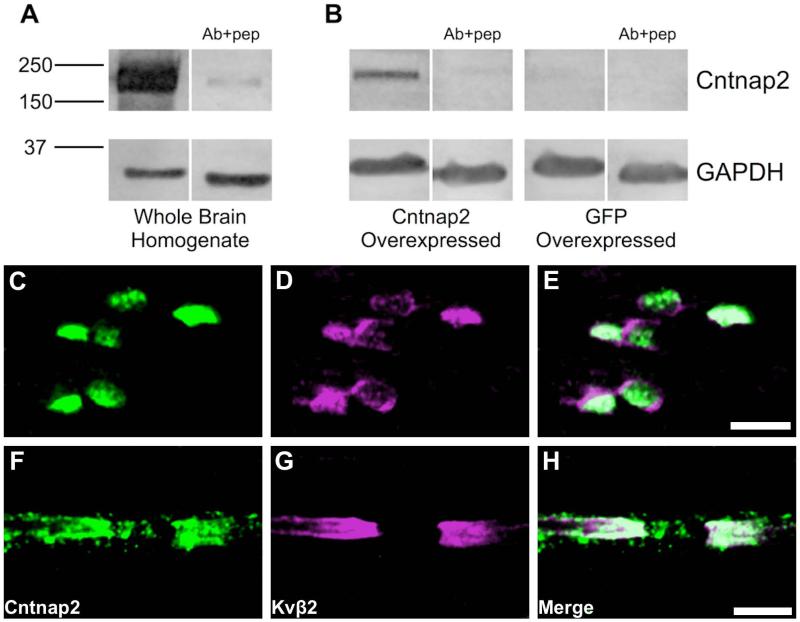
Figure 2. Antibody detection of zebra finch Cntnap2.
A) Western blot of zebra finch whole brain homogenate. Anti-Cntnap2 primary antibody detects a single prominent protein band at the predicted molecular weight (~180 kDa) for endogenous zebra finch Cntnap2. B) Western blots of the ZFTMA zebra finch established cell line with a plasmid expressing zebra finch Cntnap2 or GFP. Transfection of the Cntnap2 construct results in a detectable signal at the predicted molecular weight for Cntnap2 (left). In contrast, transfection of GFP results in no detectable signal at the same molecular weight, confirming no endogenous Cntnap2 expression in this skin-derived cell line (right). For each condition, preadsorption of the primary antibody with its antigenic peptide (Ab+pep) dramatically reduces or removes the signal. Molecular weight markers are given in kDa. C-E) Zebra finch optic and F-H) sciatic nerves double labeled with Cntnap2 and potassium channel subunit Kvβ2 antibodies. Cntnap2 signal colocalizes with putative signals for potassium channel subunit Kvβ2 in both nerves, consistent with its expression in rodents (Poliak et al., 1999; 2003). Overlap of these signals in zebra finch nerves further validates the Cntnap2 antibody for use in this model. Scale bars = 10 μm C-E; 5 μm F-H.
Gapdh
Used here to measure relative levels of glyceraldehyde 3-phosphate dehydrogenase (Gapdh) as a loading control in Western analysis, the antibody (Table 2) detects a 38kDa band in mammalian lysates, according to the manufacturer. It has been previously used in Western analysis in mice (Jones et al., 2008; Fortune and Lurie 2009) and in zebra finch (Miller et al., 2008; Hilliard et al., 2012), detecting a protein band ~36kDa in the latter animal.
Kvβ2
The Kvβ2 antibody (Table 2) was selected for use in zebra finch due to perfect homology of the antigenic site, amino acids 17-22 (TGSPG) of rat (accession number NP_034728) and zebra finch (NCBI RefSeq NC_011485.1). A translated nucleotide BLAST search revealed no other plausible targets of the antibody in zebra finch. Specificity of this antibody is described by the manufacturer. In Western analysis, the antibody detects a major protein band at 38kDa and a minor band at 41kDa in brain lysates from wild type mice, but no bands in lysates from knockout mice (http://neuromab.ucdavis.edu/datasheet/K17_70.pdf). Though the specificity of this antibody has not been confirmed for use in zebra finch, a recent study using this antibody found significant overlap of Kv1.1, Kv1.2, and Kvβ2 (Ovsepian et al., 2013) suggesting that even if the antibody is not specific to Kvβ2, it will at least have a similar immunostaining pattern. We use this antibody only to show that Cntnap2 colocalizes with potassium channel subunits and do not make any claims as to its specificity.
NeuN
The anti-NeuN antibody (Table 2) is used in this study to identify morphology in the zebra finch brain, as it was in Scott and Lois, 2007. According to the manufacturer, the antibody detects protein bands at 46 and 48kDa in Western analysis.
Parvalbumin
The anti-parvalbumin antibody (Table 2) was characterized in Celio et al., (1988). It has since been used to detect the zebra finch isoform in immunohistochemistry to identify parvalbumin-positive neurons in song control nuclei (Wild et al., 2001; 2005; 2009; Roberts et al., 2007), as it is used in this study.
Cell Culture
Whole brain homogenate was obtained from an adult male zebra finch. Following overdose with inhalation anesthetic (isoflurane, Phoenix Pharmaceutical Inc., St. Joseph, MO), the brain was dissected without fixation and homogenized with a hand-held homogenizer (Kontes, Thermo Fisher Scientific, Pittsburgh, PA) in ice-cold modified RIPA lysis buffer (pH 7.6) with protein inhibitors (No. P8340, Sigma-Aldrich) and an RC DC Protein Assay (Bio-Rad, Hercules, CA) was performed to determine protein concentration as in Miller et al. (2008). Zebra finch ZFTMA cells (Itoh and Arnold, 2011) which do not endogenously express Cntnap2 (Fig. 2B) were transfected with either a pCR-TOPO vector (Life Technologies, Grand Island, NY) containing the complete coding sequence for zebra finch Cntnap2 (Panaitof et al., 2010) or a pGIPz vector (Thermo Scientific, Lafayette, CO) containing the GFP coding sequence only, as a negative control. Cells were transfected using a Nucleofector II and chicken nucleofector solution (Lonza, Basel, CH) and distributed on BD Falcon tissue culture dishes (100×20 mm style, Fisher Scientific). At 24 hours post-transfection, GFP expression was observed in ~70% of cells in the plate transfected with the pGIPz vector (not shown). 48 hours after transfection, cells were dissolved in ice-cold modified RIPA lysis buffer with protease inhibitors and a protein assay was performed as above.
Western analysis
Samples of both brain homogenates and cell culture lysates were prepared for immunoblotting by diluting with 2X 5% betamercaptoethanol in Laemmli buffer (pH 6.8; Bio-Rad) and storing at −80°C until use. Samples of 25 μg of brain and 100 μg of cell culture lysates were boiled for 2 minutes and then resolved on a 10% isocratic SDS-polyacrylamide gel in tris-glycine-SDS buffer (pH 8.3; Bio-Rad) at 100 V. A Precision Plus Protein™ Dual Color Standard (Bio-Rad) was included on the gel as a molecular mass marker. Protein was then transferred onto a PVDF membrane with a pore size of 0.45 μm in Tris-glycine (Bio-Rad) with 20% methanol and 1% SDS. The membrane was blocked with 5% milk in Tris-buffered saline with 0.1% tween-20 (pH 7.4; TBST) for 2 hours and then incubated with the anti-Cntnap2 antibody at 1:250 and anti-Gapdh (Table 2) at 1:100,000 in 2.5% milk-TBST overnight at 4°C. A replicate set of samples was incubated with the anti-Cntnap2 antibody that had been pre-adsorbed with antigenic peptide (Millipore, Temecula, CA) at a ratio of 1:30 by mass. Blots were then incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit and anti-mouse secondary antibodies (Table 3) at 1:2,000 and 1:10,000, respectively, in 2.5% milk-TBST for 2 hours. Blots were developed with ECL Plus, imaged on a Typhoon scanner (GE Healthcare), and signal specificity assessed.
Table 3.
Secondary Antibodies.
| Catalog Number | Manufacturer | Reactivity | Conjugate |
|---|---|---|---|
| NA931 | GE Healthcare, Piscataway, NJ |
Mouse IgG | Horseradish peroxidase (HRP) |
| NA934 | GE Healthcare | Rabbit IgG | HRP |
| A-11008 | Life Technologies, Grand Island, NY |
Rabbit IgG | Alexa-Fluor 488 |
| A-11001 | Life Technologies | Mouse IgG | Alexa-Fluor 488 |
| A-21422 | Life Technologies | Mouse IgG | Alexa-Fluor 555 |
| A-11004 | Life Technologies | Mouse IgG | Alexa-Fluor 568 |
| B-1000 | Vector Laboratories, Burlingame, CA |
Rabbit IgG | Biotin |
Tissue staining and immunohistochemistry
Dissection and preparation of tissues
Birds of known age and sex were overdosed with isoflurane, then transcardially perfused with warmed saline followed by 4% paraformaldehyde in phosphate buffered saline (pH 7.4; PBS). Brains were dissected out and cryoprotected in a 20% sucrose solution. 40 μm thick sections were cut in either the coronal or sagittal orientation on a cryostat (Leica Microsystems, Bannockburn, IL) and thaw mounted onto microscope slides (Colorfrost® Plus; Fisher Scientific, Pittsburgh, PA) in a manner that produced replicate sets of adjacent or near-adjacent sections, then stored at −80°C until use. Sciatic and optic nerves were dissected from 2 adult males following brain removal and fixed in 4% paraformaldhyde for 20 minutes, then transferred to PBS. Optic nerves were cryoprotected in a 20% sucrose solution overnight, then cryosectioned at a 10 μm thickness and mounted on microscope slides. Sciatic nerves were mechanically desheathed in PBS, teased, and dried on microscope slides.
Nerve tissue
Prior to immunostaining, sciatic nerve slides were frozen on dry ice for 5 minutes, then allowed to come back to room temperature. Slides containing nerve samples were post-fixed and permeabilized in methanol at −20°C for 20 minutes. A liquid repellent border (Liquid Blocker; Ted Pella Inc., Redding, CA) was drawn along the edges of the slide, and then the samples were rehydrated with phosphate buffer (pH 7.4; PB). Samples were blocked with 10% goat serum diluted in PB with 0.1% Triton-X and 1% glycine for 1 hour, then incubated with the anti-Cntnap2 antibody diluted to 1:500 in blocking solution overnight at 4°C. After washing with PB, samples were incubated with anti-rabbit Alexa Fluor 488 (Table 3) at 1:1,000 in blocking solution for 4 hours. The procedure was then repeated with anti-Kvβ2 (Table 2) at 1:250 and anti-mouse Alexa Fluor 568 (Table 3) at 1:1,000. Glass coverslips were mounted on slides using ProLong Gold antifade reagent (Life Technologies).
Brain sections
One of the replicate sets of brain sections from each bird was used to identify those that contained song control nuclei, using 1% thionin staining to reveal cytoarchitecture. In some cases, sections were alternatively incubated with NeuroTrace™ fluorescent Nissl stain (Life Technologies) diluted at 1:200 in 0.1M PB for 20 minutes. For quantification of Cntnap2 positive neurons in RA, slides were chosen with those sections that contained the largest cross sectional area of RA, in order to control for position within the nucleus, and thawed to room temperature. A liquid repellent border was drawn along the edges of the slide, and then the sections were rehydrated with PB. Endogenous peroxidases were quenched with 0.05% hydrogen peroxide diluted in PB for 30 minutes. Sections were incubated with 5% goat serum in PB containing 0.1% Triton-X for 1 hour. Anti-Cntnap2 antibody was diluted to 1:1,000 in PB and applied to the sections overnight at 4°C. Sections were then incubated at room temperature with a biotinylated goat anti-rabbit secondary antibody (Table 3) at 1:200 in PB for 1 hour, washed, then incubated with avidin-biotin complex (VECTASTAIN Elite ABC Kit (Standard*), Vector Laboratories) at 1:200 in PB with 0.1% Triton-X for 90 minutes. Sections were stained with fluorescein- or rhodamine-tyramide (Hopman et al., 1998) at 1:1,000 in PB with 0.1% Triton-X and 0.003% hydrogen peroxide. For double labeling, following Cntnap2 immunostaining, sections were incubated overnight at 4°C with either anti-NeuN or anti-parvalbumin antibodies (Table 2) at 1:1,000 in PB. Sections were then incubated at room temperature for 4 hours with anti-mouse Alexa Fluor 488, 555, or 568 (Table 3) diluted at 1:1,000 in PB. In the hippocampus, tyramide signal amplification was used for both labels. As above, peroxidase activity was quenched and sections were incubated with anti-NeuN at 1:500, then with anti-mouse HRP at 1:1,000 for 2 hours. These sections were then stained with rhodamine-tyramide as previously described. Peroxidases were quenched again with 0.3% hydrogen peroxide and Cntnap2 immunostaining followed as described above. Slides were mounted with glass coverslips using ProLong Gold antifade reagent (Life Technologies).
Surgical procedures
General methods
Adult male zebra finches were anesthetized with 2-4% isoflurane carried by oxygen using a Universal Vaporizer (Summit Anesthesia Support, Menlo Park, CA) for the duration of the surgery. The bird was placed on a homeothermic blanket mounted in a stereotaxic apparatus at a 45° head angle from the center of the ear bars to the tip of the beak. The cranial feathers were removed to expose the scalp, which was then cleansed using povidone-iodine. In order to preserve vascular flow to the region, a semi-circular incision was made originating and terminating at the caudal edge of the exposed scalp. The scalp was then folded back over a Gelitaspon (Gelita Medical, Amsterdam, Netherlands) moistened with sterile saline, to expose the skull. Injections and recordings, described below, were made through ~1 mm2 windows cut in the skull. After each procedure, the scalp was closed and sealed with Vetbond (Fisher Scientific).
Retrograde targeting of RA projection neurons
A ~1 mm2 window was cut into the skull over the cerebellum ~0.4 mm from the midline, bilaterally. A carbon fiber electrode (Kation Scientific, Minneapolis, MN) was lowered into the brain 4.0 mm below the surface. Multiunit activity was amplified (A-M Systems, Sequim, WA), filtered (300 Hz highpass, 5 kHz lowpass), digitized at 20 kHz (Micro1401, CED, Cambridge, England) and recorded with Spike 2 software (CED). The location of nXIIts was determined by moving the electrode until multiunit activity corresponded to respiratory expiration. The carbon fiber electrode was then replaced with a glass electrode filled with Green Retrobeads™ IX (Lumafluor Inc., Naples, FL). Retrobeads were injected into nXIIts with a picospritzer (Toohey Co, Fairfield, NJ) 3 times on each side for 30 ms at 20 psi. Six days after the procedure each bird was euthanized and perfused with paraformaldehyde as described above.
LMAN lesions
A window was cut in the skull 5.15 mm rostral and 1.7 mm lateral of the midsagittal bifurcation for a unilateral injection. A glass electrode was filled with 10 mg/mL ibotenic acid (Fisher Scientific) in PB, pH 7.0, and lowered into the brain 2.0 mm from the surface to target LMAN and 96.6 nL were injected using a Nanoject II (Drummond Scientific, Broomall, PA). Four days after injection, birds were euthanized and brains collected and sectioned as described above. Sections containing LMAN were stained with thionin as described above to verify the extent of the lesion.
Cntnap2 protein quantification and analysis
Images were acquired using an Axio Imager.A1, with an AxioCam HRm digital camera or LSM 410 laser scanning confocal imager attached to an Axiovert 100 (Carl Zeiss Inc., Oberkochen, GE). Axiovision software (Carl Zeiss Inc.) was used to optimize photomicrographs to remove background, improve brightness and contrast, and to pseudocolor the images. For consistency, Cntnap2 is always represented here in green despite the true color of the fluorophore. In most cases, adjustments were made to the entire image and not to selective subregions, with the exception of the photomicrographs in Fig. 2, in which artifacts of the immunostaining were removed. Anatomical regions were identified according to the published stereotaxic zebra finch brain atlas (http://www.ncbi.nlm.nih.gov/books/NBK2348/, courtesy of Dr. Barbara Nixdorf-Bergweiler and Hans-Joachim Bischof). ImageJ (Rasband, 1997-2012) was used to quantify Cntnap2 positive cells, as follows. First, a border was drawn around RA based on the density of NeuN immunoreactivity. For areas outside of RA, a 600 pixel diameter circle was drawn laterally from RA in either the dorsal (AD) or ventral (AIV) part of the arcopallium. Within the border, all NeuN and Cntnap2 positive cells were counted. The total counts for each signal were adjusted using the Abercrombie method (Guillery 2002) to reduce errors due to the two-dimensional counting method. Each count was multiplied by the tissue thickness (T) and divided by the thickness plus the average diameter of the objects counted (T+d). This adjustment (T/(T+d)) was calculated separately for each section analyzed, and reduced the raw counts by 11-33%, with an average of 24%. To control for the different sizes of RA across sections and animals, statistical significance was determined by non-parametric resampling (bootstrapping) of the ratios of Cntnap2 to NeuN counts. This was done in two stages. First, a modified two-way analysis of variance (ANOVA) compared sex, age, and the interaction effect. A Fisher’s F statistic was calculated for each of the groups, then the groups were pooled and data was sampled with replacement 10,000 times, generating a range of pseudo-F statistics. Statistical significance was achieved when the F statistic from the real data was greater than 95% (p<0.05) or 99% (p<0.01) of the pseudo-statistics. Then, for groups with an ANOVA p-value below 0.05, modified Student’s T-tests were performed for individual groups with the same resampling protocol as described for ANOVA, instead using a Student’s T statistic.
RESULTS
Antibody validation
Bioinformatic analysis revealed that the C-terminus of Cntnap2 is highly conserved between humans and zebra finches (Panaitof et al., 2010), and that the last 76 amino acids are identical (amino acids 1255-1331 in human, 1252-1328 in zebra finch:
GVNRNSAIIGGVIAVVIFTILCTLVFLIRYMFRHKGTYHTNEAKGAESAESADAAIMNND PNFTETIDESKKEWLI). A commercial antibody available from Millipore and raised against C-terminus amino acids 1315-1331 of human CNTNAP2 (1312-1328 of zebra finch Cntnap2) was thus selected to test its ability to specifically detect the zebra finch isoform. In Western analyses of zebra finch whole brain homogenate, this antibody detects a single prominent band at the predicted molecular weight of ~180 kDa. Preadsorption of the antibody with the antigenic peptide considerably decreases the intensity of this band (Fig. 2A). The specificity of the antibody was further validated by over-expressing zebra finch Cntnap2 (accession number NM_001193337.1) in ZFTMA cells (Itoh and Arnold, 2011), a zebra finch immortalized cell line which does not endogenously express the protein. Cultures transfected with zebra finch Cntnap2 produce the same protein band, whereas those from untransfected cultures (not shown) or transfected with a control construct containing sequences coding only for GFP do not (Fig. 2B). Specificity of the antibody was again confirmed by preadsorption (see Materials and Methods).
The Millipore antibody was subsequently tested for use in immunohistochemistry. In mammals, Cntnap2 is expressed in axons of myelinated nerves, colocalized with VGKC subunits (Poliak et al., 1999; 2001; 2003; Gu and Gu, 2011). To verify that the Millipore antibody detects zebra finch Cntnap2 in situ, we immunostained optic (Fig. 2C-E) and sciatic (Fig. 2D-F) nerves dissected from zebra finches for both Cntnap2 and Kvβ2. In both nerve preparations, the signals from the two antibodies overlap, as evidenced by the colocalization tools in ImageJ, further confirming that the antibody specifically detects zebra finch Cntnap2.
Cntnap2 protein distribution in the zebra finch brain
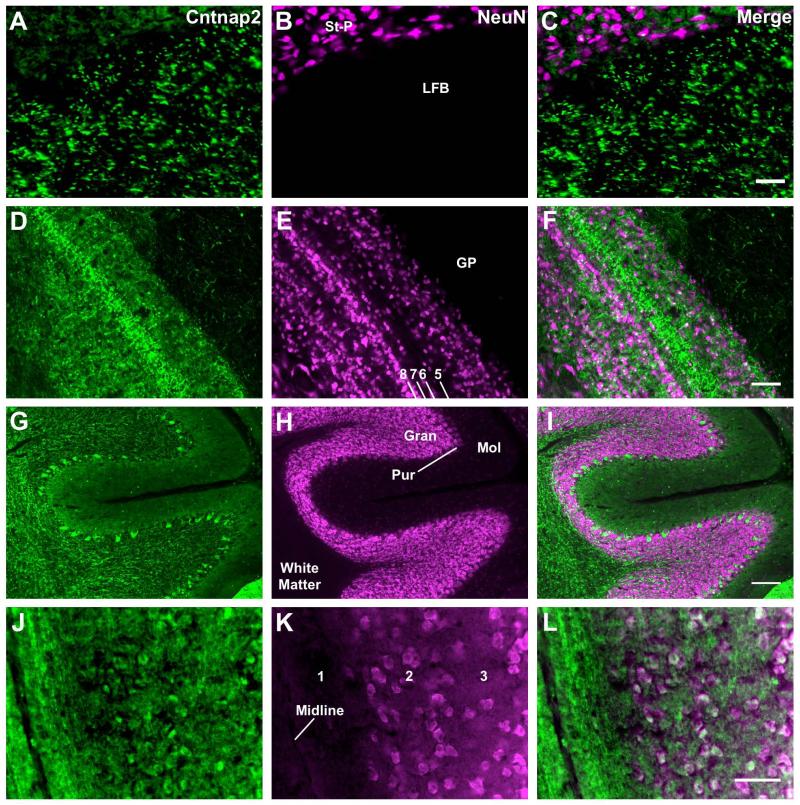
Similar to reported mammalian data (Poliak et al., 1999), Cntnap2 distribution is extensive in zebra finch brains, though not expressed to the same level in all regions. Particular enrichment is observed in myelinated areas consistent with axonal expression, such as the fronto-arcopallial tract, optic tract, optic chiasm (not shown), the lateral forebrain bundle (Fig. 3A-C), and layer 5 of the optic tectum (Fig. 3D-F). In the cerebellum, the Purkinje cell layer is marked by intense Cntnap2 immunostaining of cell bodies, and fibers containing Cntnap2 can be observed in the cerebellar white matter. Much less Cntnap2 is found in the granular and molecular layers (Fig. 3G-I). In the midbrain, Cntnap2 is found in the parvocellular portion of the isthmus nucleus (not shown). Thalamic regions containing high levels of Cntnap2 include the anterior dorsomedial nucleus, dorsal portion of the lateral mesencephalic nucleus, rotund nucleus, lateral spiriform nucleus, and pretectal nucleus. In the telencephalon, enrichment of Cntnap2 is found in the entopallium, the anterior hyperpallium, striatopallidum, globus pallidus, field L (not shown), and cell bodies resembling pyramidal neurons (Montagnese et al., 1996) in the medial hippocampus (Fig. 3J-L).
Figure 3. Cntnap2 distribution in non-song circuit brain regions.
Cntnap2 is detected in several areas outside the song circuit of the zebra finch brain, including in structures reported to express Cntnap2 in rodents (Poliak et al., 1999). Non-song circuit tissue in this figure are taken from regions depicted in Fig. 1B. Neuron specific marker NeuN (magenta) is used for reference. A-C) Axonal patterning of Cntnap2 label in the lateral forebrain bundle within the telencephalon. D-F) intense Cntnap2 (green) labeling along axons in layer 5 of the optic tectum. Numbers in 3B indicate the layers of the optic tectum according to Ramón y Cajal (1911). G-I) Purkinje cell bodies and the cerebellar white matter strongly express Cntnap2, with less in the molecular layer, and fibrous signal in the granular layer and white matter. J-L) Coronal section of the medial hippocampus; numbers indicate layers (Montagnese et al., 1996). Cntnap2 marks neuronal somata in the pyramidal cell region (white arrows). Scale bars = 50 μm A-C; 200 μm D-L.
Within the song circuit of an adult male zebra finch, Cntnap2 protein distribution generally follows the mRNA distribution reported in Panaitof et al., 2010, with some exceptions. Though cortical nucleus HVC does contain Cntnap2 positive cells, expression is not enriched relative to the surrounding nidopallium (Fig. 4A-C). As with the mRNA, cortical nuclei RA (Fig. 4D-F) and LMAN (Fig 4G-I) have elevated Cntnap2 levels relative to the surrounding nido- and arcopallium, respectively. In contrast with the reported mRNA levels, the basal ganglia song control region, area X, exhibits greater Cntnap2 protein expression than the surrounding striatopallidum (Fig. 4J-L). The Cntnap2 protein in the aforementioned areas is present not only on cell bodies, but also in the neuropil. The thalamic song nucleus DLM, however, has Cntnap2 positive cell bodies, but relatively less protein in the neuropil than the surrounding thalamic regions (Fig. 4M-O).
Figure 4. Cntnap2 protein in song circuit nuclei.
Fluorescent photomicrographic images of song control nuclei. Cntnap2 signals are in green, and NeuN signals in magenta. A-C) HVC in the nidopallium; D-F) RA in the arcopallium; G-I) LMAN in the nidopallium; J-L) Area X in the striatopallidum, inset: higher magnification inside X; M-O) DLM in the thalamus, along with the ovoid nucleus, the dorsomedial nucleus of the posterior thalamus, and the lateral forebrain bundle. Each nucleus is indicated by dashed line traces on the NeuN (middle column) panels. Greater Cntnap2 labeling is found within RA, LMAN, and area X relative to surrounding brain regions on both cell bodies and in the neuropil. HVC and DLM contain Cntnap2-expressing cells, but with expression levels comparable to their surrounding tissues. Scale bars = 200 μm A-I; 100 μm J-L (50 μm inset); 200 μm M-O.
Sexually dimorphic expression of Cntnap2 in RA
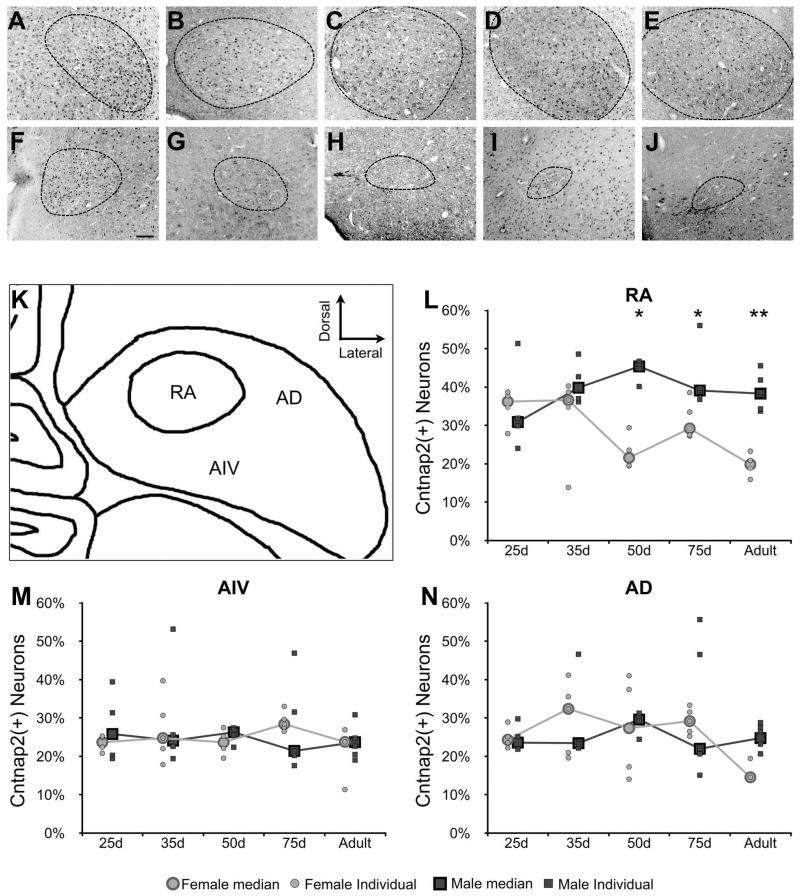
Cntnap2 mRNA expression is sexually dimorphic in LMAN and RA in developing zebra finches. Males have slightly more Cntnap2 in LMAN than females throughout development, though the level of expression increases in both sexes with age. There is a more striking difference in expression in RA at 50d. Similar Cntnap2 levels are detected in both sexes prior to 35d. Between the two time points, expression in females begins to decrease, while males maintain a high level through adulthood (Panaitof et al., 2010). We therefore compared levels of Cntnap2 immunostaining in RA in both sexes at developmental time points within sensory acquisition and sensorimotor learning periods, and after song crystallization (males, Fig. 5A-E; females, Fig. 5F-J). At 25 and 35d leading up to the onset of sensorimotor learning, the fraction of RA neurons that are positive for Cntnap2 are comparably enriched in both sexes relative to the surrounding dorsal and ventral intermediate arcopallium (AD and AIV, respectively). However, by 50d, the fraction of Cntnap2 positive neurons in female RA significantly decreases (Fig. 5L), and falls to levels comparable to those in AD and AIV (Fig. 5M and N). This time point falls within the sensorimotor phase of vocal learning, during which males practice their memorized song (Eales, 1985). Male Cntnap2 enrichment in RA is maintained throughout development and into adulthood and crystallization of song, whereas in females it is significantly reduced. The difference in Cntnap2 protein expression within the arcopallium between males and females and at different developmental stages appears unique to RA. A comparison of the number of Cntnap2 enriched cells in AD and AIV reveals no significant effect of age or sex (Fig. 5M and N).
Figure 5. Cntnap2 within RA in both sexes at developmental time points during male song learning.
A-J) Representative images of Cntnap2 immunolabeling of cells in male (A-E) and female (F-J) RA at time points during development encompassing the onset of sensory acquisition, sensorimotor learning, and crystallization of song. Anti-NeuN signals (not shown) were used to trace the border of RA in each image. As previously reported (Konishi and Akutagawa, 1985; Nixdorf-Bergweiler, 1996), the size of RA begins to decrease in females and increase in males starting around 35d and continues through development until maturity. K) A diagram of RA and the two arcopallial regions in which labeled cells were counted: the ventral intermediate arcopallium (AIV) and the dorsal arcopallium (AD). L-N) Graphs representing the percentage of Cntnap2 positive neurons out of the total number of NeuN positive cells found in RA, AIV, and AD, respectively, for 3-6 birds of each sex at each time point. Statistical significance was determined by resampling ANOVA, followed by individual Student’s T-tests *p<0.05, **p<0.01. Scale bar = 100 μm.
LMAN projections contribute to Cntnap2 expression in RA
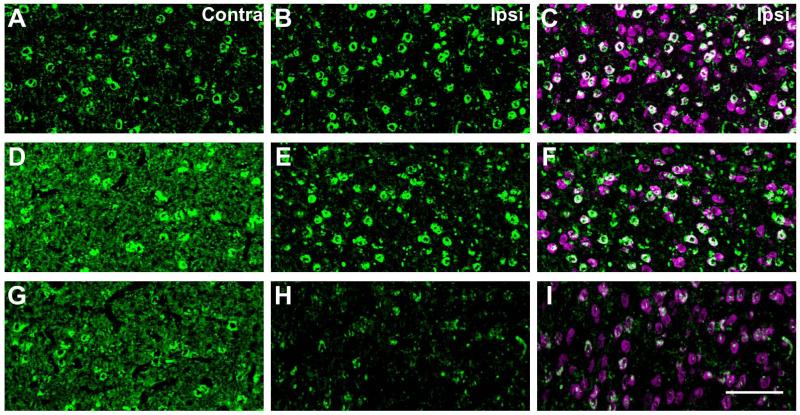
To test the possible contribution of LMAN terminals to Cntnap2 in RA, LMAN was unilaterally lesioned using ibotenic acid in three adult males (Fig. 6A-C; Fig. 6D-F; Fig. 6G-I). The resulting Cntnap2 protein expression was observed in the ipsilateral RA and compared to that in the non-lesioned contralateral side. Somatic expression of Cntnap2 remained unaffected in the ipsilateral RA, but there was a decrement in immunostaining intensity in the neuropil compared with the contralateral RA, suggesting that some of the Cntnap2 is indeed from LMAN projections. In summary, within the vocal production circuit, Cntnap2 enrichment appears to be most prominent in RA and due to expression in both neuronal somata and neuropil, including that arising from within LMAN nerve terminals.
Figure 6. Unilateral LMAN lesions result in an ipsilateral decrease of Cntnap2 in RA.
Representative photomicrographic images of Cntnap2 labeling (green) in RA from three adult male zebra finches (A-C, D-F, G-I) in which LMAN was lesioned unilaterally by injection of ibotenic acid. Double labeling with NeuN (magenta; C,F,I) indicates neuronal cell bodies. In all cases, the lesion reduces the amount of Cntnap2 in the neuropil, but not cell bodies, in ipsilateral RA relative to the contralateral nucleus, indicating that some of the Cntnap2 in the neuropil originates from LMAN projections. Scale bar = 25 μm.
Cntnap2 is expressed in RA projection neurons
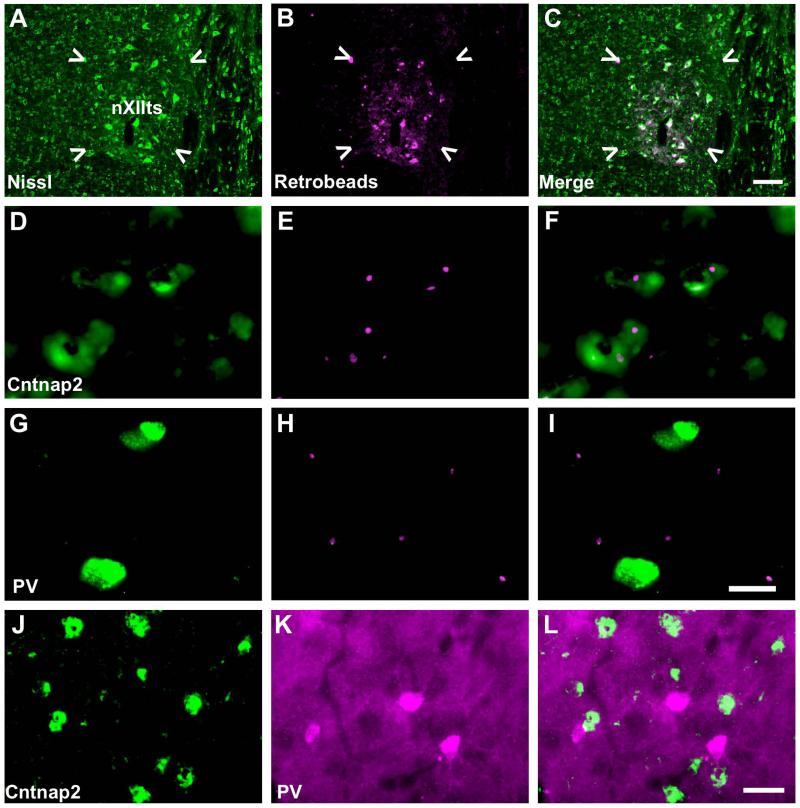
Within RA, Cntnap2 somal expression is restricted to a subset of the neuronal phenotypes. At least two distinct neuronal populations in RA have been defined based on their electrophysiological signatures and morphology: GABAergic interneurons, and glutamatergic projection neurons (Spiro et al., 1999). The latter directly synapse onto neurons within nXIIts, which directly innervates the syrinx. Parvalbumin staining has been used to differentiate these two types. Whereas some interneurons stain intensely for parvalbumin, projection neurons stain relatively weakly (Wild et al., 2001). To determine whether Cntnap2 is expressed in projection neurons, fluorescent retrobeads were injected into nXIIts (Fig. 7A-C). Following retrograde transport, fluorescent signals colocalized with Cntnap2 signals in RA (Fig. 7D-F), but not in cells that expressed a high level of parvalbumin (Fig. 7G-I). Rather, we found that Cntnap2 signals overlapped only with weakly parvalbumin positive neurons, consistent with the interpretation that RA projection neurons express Cntnap2 (Fig. 7J-L). The overlap of retrobeads with Cntnap2 signals further supports the hypothesis that Cntnap2 is expressed in RA neurons which project to nXIIts.
Figure 7. Cntnap2 is expressed in RA projection neurons, not parvalbumin positive interneurons.
A-C) Injection site of retrobeads (magenta) in nXIIts (indicated by white arrows), identified by Nissl stain (green). D-F) Retrobeads overlap with Cntnap2 (green) expressing cells in RA. G-I). Retrobeads do not overlap with strongly parvalbumin positive interneurons. J-L) Cntnap2 immunolabeling (green) does not overlap with RA inhibitory interneurons intensely labeled with parvalbumin (magenta). Retrograde labeling reveals that RA projection neurons express Cntnap2 and confirms its absence in parvalbumin positive interneurons. Scale bar = 50 μm A-C; 20 μm D-I; 25 μm J-L.
DISCUSSION
Here we have characterized the protein distribution of Cntnap2, a molecule linked to human language disorders, in the brain of a non-human vocal learner, the zebra finch species of songbird. Because the neurons that are dedicated to vocal learning are clustered together in the songbird brain (Fig. 1), this analysis enables direct comparison of Cntnap2 levels within song-dedicated neurons relative to their levels in surrounding tissues, which, though made up of similar cell types, contribute to non-vocal related functions. Moreover, the sexual dimorphism of vocal learning and the underlying song control circuitry allow us to compare protein expression between vocal and non-vocal learners within the same species. We can further draw parallels between humans and songbirds by investigating the cell types within a song nucleus in which we detect Cntnap2 expression.
Outside the song circuit, immunoreactivity is widespread throughout the telencephalon with areas of particularly high expression, such as in myelinated regions, and in the Purkinje cell layer of the cerebellum, and in pyramidal-like cells (Montagnese et al., 1996) in layers 2 and 3 of the hippocampus (Fig. 3), similar to that described for mammals (Poliak et al., 1999). Notably, however, expression within several nuclei of the song circuit in the adult brain is strikingly different than in their respective surrounding regions, which are not part of the song control circuitry (Fig. 4). In the AFP, Cntnap2 protein is enriched in cortical LMAN relative to the anterior nidopallium, in area X relative to the striatopallidum, and in the somata of DLM relative to the anterior thalamus. Though enrichment in LMAN and DLM is expected based on the mRNA data, the enrichment in area X is surprising given the lower transcript levels in this region relative to the surrounding striatopallidum (Panaitof et al., 2010). Cntnap2 protein is found in the neuropil of area X leaving open the possibility that some of the protein arises from HVC and/or LMAN terminals, similar to the contribution of LMAN to Cntnap2 expression in RA (Fig. 6). There is also somal Cntnap2 expression suggesting at least some protein originates in area X. The difference between the observed mRNA and protein data may reflect state-dependent regulation of the protein, perhaps by transcription factors such as FoxP2 (Teramitsu and White, 2006; Miller et al., 2008). Whether Cntnap2 is a direct target of FoxP2 in zebra finches, as it is in humans (Vernes et al., 2008), remains to be tested. The zebra finch genomic Cntnap2 sequence (RefSeq assembly ID GCF_000151805.1) contains many potential FoxP2 binding sites (Stroud et al., 2006), mostly in the first intron. The FOXP2 binding site in humans was confirmed to be in the first intron by chromatin immunoprecipitation (Vernes et al., 2008). The lower mRNA levels and higher protein in area X thus likely reflect a combination of cellular trafficking, transcriptional and post-transcriptional regulation. Whatever the mechanism, Cntnap2 mRNA and protein expression within the nucleus differs from levels in the surrounding tissue, despite the similar cell type composition of these subregions.
Cntnap2 protein distribution in the posterior pathway is similar to that for the mRNA. The amount within HVC is comparable to the surrounding nidopallium, whereas it is enriched in RA of males and juvenile females (Panaitof et al., 2010). The connectivity of the posterior vocal pathway in males suggests that RA-projecting neurons in HVC are analogous to mammalian neurons in cortical layer 2/3, which do not show prominent Cntnap2 staining, whereas RA projection neurons are analogous to mammalian cortical layer 5 pyramidal neurons (Jarvis, 2004), which exhibit prominent Cntnap2 levels (Poliak et al., 1999). The projection from RA to nXIIts is a corticospinal connection shared with mammalian motor cortex and is hypothesized to allow direct activation of individual muscles necessary for fine motor control (Vicario, 1991). Notably, these direct connections onto motor neurons controlling the muscles involved in phonation are posited to enable the vocal learning capacity of select species such as humans and songbirds (Jürgens, 2009; Arriaga et al., 2012). Overlap of retrobeads injected into nXIIts in RA and Cntnap2 positive neurons (Fig. 7) indicates that Cntnap2 is present in this connection, raising the possibility that Cntnap2 is required for its establishment and/or proper function. Additionally, the reduction of Cntnap2 in the neuropil of RA following an ipsilateral LMAN lesion (Fig. 6) suggests that some of the enrichment in RA is provided from LMAN projections. This long-range connection is reminiscent of the connectivity that is altered in humans bearing the CNTNAP2 risk alleles for ASD and SLI who exhibit increased local and decreased long-range connectivity of the medial prefrontal cortex (mPFC), and less lateralization than their non-risk allele counterparts (Scott-Van Zeeland et al., 2010). In fact, LMAN is postulated to be homologous to human PFC based on shared physiologic and anatomic features including connectivity (Kojima et al., 2013). Taken together, these parallel observations in humans and songbirds support the idea that Cntnap2 affects neural connectivity critical for vocal learning across taxonomic classes.
This hypothesis is further supported by the sexually dimorphic expression in zebra finch brain. Similar to that reported for Cntnap2 mRNA, males and females share protein enrichment in RA early in development. However, by 50d the enrichment in female RA wanes, whereas it persists in males throughout adulthood. Since Cntnap2 is similarly enriched in RA in males and females prior to 50 days, the sexual dimorphism may reflect a change in cell composition in RA or a sex-based difference in gene expression within each cell. These data demonstrate a loss of Cntnap2 labeled cells in female RA with age. This may be due to preferential apoptosis (Konishi and Akutagawa, 1990) of neurons that express Cntnap2 or down regulation of both Cntnap2 mRNA and protein in female zebra finches, who do not use this nucleus for producing learned vocalizations. In mammals, some sex typical behaviors have been associated with sexually dimorphic expression of individual genes, supporting the hypothesis that sex-related behaviors driven by hormones are mediated in part by genes (Xu et al., 2012) or in fact by genes independent of hormones (Arnold et al., 2013). In the case of the zebra finch, genes that exhibit sexually dimorphic expression in song circuitry are likely to be involved, perhaps even crucial, for singing. These same genes may also be involved in human speech and language. This hypothesis was the basis for the prediction that FOXP1 mutations would impair human speech. FOXP1 is a transcription factor closely related to FOXP2, and the two form heterodimers to control gene expression. Sexually dimorphic expression of FoxP1, but not FoxP2, was found in the song circuit of quiescent zebra finches, leading to the aforementioned prediction (Teramitsu et al., 2004). Subsequently, several cases were described of FOXP1 mutations in people with language disorders (Pariani et al., 2009; Carr et al., 2010; Hamdan et al., 2010; Palumbo et al., 2012). The sexually dimorphic expression of Cntnap2 in RA also fits this pattern, and may in fact be regulated by FoxP1 in tandem or independent from FoxP2.
What might be the mechanistic function of Cntnap2 in the song circuit, or RA specifically? Cntnap2 is closely related to the neurexins, which have also been implicated in ASD (Südhof, 2008). Though neurexins function at the synapse, Cntnap2 is found at the juxtaparanodes of myelinated axons. There, it is responsible for the clustering of Shaker-type VGKCs (Poliak et al., 1999; 2003; Horresh et al., 2008). Selective blockade of these channels on axons from rat central nervous system during myelination early in development leads to aberrant action potential waveforms. However, when the animal becomes mature, application of the blocker no longer affects the waveform (Devaux et al., 2002). In songbirds, all song circuit nuclei send and receive long-range connections, which may require Cntnap2 at a macrocircuit level to cluster VGKCs at juxtaparanodes in order to establish and/or maintain synaptic connections required for learning and producing vocalizations. Loss of Cntnap2 in the neuropil of RA following LMAN lesion is evidence for a macrocircuit role for Cntnap2 in this cortical-cortical connection. This suggests that if the role of Cntnap2 in clustering VGKCs is important for vocal learning, it will have the greatest impact early in development, while the process of myelination is still ongoing. Cntnap2 may have additional, yet unknown functions, suggested by CNTNAP2 enrichment in human embryonic cortex well before myelination (Abrahams et al., 2007). Recent evidence suggests that Cntnap2 may influence synaptic connectivity, increasing cell-autonomous dendritic arborization and the number of synaptic sites in cultured neurons. Contactin 2, the binding partner of Cntnap2, appears to have the opposite effect on synaptic connectivity (Anderson et al., 2012). Contactin 2 and Cntnap2 together may affect the development of brain areas related to vocal learning and language. Cntnap2 may be important for microcircuit connectivity in song nuclei of the adult zebra finch brain as well, by establishing and maintaining local connections within each nucleus through increasing dendritic arborization and the number of active postsynaptic connections. According to this hypothesis, we expect that loss of Cntnap2 in male RA before the onset of sensorimotor learning would lead to fewer connections with HVC and an impaired ability to mimic the tutor’s song.
Further investigation into the role of Cntnap2 in vocal learning in songbirds will certainly benefit our understanding of human speech disorders associated with risk variants of the gene, as well as the neurobiology of language as a whole. Taking advantage of the well-characterized song circuitry, an individual song nucleus could be targeted for Cntnap2 RNA interference. If Cntnap2 is involved in song learning, as it seems to be in human speech, we expect knockdown to impair vocal learning in juvenile males, whose songs have not yet crystallized. This system may also be used to parse activational versus organizational effects of Cntnap2 in vocal learning by manipulating Cntnap2 levels at different times during development. Besides behavior, we additionally expect to find neurophysiological changes. Knocking down Cntnap2 in RA may result in a mislocalization of potassium channels, which could slow the repolarization phase of an action potential similar to the effects of blocking those channels, particularly prior to the completion of myelination (Devaux et al., 2002). There may also be changes to synaptic connectivity between RA and HVC or LMAN concurrent with decreased dendritic arborization of projection neurons originating in RA, similar to the effects reported in vitro reported by Anderson et al. (2012). Reducing Cntnap2 levels in LMAN may augment its local connectivity and decrease its long-range connectivity to RA, similar to the altered connectivity in forebrains of humans with risk variants of Cntnap2 (Scott-Van Zeeland et al., 2010). The balance between inhibition and excitation is also likely to be affected, as it is in cases of autism (Cline, 2005) and Cntnap2 knockout mouse models (Peñagarikano et al., 2011). The present and future investigation into the role of Cntnap2 in vocal learning using songbirds complements studies in mammals moving toward a better understanding of its associated disorders in humans.
Table 1.
List of Abbreviations Used in Figures
| AD | dorsal arcopallium |
| AFP | anterior forebrain pathway |
| AIV | ventral intermediate arcopallium |
| Arco | arcopallium |
| Cntnap2 | contactin associated protein-like 2 |
| d | days of age |
| DLM | medial portion of the dorsolateral nucleus of the anterior thalamus |
| DMP | dorsomedial nucleus of the posterior thalamus |
| Gapdh | glyceraldehyde 3-phosphate dehydrogenase |
| GFP | green fluorescent protein |
| GP | globus pallidus |
| Gran | granule cell layer of the cerebellum |
| Hyper | hyperpallium |
| Kvβ2 | potassium channel beta subunit |
| LFB | lateral forebrain bundle |
| LMAN | lateral magnocellular nucleus of the anterior nidopallium |
| Meso | mesopallium |
| Mol | molecular cell layer of the cerebellum |
| NeuN | neuronal nuclei |
| Nido | nidopallium |
| nXIIts | hypoglossal nucleus |
| Ov | ovoid nucleus |
| Pur | purkinje cell layer of the cerebellum |
| PV | parvalbumin |
| RA | robust nucleus of the arcopallium |
| St-P | striatopallidum |
| VGKC | voltage-gated potassium channel |
| X | area X |
| ZFTMA | zebra finch immortalized cell line |
OTHER ACKNOWLEDGEMENTS
The authors thank Melissa Coleman and Felix Schweizer for assistance in the use of their equipment for retrograde labeling, and confocal fluorescence imaging, respectively. Brett Abrahams and Hongmei Dong provided the zebra finch Cntnap2 cDNA construct used in tests of antibody specificity. Yuichiro Itoh and Arthur Arnold provided the ZFTMA cell line. Vijayendran Chandran identified potential FoxP2 binding sites in zebra finch Cntnap2. Thanks to Alice Fleming for advice on immunohistochemistry and to Julie Miller for revising drafts of this manuscript and guidance on the methodology used within. Thanks also to Dorsa Beroukhim, Guillermo Milian, and Diana Sanchez for their assistance with collecting tissue samples. The authors wish to acknowledge two anonymous reviewers for their constructive comments.
Grant Sponsors: NIH 5 T32 NS058280, UCLA Eureka Scholarship, UCLA Edith Hyde Fellowship (MCC); NIH R21 HD065271, US Army AR093327 (SAW)
Footnotes
CONFLICT OF INTEREST
The authors declare they have no conflict of interest.
LITERATURE CITED
- Abrahams BS, Tentler D, Perederiy JV, Oldham MC, Coppola G, Geschwind DH. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc Natl Acad Sci. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Galfin T, Xu W, Aoto J, Malenka RC, Südhof TC. Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proc Natl Acad Sci. 2012;109(44):18120–5. doi: 10.1073/pnas.1216398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Chen X, Link JC, Itoh Y, Reue K. Cell-autonomous sex determination outside of the gonad. Dev Dyn. 2013;242:371–379. doi: 10.1002/dvdy.23936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriaga G, Zhou EP, Jarvis ED. Of mice, birds, and men: the mouse ultrasonic song system has some features similar to humans and song-learning birds. PLoS ONE. 2012;7:e46610. doi: 10.1371/journal.pone.0046610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brainard MS, Doupe AJ. Auditory feedback in learning and maintenance of vocal behaviour. Nat Rev Neurosci. 2000;1:31–40. doi: 10.1038/35036205. [DOI] [PubMed] [Google Scholar]
- Carr CW, Moreno-De-Luca D, Parker C, Zimmerman HH, Ledbetter N, Martin CL, Dobyns WB, Abdul-Rahman OA. Chiari I malformation, delayed gross motor skills, severe speech delay, and epileptiform discharges in a child with FOXP1 haploinsufficiency. Eur J Hum Genet. 2010;18:1216–1220. doi: 10.1038/ejhg.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celio MR, Baier W, Schärer L, de Viragh PA, Gerday C. Monoclonal antibodies directed against the calcium binding protein parvalbumin. Cell Calcium. 1988;9:81–86. doi: 10.1016/0143-4160(88)90027-9. [DOI] [PubMed] [Google Scholar]
- Cline H. Synaptogenesis: a balancing act between excitation and inhibition. Curr Biol. 2005;15:R203–5. doi: 10.1016/j.cub.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Devaux J, Gola M, Jacquet G, Crest M. Effects of K+ channel blockers on developing rat myelinated CNS axons: identification of four types of K+ channels. J Neurophysiol. 2002;87:1376–1385. doi: 10.1152/jn.00646.2001. [DOI] [PubMed] [Google Scholar]
- Eales LA. Song learning in zebra finches: some effects of song model availability on what is learnt and when. Anim Behav. 1985;33:1293–1300. [Google Scholar]
- Fitch WT. Evolutionary Developmental Biology and Human Language Evolution: Constraints on Adaptation. Evol Biol. 2012;39:613–637. doi: 10.1007/s11692-012-9162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune T, Lurie DI. Chronic low-level lead exposure affects the monoaminergic system in the mouse superior olivary complex. J Comp Neurol. 2009;513:542–558. doi: 10.1002/cne.21978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SA, Fisher SE. Decoding the genetics of speech and language. Curr Opin Neurobiol. 2012 doi: 10.1016/j.conb.2012.11.006. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Gu C, Gu Y. Clustering and activity tuning of Kv1 channels in myelinated Hippocampal axons. J Biol Chem. 2011;286(29):25835–25847. doi: 10.1074/jbc.M111.219113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillery RW. On counting and counting errors. J Comp Neurol. 2002;447:1–7. doi: 10.1002/cne.10221. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Daoud H, Rochefort D, Piton A, Gauthier J, Langlois M, Foomani G, Dobrzeniecka S, Krebs M-O, Joober R, et al. De novo mutations in FOXP1 in cases with intellectual disability, autism, and language impairment. Am J Hum Genet. 2010;87:671–678. doi: 10.1016/j.ajhg.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard AT, Miller JE, Fraley ER, Horvath S, White SA. Molecular microcircuitry underlies functional specification in a Basal Ganglia circuit dedicated to vocal learning. Neuron. 2012;73:537–552. doi: 10.1016/j.neuron.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopman AHN, Ramaekers FCS, Speel EJM. Rapid Synthesis of Biotin-, Digoxigenin-, Trinitrophenyl-, and Fluorochrome-labeled Tyramides and Their Application for In Situ Hybridization Using CARD Amplification. Journal of Histochemistry & Cytochemistry. 1998;46:771–777. doi: 10.1177/002215549804600611. [DOI] [PubMed] [Google Scholar]
- Horresh I, Poliak S, Grant S, Bredt D, Rasband MN, Peles E. Multiple molecular interactions determine the clustering of Caspr2 and Kv1 channels in myelinated axons. J Neurosci. 2008;28:14213–14222. doi: 10.1523/JNEUROSCI.3398-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immelmann K. Song development in the zebra finch and other estrildid finches. In: Hinde RA, editor. Bird vocalizations. Cambridge Univ Pr.; 1969. pp. 61–74. [Google Scholar]
- Itoh Y, Arnold AP. Zebra finch cell lines from naturally occurring tumors. In Vitro Cell Dev Biol Anim. 2011;47(4):280–2. doi: 10.1007/s11626-011-9392-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED. Learned Birdsong and the Neurobiology of Human Language. Ann N Y Acad Sci. 2004;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LG, Prins J, Park S, Walton JP, Luebke AE, Lurie DI. Lead exposure during development results in increased neurofilament phosphorylation, neuritic beading, and temporal processing deficits within the murine auditory brainstem. J Comp Neurol. 2008;506:1003–1017. doi: 10.1002/cne.21563. [DOI] [PubMed] [Google Scholar]
- Jürgens U. The neural control of vocalization in mammals: a review. J Voice. 2009;23:1–10. doi: 10.1016/j.jvoice.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Knornschild M, Nagy M, Metz M, Mayer F, Helversen von O. Complex vocal imitation during ontogeny in a bat. Biology Letters. 2010;6:156–159. doi: 10.1098/rsbl.2009.0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Kao MH, Doupe AJ. Task-related “cortical” bursting depends critically on basal ganglia input and is linked to vocal plasticity. Proc Natl Acad Sci. 2013;110:4756–4761. doi: 10.1073/pnas.1216308110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Akutagawa E. Neuronal growth, atrophy and death in a sexually dimorphic song nucleus in the zebra finch brain. Nature. 1985;315:145–147. doi: 10.1038/315145a0. [DOI] [PubMed] [Google Scholar]
- Konishi M, Akutagawa E. Growth and atrophy of neurons labeled at their birth in a song nucleus of the zebra finch. Proc Natl Acad Sci. 1990;87:3538–3541. doi: 10.1073/pnas.87.9.3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Li X, Hu Z, He Y, Xiong Z, Long Z, Peng Y, Bu F, Ling J, Xun G, Mo X, et al. Association analysis of CNTNAP2 polymorphisms with autism in the Chinese Han population. Psychiatr Genet. 2010;20:113–117. doi: 10.1097/YPG.0b013e32833a216f. [DOI] [PubMed] [Google Scholar]
- Miller JE, Spiteri E, Condro MC, Dosumu-Johnson RT, Geschwind DH, White SA. Birdsong decreases protein levels of FoxP2, a molecule required for human speech. J Neurophysiol. 2008;100:2015–2025. doi: 10.1152/jn.90415.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnese CM, Krebs JR, Meyer G. The dorsomedial and dorsolateral forebrain of the zebra finch, Taeniopygia guttata: a Golgi study. Cell Tissue Res. 1996;283:263–282. doi: 10.1007/s004410050537. [DOI] [PubMed] [Google Scholar]
- Newbury DF, Paracchini S, Scerri TS, Winchester L, Addis L, Richardson AJ, Walter J, Stein JF, Talcott JB, Monaco AP. Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. Behav. Genet. 2011;41:90–104. doi: 10.1007/s10519-010-9424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixdorf-Bergweiler BE. Divergent and parallel development in volume sizes of telencephalic song nuclei in male and female zebra finches. J Comp Neurol. 1996;375:445–456. doi: 10.1002/(SICI)1096-9861(19961118)375:3<445::AID-CNE7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- Ovsepian SV, Steuber V, Le Berre M, O’Hara L, O’Leary VB, Dolly JO. A defined heteromeric KV1 channel stabilizes the intrinsic pacemaking and regulates the output of deep cerebellar nuclear neurons to thalamic targets. J Physiol (Lond) 2013;591:1771–1791. doi: 10.1113/jphysiol.2012.249706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaitof SC, Abrahams BS, Dong H, Geschwind DH, White SA. Language-related Cntnap2 gene is differentially expressed in sexually dimorphic song nuclei essential for vocal learning in songbirds. J Comp Neurol. 2010;518:1995–2018. doi: 10.1002/cne.22318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariani MJ, Spencer A, Graham JM, Rimoin DL. A 785kb deletion of 3p14.1p13, including the FOXP1 gene, associated with speech delay, contractures, hypertonia and blepharophimosis. Eur J Med Genet. 2009;52:123–127. doi: 10.1016/j.ejmg.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, et al. Absence of CNTNAP2 Leads to Epilepsy, Neuronal Migration Abnormalities, and Core Autism-Related Deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter B, Raskind WH, Matsushita M, Lisowski M, Vu T, Berninger VW, Wijsman EM, Brkanac Z. Replication of CNTNAP2 association with nonword repetition and support for FOXP2 association with timed reading and motor activities in a dyslexia family sample. J Neurodev Disord. 2011;3:39–49. doi: 10.1007/s11689-010-9065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliak S, Gollan L, Martinez R, Custer A, Einheber S, Salzer JL, Trimmer JS, Shrager P, Peles E. Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron. 1999;24:1037–1047. doi: 10.1016/s0896-6273(00)81049-1. [DOI] [PubMed] [Google Scholar]
- Poliak S, Gollan L, Salomon D, Berglund EO, Ohara R, Ranscht B, Peles E. Localization of Caspr2 in myelinated nerves depends on axon-glia interactions and the generation of barriers along the axon. J Neurosci. 2001;21:7568–7575. doi: 10.1523/JNEUROSCI.21-19-07568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, Stewart CL, Xu X, Chiu S-Y, Shrager P, et al. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. 2003;162:1149–1160. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price PH. Developmental determinants of structure in zebra finch song. J Comp Physiol Psych. 1979;93:260–277. [Google Scholar]
- Ramón y Cajal S. Le lobe optique des vertébrés inférieurs, toit optique des oiseaux. In: Ramón y Cajal, editor. Histologie du systéme nerveux de l’homme e des vertébrés. Maloine; Paris: 1911. [Google Scholar]
- Rasband WS. ImageJ. U. S. National Institutes of Health; Bethesda, Maryland, USA: 1997-2012. http://imagej.nih.gov/ij/ [Google Scholar]
- Reiner A, Perkel DJ, Bruce LL, Butler AB, Csillag A, Kuenzel W, Medina L, Paxinos G, Shimizu T, Striedter G, et al. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J Comp Neurol. 2004;473:377–414. doi: 10.1002/cne.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TF, Wild JM, Kubke MF, Mooney R. Homogeneity of intrinsic properties of sexually dimorphic vocal motoneurons in male and female zebra finches. J Comp Neurol. 2007;502:157–169. doi: 10.1002/cne.21310. [DOI] [PubMed] [Google Scholar]
- Scott BB, Lois C. Developmental origin and identity of song system neurons born during vocal learning in songbirds. J Comp Neurol. 2007;502:202–214. doi: 10.1002/cne.21296. [DOI] [PubMed] [Google Scholar]
- Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Sonnenblick LI, Rudie JD, Ghahremani D, Mumford JA, Poldrack RA, Dapretto M, Geschwind DH, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010;2:56ra80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Horwitz B, Jarvis ED. Dopamine regulation of human speech and bird song: a critical review. Brain Lang. 2012;122:142–150. doi: 10.1016/j.bandl.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro JE, Dalva MB, Mooney R. Long-range inhibition within the zebra finch song nucleus RA can coordinate the firing of multiple projection neurons. J Neurophysiol. 1999;81:3007–3020. doi: 10.1152/jn.1999.81.6.3007. [DOI] [PubMed] [Google Scholar]
- Stoeger AS, Mietchen D, Oh S, de Silva S, Herbst CT, Kwon S, Fitch WT. An asian elephant imitates human speech. Curr Biol. 2012;22:2144–2148. doi: 10.1016/j.cub.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006;14:159–166. doi: 10.1016/j.str.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci. 2004;24:3152–3163. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramitsu I, White SA. FoxP2 regulation during undirected singing in adult songbirds. J Neurosci. 2006;26:7390–7394. doi: 10.1523/JNEUROSCI.1662-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcón M, Oliver PL, Davies KE, Geschwind DH, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario DS. Organization of the zebra finch song control system: II. Functional organization of outputs from nucleus Robustus archistriatalis. J Comp Neurol. 1991;309:486–494. doi: 10.1002/cne.903090405. [DOI] [PubMed] [Google Scholar]
- White SA. Genes and vocal learning. Brain Lang. 2010;115:21–28. doi: 10.1016/j.bandl.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse AJO, Bishop DVM, Ang QW, Pennell CE, Fisher SE. CNTNAP2 variants affect early language development in the general population. Genes, brain, and behavior. 2011;10:451–456. doi: 10.1111/j.1601-183X.2011.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney O, Johnson F. Motor-induced transcription but sensory-regulated translation of ZENK in socially interactive songbirds. J Neurobiol. 2005;65:251–259. doi: 10.1002/neu.20187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild JM, Williams MN, Suthers RA. Parvalbumin-positive projection neurons characterise the vocal premotor pathway in male, but not female, zebra finches. Brain Res. 2001;917:235–252. doi: 10.1016/s0006-8993(01)02938-9. [DOI] [PubMed] [Google Scholar]
- Wild JM, Williams MN, Howie GJ, Mooney R. Calcium-binding proteins define interneurons in HVC of the zebra finch (Taeniopygia guttata) J Comp Neurol. 2005;483:76–90. doi: 10.1002/cne.20403. [DOI] [PubMed] [Google Scholar]
- Wild JM, Kubke MF, Mooney R. Avian nucleus retroambigualis: cell types and projections to other respiratory-vocal nuclei in the brain of the zebra finch (Taeniopygia guttata) J Comp Neurol. 2009;512:768–783. doi: 10.1002/cne.21932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Coats JK, Yang CF, Wang A, Ahmed OM, Alvarado M, Izumi T, Shah NM. Modular Genetic Control of Sexually Dimorphic Behaviors. Cell. 2012;148:596–607. doi: 10.1016/j.cell.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]