Abstract
Background
Turner syndrome (TS) is a developmental disorder caused by partial or complete monosomy for the X chromosome in 1:2500 females. We hypothesized that single nucleotide polymorphism (SNP) array genotyping can provide superior resolution in comparison to metaphase karyotype analysis to facilitate genotype-phenotype correlations.
Methods
We genotyped 187 TS patients with 733,000 SNP marker arrays. All cases met diagnostic criteria for TS based on karyotypes (60%) or characteristic physical features. SNP array results confirmed the diagnosis of TS in 100% of cases.
Results
We identified a single X chromosome (45,X) in 113 cases. In 58 additional cases (31%), other mosaic cell lines were present including isochromosomes (16%), rings (5%) and Xp deletions (8%). The remaining cases were mosaic for monosomy X and normal male or female cell lines. Array-based models of X chromosome structure were compatible with karyotypes in 104 of 116 comparable cases (90%). We found that SNP array data did not detect X;autosome translocations (3 cases), but did identify 2 derivative Y chromosomes and 13 large copy number variants that were not detected by karyotyping.
Conclusions
Our data is the first systematic comparison between the two methods and supports the utility of SNP array genotyping to address clinical and research questions in TS.
Keywords: Turner Syndrome, Array, sex chromosomes, Karyotype, Cytogenetics, diagnosis
INTRODUCTION
Turner Syndrome (TS) is a developmental disorder caused by partial or complete loss of one X chromosome that occurs in approximately 1 in 2500 live female births. TS women present with multiple developmental defects, including skeletal abnormalities, cardiac defects and premature ovarian failure.1 Approximately half of TS patients have a single X chromosome in all somatic cells that are analyzed clinically, mainly lymphocytes. The others are mosaics of 45,X and 46,XX cells or structural derivatives including isochromosomes, rings and deletions. The severity of TS features are generally correlated with the percentage of cells that harbor a single copy of Xp, but only one gene has been implicated in a specific phenotype. Decreased expression of the SHOX gene in the pseudoautosomal region of Xp is associated with short stature and skeletal deformities.2,3
Karyotype analysis of phytohemagglutinin-stimulated lymphocyte cultures has been the gold standard for the diagnosis of TS. In a metaphase spread, clinical laboratories typically evaluate 20–50 Giemsa-banded cells, and follow up abnormal results using fluorescence in situ hybridization (FISH). However this may not be sufficient to detect low-level (<10%) mosaicism. Differential growth of aneuploid cells may lead to their abnormal representation in culture. In some cases, lymphocytes have even been shown to accumulate chromosomal aberrations while in culture that are not detectable in whole blood.4,5 The likelihood that these artifacts will occur depends on the manner in which the samples are prepared. Cytogenetic analysis is a slow, labor intensive, multistep process that is difficult to standardize and subject to considerable variability. As the cost declines and speed of analysis increases, whole-genome microarray methods have become the diagnostic standard for many chromosomal disorders, but not for TS.
We hypothesized that single nucleotide polymorphism (SNP) array genotyping can provide superior resolution in comparison to karyotyping to facilitate rapid diagnosis and genotype-phenotype correlations in TS. Here we present the first systematic comparison between the two methods and provide evidence for the utility of SNP array genotyping to address clinical and research issues in TS.
MATERIALS AND METHODS
Subject selection
All women who were diagnosed with Turner syndrome (TS) based on suggestive phenotypic features were included in the study. For patients with peripheral blood cell karyotypes, an X chromosomal abnormality was required to be present for inclusion. After reviewing the clinical data, five patients were excluded from further study because they did not meet diagnostic criteria for TS. These subjects had been labeled as ‘possible TS’ and had not previously been genotyped. All other patients, including those with ring X chromosomes, had typical TS features and X chromosome abnormalities consistent with TS.
Samples
We studied 192 samples from females of European ancestry. We obtained 111 samples from the National Registry of Genetically Triggered Aortic Aneurysms and Other Cardiovascular Conditions (GenTAC, Rockville, MD). GenTAC is a consortium of eight institutions that is coordinated by the National Heart, Lung and Blood Institute, which maintains a central repository of tissue, DNA and limited phenotypic data on patients with congenital heart disease, including TS. We also obtained 75 samples from the National Institute of Child Health and Human Development (NICHD). An additional six samples were from Baylor College of Medicine (two) and University of Texas Health Science Center at Houston (UTHSC-H) (four). Reports of karyotypes were available for 116 cases (60%). All karyotypes were post-natal. Of 44 reports with mosaic cell lines, 27 reported 50-cell karyotypes and 9 reported karyotypes of 30 or fewer cells. The number of karyotyped cells was not reported in the other cases. Samples were collected simultaneously from NIH patients for karyotyping and genotyping. The relative timing of sample collections from GenTAC patients was not known to the investigators.
Genotyping and Copy Number Analysis
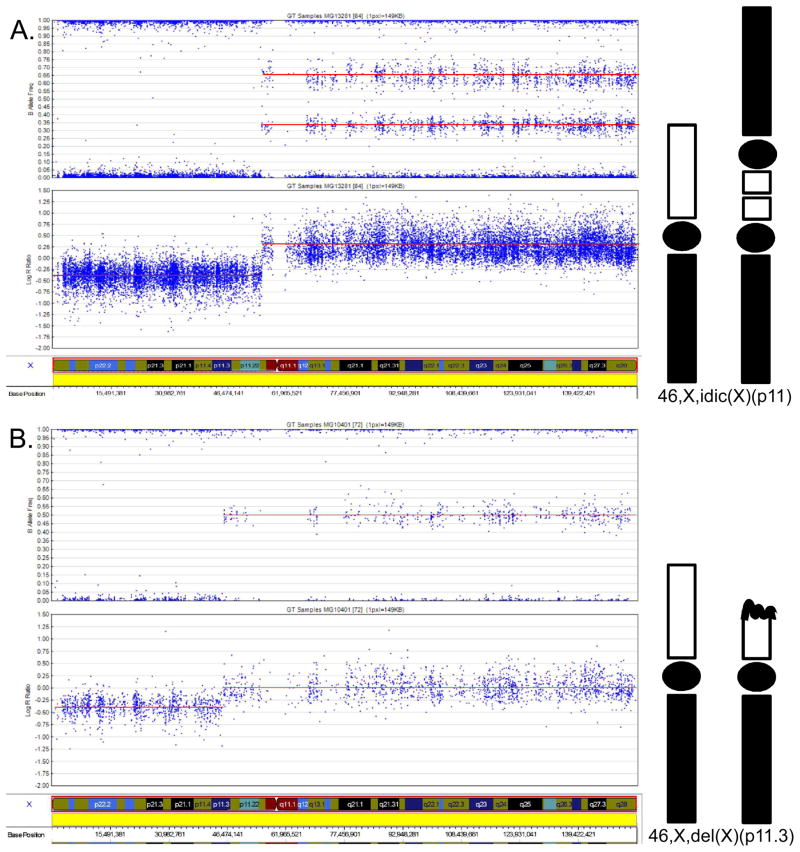
All DNAs were obtained by extraction from whole blood. Sample processing, DNA purification and hybridization were performed as previously described.6 Data for each Omni-Express BeadChip (Illumina, Inc.) were normalized in Genome Studio using information contained within the array. After allele detection and genotype calling, B allele frequencies (BAF) and logR ratios (LRR) were exported as text files for analysis with PennCNV software (www.openbioinformatics.org/penncnv).7 A second CNV detection algorithm, CNVPartition, was run as a plug-in within the GenomeStudio browser. Confidence thresholds and minimum number of probes per CNV were set to default values. Control genotypes from unrelated individuals of European ancestry in the database of Genotypes and Phenotypes were analyzed for copy number variation using the same methods.8 DNA copy number and percent mosaicism were calculated using BAF and LRR values for 18,239 SNPs along the length of the X chromosome. Non-mosaic 45,X genotypes were identified by loss of heterozygosity (LOH) across the entire X chromosome. BAF values were also used to determine the number of haplotypes present in isochromosome cell lines and map crossover events. Calculation of percent mosaicism for X and Y chromosome SNPs was based on the deviation of allele frequencies from expected values for copy losses or copy gains according to the method of Conlin et al.9 To deduce the most likely genotypes and estimate the percentages of mosaic cell lines, we compared mean BAF and LRR values of segmental aneuploidies to expected values for monosomies and trisomies. Figure 1 illustrates these computations for a TS case with a non-mosaic, isodicentric X chromosome. Array data and karyotypes were judged to be compatible if cell lines that were observed in more than one G-banded cell were also present in the genotypes.
Figure 1.
SNP array genotypes of two TS cases illustrate the computation of X chromosome structure. B allele frequencies (BAF) are plotted on the upper panel and corresponding LogR ratios (LRR) on the lower panel. Mean values are indicated by red lines. Positive LRR values represent copy gains and negative LRR values represent copy losses. The X chromosome model corresponding to these values is illustrated on the right. Xp is white, Xq is black and centromeres are oval. A. A TS case with 46,X,idic(X)(p11). Homozygosity and mean LRR of −0.41 indicate segmental monosomy of Xpter-Xp11.22. The abrupt increase of LRR to 0.26 and division of BAF into four tracks (1.00, 0.66, 0.34, 0) indicate trisomy of Xp11-Xqter. B. A TS case with 46,X,del(X)(p11.3). Homozygosity and mean LRR of −0.39 indicate segmental monosomy of Xpter-Xp11.3. The abrupt increase of LRR to and division of BAV into three tracks (1.00, 0.50, 0) indicate two copies of Xp11.3-Xqter. A mosaic such as 45,X[50]/46,X,i(X)(p11.3)[50] would appear similar to 46,X,del(X)(p11.3) in the array data, but could be distinguished from the deletion by combined analysis of B-allele frequencies and LogR ratios.
Statistical methods
Chi-squared or Fisher’s exact tests were applied to test quantitative parameters between the different study groups. All tests applied were two-tailed, and a P value of 5% or less was considered statistically significant.
Human subjects
Studies were carried out with the approval of the Committee for the Protection of Human Subjects at the UTHSC-H, the GenTAC Scientific Advisory Committee (Rockville, MD) and the NICHD Institutional Review Board. All samples were from de-identified individuals who had been referred for clinical genetic testing. Karyotype information was provided by the treating physicians. After collection, all samples were de-identified to preserve patient confidentiality.
RESULTS
DNA samples from 187 TS patients (European American, average age 32 years) were genotyped using 733,000 SNP marker arrays. 116 cases had karyotypes and 71 cases were thought to have TS based on characteristic physical features including short stature, premature ovarian failure and congenital cardiovascular defects such as bicuspid aortic valve or coarctation (Table 1). Importantly, genotyping confirmed the presence of X chromosome aneuploidy consistent with the diagnosis of TS in all patients with a strong index of clinical suspicion for whom karyotypes were not available.
Table 1.
Characteristics of 187 Turner Syndrome (TS) Cases
| Mean | Quartile 1 | Quartile 3 | |
|---|---|---|---|
| Age (yrs) | 31.6 | 17.9 | 44.8 |
| Height (cm) | 145.2 | 140 | 152 |
| Weight (kg) | 58.9 | 47.3 | 69.6 |
| BSA (m2) | 1.52 | 1.37 | 1.69 |
| Number | Percentage | ||
| BAV | 68 | 35 | |
| Coarctation | 29 | 15 | |
| Genotype | |||
| 45,X | 113 | 60 | |
| 45,X/46,X,iso(Xq) | 31 | 16 | |
| 45,X/46,X,del(X) | 16 | 8 | |
| 45,X/46,XY | 9 | 5 | |
| 45,X/46,X,der(X) | 8 | 4 | |
| 45,X/46,XX | 7 | 4 | |
| 46,X,der(X)t(X;A) | 3 | 2 | |
Values are presented as means and interquartile ranges except for BAV and coarctation. There were no missing values for age, 3 missing values for height and 2 missing values for weight. Der(X) indicates probable ring chromosome. Total of genotypes does not include translocations. BSA: body surface area; BAV: bicuspid aortic valve.
We identified a single X chromosome (45,X) in 113 cases (60%) and mosaic 45,X and 46,XX or 46,XY cell lines in 16 cases (9%). In 58 additional cases (31%), other structural variants were present, including isochromosomes (16%), rings (5%) and Xp deletions (8%) (Table 1). Arrays did not detect the presence of 3 or more cell lines in any sample, whereas karyotyping identified a third cell line in a single case. Arrays did detect Y chromosome material in 10 TS cases (5%), including all six cases with Y-positive karyotypes and two cases that were not detected by karyotyping. This distribution of genotypes is not significantly different from that reported in the literature.10
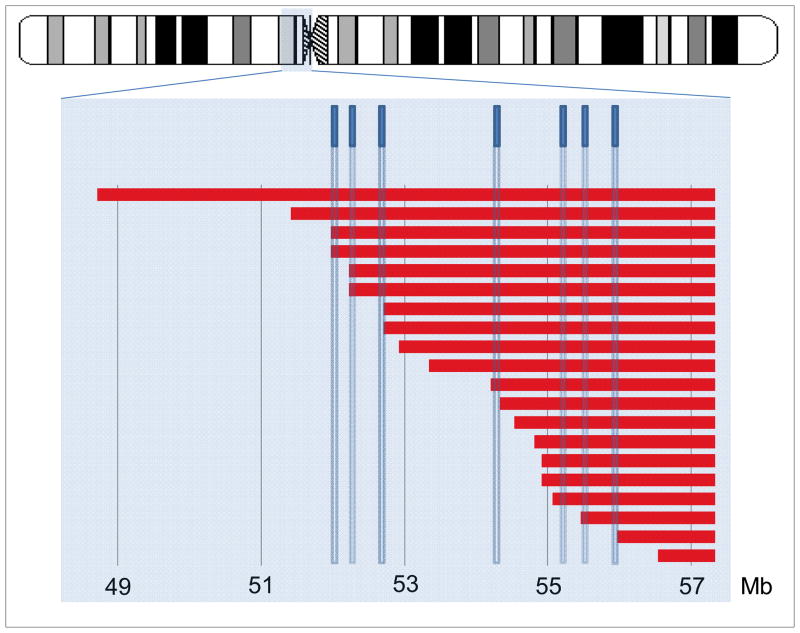
Array-based models of X chromosome structure were compatible with karyotypes in 104 of 116 cases (90%) with available data (Table S1). Sixty-three of these cases (55%) had 45, X genotypes, compared with 60% (113/187) of all cases (P>0.05). Array analysis showed that 19 of the 31 X isochromosomes include proximal Xp material and are actually isodicentric. The breakpoints of these isochromosomes are clustered within a 5 Mb region of Xp11 that is enriched for low copy palindromic repeats (Figure 1). Recurrent Xp deletions and X;autosome translocations were also mapped to this interval.11,12 Our findings provide further evidence that Xp11 is uniquely susceptible to chromosomal rearrangements.
Eight of 12 cases that were discordant between the SNP and karyotyped data involved mosaic cell lines that were detected in one but not both assays. Four cases with 45,X karyotypes were reclassified as mosaics according to SNP data, with a second cell line containing an abnormal sex chromosome, or a normal cell line. Three cases had mosaic karyotypes, but were found to be non-mosaic for monosomy X using SNP arrays. In five cases, arrays and karyotypes identified discrepant X chromosome abnormalities. In four of these cases, mosaicism was demonstrated in both arrays and karyotypes. These mosaic lines comprised between 4 and 30% of sampled cells and included an intact X chromosome in 1 case and derivative chromosomes in 7 cases (Table 2). In 4 of these 8 cases, samples for arrays and karyotypes were known to have been collected simultaneously. We also found that the percentages of aneuploid cell lines as determined by arrays and karyotypes differed by more than two-fold in 11 of 34 mosaic cases (32%).
Table 2.
Discordant Cytogenetic and Microarray Findings in 12 TS Cases
| Cyto | Array |
|---|---|
| 45,X | 45,X[83%]/46,X,der(X)(p22.12-q27.3)[17%] |
| 45,X | 45,X[75%]/46,X,del(Y)(q11.21)[25%] |
| 45,X | 45,X[85%]/46,X,idic(X)(p11.22)[15%] |
| 45,X | 45,X[83%]/46XX[17%] |
| 45,X[80%]/46,X,r(X)[20%]c | 45,X |
| 45,X[96%]/46,X,r(X)[4%]b | 45,X |
| 45,X/46,X,iso(Xq)/47,X,iso(Xq),iso(Xq)c | 45,X |
| 46,X,del(X)(p11.1) | 45,X |
| 45,X[80%]/46,X,idic(Y)(q12)[20%]a | 45,X[90%]/46,XY[10%] |
| 45,X[88%]/46,X,psuidic(X)(q21)[12%]b | 45,X[97%]/46,X,del(X)(q21.31)[3%] |
| 45,X[98%]/47,XXX[2%]b | 45,X[98%]/46,XX[2%] |
| 45,X[85%]/46,XX[15%]a | 45,X[91%]/46,X,del(X)(q21.1)[9%] |
Clinical karyotypes (Cyto) were compared with the most likely karyotypes inferred from SNP array analysis (Array). The percentages of mosaic cell lines in the sample are bracketed. For Cyto cases, these were extrapolated from the number of karyotyped cells: 20a, 50b or unknownc. Breakpoints are in parentheses. iso: isochromosomes; idic: isodicentric chromosomes; der: derivative chromosomes, probably rings; del: deleted chromosomes.
In four cases, the SNP data were compatible with two possible genotypes that result in identical array patterns: mosaics with equal 50% contributions of 46, X, i(X) and 45, X cell lines, or non-mosaic Xp deletions (Table S1). To distinguish between these possibilities, we evaluated the chromosomal regions of the breakpoints, searched for additional haplotypes indicative of meiotic non-disjunction and precisely quantitated the logR ratio deviations. Further analysis confirmed that the structural variants in all four of these cases were isochromosomes.
Array data successfully identified four marker chromosomes whose chromosomal origins were not identified in karyotypes as a Y chromosome, a derivative Y chromosome, a deleted X chromosome and a ring X chromosome. SNP analysis of three TS cases with reported X;autosome translocations (2%) showed that the translocations were unbalanced and resulted in copy losses with breakpoints in Xp. Two of these translocations did not cause autosomal copy changes. However, in the other case, unbalanced t(X;9) resulted in trisomy 9p. We were unable to determine whether typical features of the trisomy 9p syndrome (dysmorphic facial features and mental retardation) are present in this individual.
Arrays also identified 13 large X chromosome copy number variants (CNVs > 100 Kb in size) in 12 different individuals that were not detected by karyotyping (Table 3). Five of these CNVs were extremely rare and were present in fewer than 0.1% of controls. None of these CNVs involved genes with known roles in TS phenotypes.
Table 3.
Large X Chromosome CNVs Detected by SNP Arrays in 13 TS Cases
| Start | End | Size | Genes | Rare | Karyotype |
|---|---|---|---|---|---|
| 47,764,051 | 47,929,059 | 165,008 | ZNF630,SS6,SPACA5 | Yes | 45,X |
| 56,848,420 | 58,499,972 | 1,651,552 | SPIN3,SPIN2B,SPIN2A,FAAH2, ZXDB,ZXDA | Yes | 45,X |
| 139,827,866 | 139,903,440 | 75,574 | None | Yes | 45,X |
| 80,418,197 | 81,836,210 | 1,418,013 | SH3BGRL | Yes | 45X/46X,der(X)(p11.3-q21.1) |
| 52,837,999 | 72,818,464 | 19,980,465 | 109 genes | Yes | 45,X/46,X,idic(X)(p11.22) |
| 23,280,606 | 25,087,076 | 1,806,470 |
PTCHD1,PRDX4,ACOT9,SAT1, APOO,CXORF58,KLHL15 EIF2S3,ZFX1,PDK3,POLA1, EEF1B2,ARX |
Yes | 45,X/46,X,idic(X)(p11.21) |
| 154,592,212 | 154,913,754 | 321,542 | SPRY3,VAMP7,IL9R,WASH1 | Yes | 46,X,der(Y)(p11.2-qter) |
| 81,067,157 | 89,536,657 | 8,469,500 | POU3F4,CYLC1,RPS6KA6,HDX, APOOL,SATL1,ZNF711,CHM, DACH2,KLHL4,CPXCR1,TGIF2LX | Yes | 45,X/46X,der(X)(p11.3-q21.1) |
| 114,122,419 | 114,301,487 | 179,068 | IL13RA2,LRCH2 | Yes | 45,X |
| 31,946,712 | 32,012,647 | 65,935 | DMD | Yes | 45,X/47,XXX |
| 123,864,615 | 154,913,754 | 31,049,139 | 214 genes | Yes | 46,X,del(X)(p11.23) |
| 6,452,425 | 8,095,053 | 1,642,628 | VCX3A,HDHD1A,STS,VCX, PLPLA4 | No | 45,X |
| 115,825,108 | 115,929,301 | 104,193 | None | No | 45,X/46,XY |
Start, and end base pair values are in hg18 coordinates. All CNVs were copy gains except for 139,827,866–139,903,440, which was a copy loss. Genes: genes included in the CNV. Rare: Yes if CNVs overlapped indicated regions in less than 1% of 5,108 female controls from the database of Genotypes and Phenotypes (dbGAP). Karyotype: karyotype of individual with CNV.
DISCUSSION
Early diagnosis of TS is essential for interventions to restore normal or near-normal adult stature and for management of complications, including aortic coarctation, renal disease or gonadoblastomas, which are prevalent in the subgroup with Y chromosome material.13 Four NIH participants in this study underwent relevant evaluations, genetic counseling and prophylactic gonadectomies according to the current standard of care after karyotypes disclosed cryptic Y chromosomes.14 TS patients without 45,X genotypes tend to be diagnosed later in life but have similar rates of cardiovascular complications.15 Current PCR-based diagnostic methods have limited ability to detect the 40% of cases that involve mosaicism or X chromosome structural variants.16 Cytogenetic analysis by karyotyping is too labor intensive for rapid population-based screening and may not detect small fragments of Y chromosome material. Our data show that SNP array genotyping is a feasible alternative to karyotype testing for diagnosing TS. Moreover, the sensitivity and precision of SNP arrays may lead to improved genotype-phenotype correlations in TS.
TS may be caused by loss of an entire X chromosome (45,X) in all cells, partial deletion of one X chromosome in all cells, or X chromosome deletions in a subset of cells (TS mosaicism). In almost all cases, the deletions cause partial or complete monosomy for Xp. Variation of Xq dosage may be extensive and includes duplications of Xq due to isochromosome formation in 15% of patients. SNP arrays were robustly able to detect all four categories of TS cases. We also showed that SNP data led to the reclassification of four reported 45,X karyotypes (8%) to mosaic TS cases and identified Y chromosome material that was not detected in two karyotypes. Ten of the large X chromosome CNVs found in 6% of cases are rare or absent in controls and involve genes that may modify TS outcomes. It is important to note that rare CNVs were previously implicated in neurodevelopmental disorders and thoracic aortic aneurysms and dissections, phenotypes that are both relevant to TS.8,17 These data show that SNP arrays can provide additional prognostic value beyond karyotyping alone in the evaluation of TS patients.
Arrays and karyotypes were most frequently discordant due to differences in the detection of mosaic cell lines. In 4 of the 12 discordant cases, arrays led to incorrect interpretations of rare cell lines that were present in fewer than 5% of sampled cells. This was most likely caused by technical limitations of the array data. However, 6 cases involved much larger variations in relatively abundant cell lines (greater than 10% mosaicism) that could impact prognosis. We found no correlation between differences in the timing of sample collection and these discrepancies. Because both techniques are sensitive enough to detect mosaicism down to a level of 5%, we propose that these larger differences most likely arose or were amplified during cell culture.9,18 Karyotypes are routinely produced after culturing peripheral blood lymphocytes for up to 72 hours. Under similar conditions, aneuploid cells have been shown to occur spontaneously in other human cell types, with the potential for multiple different mosaic lines to arise in consecutive cultures from the same individual.19–21 Aneuploid cells may also have a selective growth advantage or disadvantage compared with co-cultured euploid cells from mosaic patients and may be amplified or suppressed by current culture methods. Moreover, in comparison with euploid cells, chromosomal instability appears to be more pronounced in TS patients.22–24 Mosaicism differences between karyotypes and arrays may also be accentuated due to sampling bias, especially if fewer than 50 cells are counted. SNP array analysis of whole blood eliminates the potential for selective pressure due to cell culture and may therefore provide a more accurate representation of mosaicism in peripheral blood.
One important limitation of this study is our inability to verify our findings using an independent method such as FISH or by counting additional karyotyped cells. We found that array data lacks sufficient resolution to identify some low abundance mosaic cell lines, which may be more accurately assessed in karyotypes. We also confirmed that karyotyping retains an important advantage over arrays to identify complex mosaicism, including translocations and rare X chromosome structural variants. SNP genotyping is unable to detect fully balanced X-autosome translocations, but these were not present in our series.
In summary, the findings presented here indicate that SNP array analysis can be used to diagnose TS and may provide distinct advantages over karyotypes in the evaluation of TS.
Supplementary Material
Figure 2.
Location of 20 isochromosome breakpoints in Xp11.22-Xp11.1 in relation to an ideogram of the X chromosome with the highlighted region and previously mapped low copy repeat sequences (blue rectangles). The breakpoints cluster in a region between 52 and 56 Mb as previously shown by Koumbaris et al. The array that was used in this study has an average maximum resolution of 8000 base pairs.
Acknowledgments
GenTAC participating centers are as follows: Johns Hopkins University, Kathryn W. Holmes, MD, Harry C. Dietz, MD, Williams Ravekes, MD, Kira Lurman, RN; University of Texas Health Science Center at Houston, Dianna M. Milewicz, MD, PhD, Meghan Terry, Alana Cecchi, MS, CGC; Baylor College of Medicine, Scott A. LeMaire, MD, Irina Volguina, PhD; Oregon Health and Science University, Cheryl L. Maslen, PhD, Howard K. Song, MD, PhD, Victor Menashe, MD, Jessica D. Kushner, MS, CGC; University of Pennsylvania, Reed E. Pyeritz, MD, PhD, Joseph E. Bavaria, MD, Megan Morales; Weill Medical College of Cornell University, Craig T. Basson, MD, PhD, Richard Devereux, MD, Jonathan W. Weinsaft, MD, Deborah McDermott, MS, CGC; University of Michigan, Kim Eagle, MD; National Heart, Lung, and Blood Institute, H. Eser Tolunay, PhD, Patrice Desvigne-Nickens, MD, Mario P. Stylianou, PhD, Megan Mitchell, MPH; RTI International, Barbara L. Kroner, PhD, Donald Brambilla, PhD, Tabitha Hendershot, Danny Ringer, Meg Cunningham, Mark Kindem.
The authors are extremely grateful to patients for providing clinical data and samples for this study. The following sources provided funding to D.M.M. for these studies: RO1 HL62594 (D.M.M.), P50HL083794-01 (D.M.M.), Vivian L. Smith Foundation, Richard T. Pisani Funds. and TexGen Foundation. The NIH UL1 RR024148 (CTSA) grant also provided funds for these studies. The GenTAC Registry has been supported by US Federal Government contracts HHSN268200648199C and HHSN268201000048C from the NHLBI (Bethesda, MD) with RTI International (Research Triangle Park, NC). Additional support was provided by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, the Oregon Clinical and Translational Research Institute (Portland, OR), grant UL1 RR024140 from the National Center for Research Resources (Bethesda, MD), and by the Weill Cornell Medical College Clinical Translational Science Center (New York, NY), grant UL1RR024996.
Footnotes
CONFLICTS OF INTEREST
S.P. Nothing to disclose
D.G. Nothing to disclose
D.M.M. Nothing to disclose
C.L.M Nothing to disclose
M.S Nothing to disclose
C.A.B Nothing to disclose
Supplementary information is available at the Genetics in Medicine website.
References
- 1.Matura LA, Ho VB, Rosing DR, Bondy CA. Aortic dilatation and dissection in Turner syndrome. Circulation. 2007 Oct 9;116(15):1663–1670. doi: 10.1161/CIRCULATIONAHA.106.685487. [DOI] [PubMed] [Google Scholar]
- 2.Rao E, Weiss B, Fukami M, et al. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nature genetics. 1997 May;16(1):54–63. doi: 10.1038/ng0597-54. [DOI] [PubMed] [Google Scholar]
- 3.Hirschfeldova K, Solc R, Baxova A, et al. SHOX gene defects and selected dysmorphic signs in patients of idiopathic short stature and Leri-Weill dyschondrosteosis. Gene. 2012 Jan 10;491(2):123–127. doi: 10.1016/j.gene.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 4.El-Zein R, Gu Y, Sierra MS, Spitz MR, Strom SS. Chromosomal instability in peripheral blood lymphocytes and risk of prostate cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive. Oncology. 2005 Mar;14(3):748–752. doi: 10.1158/1055-9965.EPI-04-0236. [DOI] [PubMed] [Google Scholar]
- 5.Salas C, Niembro A, Lozano V, et al. Persistent genomic instability in peripheral blood lymphocytes from Hodgkin lymphoma survivors. Environmental and molecular mutagenesis. 2012 May;53(4):271–280. doi: 10.1002/em.21691. [DOI] [PubMed] [Google Scholar]
- 6.Peiffer DA, Le JM, Steemers FJ, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006 Sep;16(9):1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007 Nov;17(11):1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prakash SK, LeMaire SA, Guo DC, et al. Rare copy number variants disrupt genes regulating vascular smooth muscle cell adhesion and contractility in sporadic thoracic aortic aneurysms and dissections. American journal of human genetics. 2010 Dec 10;87(6):743–756. doi: 10.1016/j.ajhg.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlin LK, Thiel BD, Bonnemann CG, et al. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Human molecular genetics. 2010 Apr 1;19(7):1263–1275. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolff DJ, Van Dyke DL, Powell CM Working Group of the ALQAC. Laboratory guideline for Turner syndrome. Genet Med. 2010 Jan;12(1):52–55. doi: 10.1097/GIM.0b013e3181c684b2. [DOI] [PubMed] [Google Scholar]
- 11.Koumbaris G, Hatzisevastou-Loukidou H, Alexandrou A, et al. FoSTeS, MMBIR and NAHR at the human proximal Xp region and the mechanisms of human Xq isochromosome formation. Human molecular genetics. 2011 May 15;20(10):1925–1936. doi: 10.1093/hmg/ddr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott SA, Cohen N, Brandt T, Warburton PE, Edelmann L. Large inverted repeats within Xp11.2 are present at the breakpoints of isodicentric X chromosomes in Turner syndrome. Human molecular genetics. 2010 Sep 1;19(17):3383–3393. doi: 10.1093/hmg/ddq250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiktor A, Van Dyke DL. FISH analysis helps identify low-level mosaicism in Ullrich-Turner syndrome patients. Genet Med. 2004 May-Jun;6(3):132–135. doi: 10.1097/01.gim.0000127270.49902.56. [DOI] [PubMed] [Google Scholar]
- 14.Bondy CA Turner Syndrome Study G. Care of girls and women with Turner syndrome: a guideline of the Turner Syndrome Study Group. J Clin Endocrinol Metab. 2007 Jan;92(1):10–25. doi: 10.1210/jc.2006-1374. [DOI] [PubMed] [Google Scholar]
- 15.El-Mansoury M, Barrenas ML, Bryman I, et al. Chromosomal mosaicism mitigates stigmata and cardiovascular risk factors in Turner syndrome. Clin Endocrinol (Oxf) 2007 May;66(5):744–751. doi: 10.1111/j.1365-2265.2007.02807.x. [DOI] [PubMed] [Google Scholar]
- 16.Rivkees SA, Hager K, Hosono S, et al. A highly sensitive, high-throughput assay for the detection of Turner syndrome. J Clin Endocrinol Metab. 2011 Mar;96(3):699–705. doi: 10.1210/jc.2010-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto D, Pagnamenta AT, Klei L, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010 Jul 15;466(7304):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hook EB. Exclusion of chromosomal mosaicism: tables of 90%, 95% and 99% confidence limits and comments on use. American journal of human genetics. 1977 Jan;29(1):94–97. [PMC free article] [PubMed] [Google Scholar]
- 19.Ly P, Eskiocak U, Kim SB, et al. Characterization of aneuploid populations with trisomy 7 and 20 derived from diploid human colonic epithelial cells. Neoplasia. 2011 Apr;13(4):348–357. doi: 10.1593/neo.101580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu AM, Qu WW, Liu X, Qu CK. Chromosomal instability in in vitro cultured mouse hematopoietic cells associated with oxidative stress. American journal of blood research. 2012;2(1):71–76. [PMC free article] [PubMed] [Google Scholar]
- 21.Josse C, Schoemans R, Niessen NA, et al. Systematic chromosomal aberrations found in murine bone marrow-derived mesenchymal stem cells. Stem cells and development. 2010 Aug;19(8):1167–1173. doi: 10.1089/scd.2009.0264. [DOI] [PubMed] [Google Scholar]
- 22.Tyrkus M, Hoffman WH, Kraemer-Flynn KM. X chromosome instability associated with familial Turner syndrome. Clin Genet. 1989 Feb;35(2):111–115. doi: 10.1111/j.1399-0004.1989.tb02914.x. [DOI] [PubMed] [Google Scholar]
- 23.Park JP, Brothman AR, Butler MG, et al. Extensive analysis of mosaicism in a case of Turner syndrome: the experience of 287 cytogenetic laboratories. College of American Pathologists/American College of Medical Genetics Cytogenetics Resource Committee. Arch Pathol Lab Med. 1999 May;123(5):381–385. doi: 10.1043/0003-9985(1999)123<0381:EAOMIA>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawyer JR, Swanson CM, Lukacs JL, et al. Telomeric fusion and chromosome instability in multiple tissues of a patient with mosaic Ullrich-Turner syndrome. Am J Med Genet. 1997 Apr 14;69(4):383–387. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.