Abstract
Regulator of G Protein Signaling 14 (RGS14) is a multifunctional scaffolding protein that integrates G protein and MAPK signaling pathways. In the adult mouse brain, RGS14 mRNA and protein are found almost exclusively in hippocampal CA2 neurons. We have shown that RGS14 is a natural suppressor of CA2 synaptic plasticity and hippocampal-dependent learning and memory. However, the protein distribution and spatiotemporal expression patterns of RGS14 in mouse brain during postnatal development are unknown. Here, using a newly characterized monoclonal anti-RGS14 antibody, we demonstrate that RGS14 protein immunoreactivity is undetectable at birth (P0) with very low mRNA expression in the brain. However, RGS14 protein and mRNA are upregulated during early postnatal development, with protein first detected at P7, and both increasing over time until reaching highest sustained levels throughout adulthood. Our immunoperoxidase data demonstrate that RGS14 protein is expressed in regions outside of hippocampal CA2 during development including the primary olfactory areas, the anterior olfactory nucleus and piriform cortex, and the olfactory associated orbital and entorhinal cortices. RGS14 is also transiently expressed in neocortical layers II/III and V during postnatal development. Finally, we show that RGS14 protein is first detected in the hippocampus at P7 with strongest immunoreactivity in CA2 and fasciola cinerea and sporadic immunoreactivity in CA1; labeling intensity in hippocampus increases until adulthood. These results show that RGS14 mRNA and protein are upregulated throughout postnatal mouse development, and RGS14 protein exhibits a dynamic localization pattern that is enriched in hippocampus and primary olfactory cortex in the adult mouse brain.
Keywords: RGS14, hippocampus, hippocampal CA2, synaptic plasticity, RGS proteins
INTRODUCTION
Regulator of G Protein Signaling 14 (RGS14) is a highly unusual signaling protein that integrates G protein and MAPkinase signaling pathways to regulate synaptic plasticity and hippocampal-based learning and memory (Lee et al., 2010; Shu et al., 2010; Vellano et al., 2013). RGS14 was first identified as a complex scaffolding protein with an unconventional domain structure that allows it to interact with various protein binding partners (Snow et al., 1997; Traver et al., 2000). Like other RGS proteins (Hollinger and Hepler, 2002), RGS14 contains an RGS domain, which binds to and accelerates the intrinsic GTPase activity of activated Gαi/o-GTP subunits to limit heterotrimeric G protein signaling. However, unlike most other RGS proteins, RGS14 also contains two tandem Ras/Rap-binding domains (RBDs), and a G protein regulatory (GPR) motif (Cho et al., 2000; Hollinger et al., 2001; Traver et al., 2000). RGS14 preferentially binds activated H-Ras-GTP as well as Raf kinases through the first RBD (RBD1) (Kiel et al., 2005; Shu et al., 2010; Vellano et al., 2013) and selectively binds inactive Gαi1/3-GDP subunits through the GPR motif to inhibit guanine nucleotide exchange and localize to cellular membranes (Kimple et al., 2001; Mittal and Linder, 2004; Shu et al., 2007). RGS14 has been shown to suppress ERK1/2 activity through a Giα1- and H-Ras-dependent mechanism, indicating that RGS14 functionally integrates G protein and MAP kinase signaling pathways (Shu et al., 2010).
Northern blot experiments (Snow et al., 1997), in situ hybridization studies (Grafstein-Dunn et al., 2001), and quantitative PCR (Larminie et al., 2004) have independently reported that RGS14 mRNA is present in rat and human brain tissue. Similarly, immunohistochemical studies (Lopez-Aranda et al., 2006) and immunoblot experiments (Hollinger et al., 2001) have found that RGS14 protein is enriched in rat and monkey brain. In the adult mouse brain, RGS14 mRNA and protein are predominantly found in CA2 hippocampal neurons, specifically within spines and dendrites (Lee et al., 2010). CA2 neurons uniquely exhibit a marked resistance to long-term potentiation (LTP) of excitatory glutamatergic synaptic transmission in response to stimulation of incoming CA3 Schaffer collaterals, which reliably induce LTP in neighboring CA1 neurons (Zhao et al., 2007). However, we have found that RGS14 knockout (RGS14-KO) mice display nascent and robust LTP in CA2 neurons, indicating that RGS14 is a natural suppressor of CA2 synaptic plasticity (Lee et al., 2010). Consistent with our previous report of RGS14 as a suppressor of MAP kinase signaling (Shu et al., 2010), CA2 LTP in RGS14-KO mice was blocked by a MEK/ERK inhibitor suggesting that RGS14 inhibits this pathway to limit synaptic plasticity in area CA2 (Lee et al., 2010; Caruana et al., 2012). Further, RGS14-KO mice have enhanced performance on tests of spatial and novel object memory, with no differences in nonhippocampal-dependent behaviors (Lee et al., 2010). Taken together, these data indicate that RGS14 is a natural suppressor of both synaptic plasticity and hippocampal-dependent learning and memory (Vellano et al., 2011).
Although previous studies have examined the localization of RGS14 mRNA and protein in adult rat and monkey brain (Grafstein-Dunn et al., 2001; Lopez-Aranda et al., 2006), a detailed anatomical analysis of RGS14 localization in mouse brain has not yet been reported. Further, no studies have examined the spatiotemporal expression pattern of RGS14 in the brain of any animal during postnatal development, a period in which hippocampal-dependent processes are required for adaptation and survival. Thus, to gain a better understanding of how RGS14 expression is regulated during postnatal mouse brain development, we examined total mRNA and protein levels using quantitative real-time PCR and immunoblotting. We also determined the localization and distribution of RGS14 protein during postnatal development by performing a detailed light microscopic immunohistochemical analysis of RGS14 localization in the mouse brain. We present the first comprehensive characterization of a recently described anti-RGS14 monoclonal antibody (Lee et al., 2010). Here we report that RGS14 protein expression is absent from brain at birth, but is upregulated and differentially expressed across brain regions during postnatal development (P0-P21) into adulthood where expression is restricted to hippocampal regions CA2 and fasciola cinerea, anterior olfactory nucleus, and piriform cortex. These findings suggest new roles for RGS14 in regulating physiology and behavior during mouse brain development.
MATERIALS AND METHODS
Animals and tissue preparation
Male and female wild-type C57BL/6J and RGS14 knockout (RGS14-KO) mice were used in this study. All procedures were approved by the animal care and use committee of Emory University and conform to the U.S. National Institutes of Health guidelines. Adult (older than 2 months) wild-type and RGS14-KO mice were obtained from mouse colonies maintained at Emory University. RGS14-KO mice were created by insertion of a LacZ/Neo cassette that deletes exons 2–7 of the RGS14 gene, and these mice were backcrossed to the C57BL/6J background as previously described (Lee et al., 2010). Timed pregnant wild-type C57BL/6J female mice were purchased from Jackson Laboratories (Bar Harbor, Maine), and mice from these litters were collected at postnatal stages P0, P3, P5, P7, P10, P14, and P21. Mouse pups (P0, P3, and P5) were deeply anesthetized on ice in combination with isoflurane. At all other stages, mice were deeply anesthetized with isoflurane.
For immunohistochemical studies, P7 mice and older were deeply anesthetized with isoflurane and transcardially perfused with cold 0.9% saline solution, followed by 4% paraformaldehyde (PFA, w/v) in phosphate-buffered saline (PBS), pH 7.4. After perfusion, the brains were removed from the skull and immersion fixed in 4% PFA in PBS, pH 7.4, for 24 hours at 4°C. P0, P3, and P5 mouse brains were fixed by immersion. Brains were embedded in paraffin and coronally sliced in 10 µm thick sections.
For immunoblotting studies, mice were deeply anesthetized and euthanized by decapitation. Brains were rapidly removed from the skull and homogenized on ice using a glass dounce homogenizer with 10 strokes in an ice-cold homogenization buffer containing 50 mM Tris, 150 mM NaCl, 1 mM EDTA, 2 mM DTT, 5 mM MgCl2, phosphatase inhibitors (1:1,000, Sigma Aldrich), and one mini protease inhibitor cocktail tablet (Roche Applied Science, Basel, Switzerland), diluted with dH2O to 10 mL, pH 7.4.
Antibody characterization
Antibodies, sources, and the concentrations at which they were used are listed in Table 1.
TABLE 1.
Primary Antibodies Used
| Antibody | Immunizing Antigen | Host Species | Source/catalog number | Dilution |
|---|---|---|---|---|
| Beta actin | Synthetic peptide derived from residues 1–100 of human beta actin | Rabbit (polyclonal) | Abcam # ab8227 | IB: 1:5,000 |
| Flag M2-HRP | Flag peptide (DYKDDDDK) fused onto the N-terminus of interleukin 2 (IL-2) | Mouse (monoclonal) | Sigma Aldrich # A8592 | IB: 1:25,000 |
| Green Fluorescent Protein (GFP) | Recombinant GFP protein | Mouse (monoclonal) | MBL International # 5892 | IB: 1:1,000 |
| HA-Peroxidase, Clone HA-7 | Synthetic peptide from residues 98–106 of human influenza virus hemagglutinin (HA) conjugated to KLH | Mouse (monoclonal) | Sigma Aldrich # H6533 | IB: 1:1,000 |
| Regulator of G Protein Signaling 14 (RGS14) | Full length rat RGS14 | Mouse (monoclonal) | Neuromabs (Clone N133/21) # 75–170 | IB: 1:1,000–5,000 IHC: 1:500 |
HEK293 cells (ATCC, Manassas, VA) were maintained in Dulbecco’s modified eagle’s medium (Mediatech) supplemented with 10% fetal bovine serum (Atlanta Biologicals, 5% after transfection), 2 mM L-glutamine (Invitrogen), 100 U/mL penicillin (Mediatech), and 100 mg/mL streptomycin (Mediatech) in a humidified environment at 37°C with 5% CO2.
The rat RGS14 cDNA used in this study (Genbank accession number U92279) was acquired as described (Hollinger et al., 2001). Flag-RGS14 truncation mutants containing residues 1–202, 205–490, 371–544, and 444–544 were created as described (Shu et al., 2007). HA-RGS2, HA-RGS4, and HA-RGS16 were created in our laboratory as described (Bernstein et al., 2004). The Flag-RGS10 cDNA used in this study was kindly provided by Drs. Malu Tansey and Jae-Kyung Lee (Emory University School of Medicine). GFP-RGS12 TS was a kind gift of Dr. Rory Fisher (University of Iowa). Thioredoxin and hexa-histidine tagged RGS14 (TxH6-RGS14) protein was purified as described (Hollinger et al., 2001).
Transfections were performed using previously described protocols with polyethyleneimine (PEI; Polysciences, Inc.) (Oner et al., 2010). After 24 hours of expression, cells were washed with ice-cold PBS and harvested in a lysis buffer containing 50 mM Tris, 150 mM NaCl, 1 mM EDTA, 2 mM DTT, 5 mM MgCl2, 1% Triton X-100 (v/v), phosphatase inhibitors (1:1,000, Sigma Aldrich), and one mini protease inhibitor cocktail tablet (Roche Applied Science, Basel, Switzerland), diluted with dH2O to 10 mL, pH 7.4. Cells were lysed for one hour at 4°C rocking end-over-end, and subsequently centrifuged to pellet cell debris.
Immunoblotting
After preparing cell lysates and mouse brain homogenates, Bradford protein assays (Thermo Scientific) were performed to assess total protein content in order to normalize protein across samples. Samples were prepared for immunoblotting by diluting with 4× Laemmli sample buffer to a final 1× concentration and heating samples to 95°C in a heating block for 5 minutes. Mouse brain homogenates were subsequently sonicated on ice. Samples from the cell lysates and mouse brain homogenates were loaded onto 11% acrylamide gels and subjected to SDS-PAGE to separate proteins. Proteins were then transferred to nitrocellulose and subjected to immunoblotting to probe for RGS14 and to test the specificity of the anti-RGS14 antibody. After blocking nitrocellulose membranes for 1 hour at room temperature in blocking buffer containing 5% nonfat milk (w/v), 0.1% Tween-20, and 0.02% sodium azide, diluted in 20 mM Tris buffered saline, pH 7.6, membranes were incubated with primary antibodies diluted in the same buffer overnight at 4°C, except for anti-Flag and anti-HA primary antibodies. Membranes were washed in Tris buffered saline containing 0.1% Tween-20 (TBST) and subsequently incubated with either an anti-mouse, anti-rabbit, or anti-goat HRP-conjugated secondary antibody diluted in TBST (1:5,000, 1:25,000, or 1:3,000, respectively) for 1 hour at room temperature. Following block, anti-Flag-HRP and anti-HA-HRP primary antibodies were diluted in TBST and incubated with membranes for 1 hour at room temperature with no secondary antibody. Protein bands were visualized using enhanced chemiluminescence and exposing membranes to films.
Reverse transcription and real-time quantitative PCR
For real-time quantitative-PCR (qRT-PCR) studies, mice were deeply anesthetized and euthanized by decapitation. Brains were rapidly removed from the skull, and total RNA was purified from whole brains using a PureLink RNA Mini Kit (Ambion). RNA yields were quantified using a Nanodrop 1000 (Thermo Scientific), and reverse transcription was performed using a SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen) with 1 µg of total RNA from each brain. qPCR was performed using a DyNAmo HS SYBR Green qPCR kit (Thermo Scientific) on an iQ5 Multicolor Real-time PCR Detection System (Biorad). All samples were diluted 1:50 in nuclease-free water, and 8 µl of these dilutions were used for each SYBR Green PCR reaction containing 10 µl SYBR Green PCR Master Mix, 5 µM each primer, and dH2O. The reactions were incubated for 30 seconds at 95°C, followed by 40 cycles with 30 seconds denaturation at 95°C, 30 seconds annealing at 60°C, and 30 seconds extension at 72°C. Following the amplification protocol, melt curves were generated for samples. RGS14 mRNA was amplified using the following oligonucleotide primers: forward primer, 5’-AAATCCCCGCTGTACCAAG-3’; reverse primer, 5’-GTGACTTCCCAGGCTTCAG-3’. The housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Genbank accession number M32599) was used as a standard to normalize levels of RGS14. GAPDH mRNA was amplified using the following oligonucleotide primers: forward primer, 5’-TGAAGCAGGCATCTGAGGG-3’; reverse primer, 5’-CGAAGGTGGAAGAGTGGGAG-3’.
qRT-PCR expression and analysis
Following qPCR amplification, the data were analyzed in Microsoft Excel. All qPCR reactions were performed on 96-well plates with all postnatal stages present and all samples tested in triplicate. For all qPCR analyses, RNA from two biological replicates was amplified in 5 separate qPCR reactions. Within each qPCR reaction, sample C(t) values were averaged and RGS14 levels were normalized to GAPDH using the ΔΔC(t) method (ΔΔC(t)=2−(RGS14-GAPDH)). ΔΔC(t) values from each experiment were averaged and expressed as percent of wild-type adult. Error bars represent the standard error of the mean.
Immunohistochemistry
Coronal sections of paraffin embedded mouse brains were manually dewaxed and dehydrated in a series of ethanol washes. Endogenous peroxidase tissue was quenched by incubating brain sections in 3% hydrogen peroxide diluted in methanol for 5 minutes at 40°C followed by 3 rinses in 0.075% Brij 35 solution Tris Brij pH 7.5 solution (0.1 M Tris-HCl pH 7.5, 0.1 M NaCl, 0.005 M MgCl2, 0.075% Brij 35). Sections were then blocked for 1 hour and 45 minutes at 4°C with goat anti-mouse IgG AffiniPure fab fragment (Jackson Immunoresearch) at a dilution of 1:250 in blocking buffer (Vectastain standard ABC kit, Vector Laboratories). Sections were incubated with the RGS14 monoclonal antibody diluted in Tau Secret Formula (1:500) for 24 hours at 4°C. The following day, sections were rinsed three times in Tris Brij, pH 7.5 and incubated with 1:200 horse anti-mouse biotinylated secondary antibody diluted in Tris Brij containing 2% horse serum for 30 minutes at 37°C. Sections were then rinsed three times in Tris Brij and incubated for 80 minutes at 37°C in the avidin-biotin peroxidase complex (ABC) solution (Vectastain standard ABC kit, Vector Laboratories). For revelation, sections were first rinsed in Tris Brij solution, then incubated in a solution containing 0.096% 3,3’-diaminobenzidine tetrahydrochloride (DAB, Vector Laboratories) for 5 minutes. Finally, sections were counterstained with hematoxylin (Biomedia), and washed in Tris Brij followed by a rinse in distilled water containing 0.075% Brij 35 solution.
Light microscopy and photomicrograph production
Mouse brain sections mounted and coverslipped on glass slides were analyzed using an Olympus BX51 light microscope. Digital images of the slides were captured and analyzed using DP Controller software on a DP70 camera (Olympus). Brain regions were identified using the Allen Brain Atlas (http://mouse.brain-map.org/) coronal mouse brain reference atlas. Representative images were cropped for presentation and assembled into montages using Adobe Photoshop CS6 Extended.
RESULTS
Antibody characterization
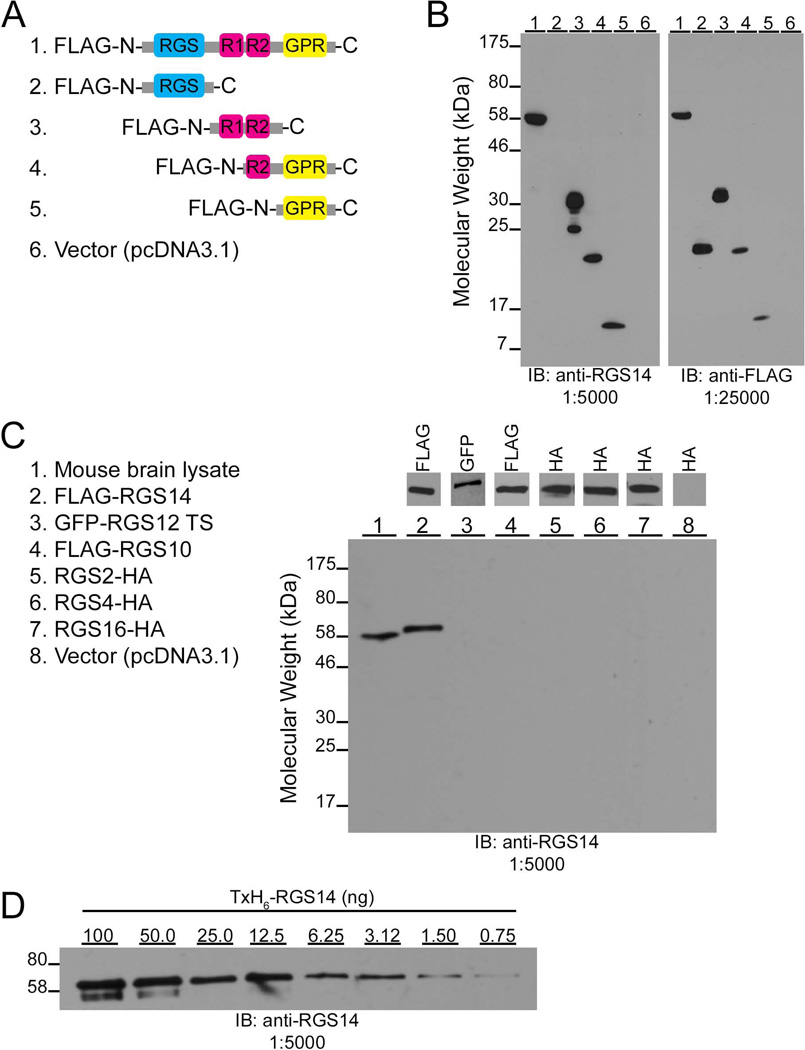
To validate the immunohistochemical findings of this study, we provide the first detailed characterization of an anti-RGS14 monoclonal antibody (clone N133/21) developed in collaboration between our laboratory and the NIH/NINDS-sponsored University of California-Davis/NeuroMab Facility (http://neuromab.ucdavis.edu/catalog.cfm). Our previous initial studies indicated that this antibody specifically recognizes native RGS14 in mouse brain (Lee et al., 2010). To identify the region on RGS14 containing the epitope recognized by the antibody, we performed a series of immunoblot experiments with this antibody probing cell lysates expressing different regions of RGS14 (Fig. 1). HEK293 cell lysates transfected with N-terminally Flag-tagged full-length rat RGS14, various N-terminally Flag-tagged RGS14 truncation mutants, or empty vector (pcDNA3.1) were immunoblotted for detection using the anti-RGS14 antibody (Fig. 1A). Rat RGS14 is 544 amino acids in length with a predicted molecular weight of 61 kDa. We detect a prominent band corresponding to full-length Flag-RGS14 at 62 kDa and also detect the presence of RGS14 truncation mutants encoding residues 205–490 (32 kDa), 371–544 (21 kDa), and 444–544 (15 kDa). The anti-RGS14 antibody does not recognize any protein in HEK293 cell lysates transfected with the RGS14 truncation mutant encoding amino acids 1–202 or empty vector alone (Fig. 1B, left). Independently probing equal amounts of the cell lysates with an anti-Flag antibody reveals prominent bands for full-length RGS14 and all truncation mutants, but does not detect any protein cell lysates transfected with empty vector (Fig. 1B, right). The recognition of the RGS14 truncation mutants containing residues 205–490, 371–544, and 444–544, but not the mutant containing residues 1–202, indicates that the epitope recognized by this anti-RGS14 antibody is located between residues 444 and 490 of RGS14.
Figure 1.
Characterization of the epitope, specificity, and sensitivity of the RGS14 mouse monoclonal antibody. A: Cartoons depicting the domain structure of Flag-tagged RGS14 truncation mutants used to map the region recognized by the antibody. RGS, Regulator of G protein signaling domain; RBD, Ras/Rap- binding domain; GPR, G protein regulatory motif. B: HEK293 cell lysates transfected with Flag-tagged RGS14 or truncation mutant cDNAs were subjected to SDS-PAGE and immunoblotting with anti-RGS14 antibody (1:5,000). Equal amounts of these samples were separately subjected to SDS-PAGE and immunoblotting with an anti-Flag antibody (1:25,000) to verify cDNA expression. C: Wild-type mouse brain homogenate (Lane 1) and HEK293 cell lysates transfected with Flag-RGS14 (Lane 2), GFP-RGS12 TS (Lane 3), Flag-RGS10 (Lane 4), RGS2-HA (Lane 5), RGS4-HA (Lane 6), RGS16-HA (Lane 7), or empty vector (Lane 8, pCDNA3.1) were subjected to SDS-PAGE and immunoblotting with the anti-RGS14 antibody (1:5,000). Equal amounts of these samples were separately subjected to SDS-PAGE and immunoblotting with either an anti-Flag antibody (Lanes 2 and 4, 1:25,000), anti-GFP antibody (Lane 3, 1:1,000), or anti-HA antibody (Lanes 5–8, 1:1,000) to confirm protein expression. GFP, green fluorescent protein; HA, hemagglutinin. D: A serial dilution of purified thioredoxin-and hexa-histidine-tagged RGS14 (TxH6-RGS14) at 100 ng, 50, 25, 12.5, 6.25, 3.12, 1.50, 0.75 ng was subjected to SDS-PAGE and immunoblotting with the anti-RGS14 antibody (1:5,000).
To demonstrate the specificity of the RGS14 antibody, we performed an immunoblot experiment probing mouse brain homogenate and cell lysates expressing various other epitope-tagged RGS proteins including RGS14’s closest relatives, RGS12 and RGS10. Wild-type mouse brain homogenate and HEK293 cell lysates transfected with Flag-tagged rat RGS14, GFP-RGS12-TS, Flag-RGS10, RGS2-HA, RGS4-HA, RGS16-HA, or empty vector (pcDNA3.1) were immunoblotted for detection with the RGS14 antibody (Fig. 1C, left). Probing with the anti-RGS14 antibody only detects a prominent single band in wild-type mouse brain lysate at 61 kDa corresponding to native RGS14 and a slightly higher molecular weight band from cell lysate transfected with Flag-tagged full-length rat RGS14, but no bands are detected in cell lysates transfected with other RGS proteins or empty vector (Fig. 1C, right). The same cell lysates were also probed with an anti-GFP, anti-FLAG, or anti-HA antibody to confirm expression of transfected cDNAs (Fig. 1C top). These results demonstrate the specificity of the RGS14 antibody as it does not detect any other RGS proteins, including RGS14’s closest protein relatives RGS12 and RGS10 (Hollinger and Hepler, 2002; Kimple et al., 2011). This antibody was also tested on human and monkey brain tissue, but does not detect any protein, either by immunoperoxidase labeling of brain sections or by immunoblot of brain tissue homogenates, suggesting that the specific epitope present in mouse and rat RGS14 recognized by this monoclonal antibody is not present in primate RGS14 (data not shown).
To determine the detection limit of this antibody, recombinant rat thioredoxin- and hexa-histidine-tagged RGS14 (TxH6-RGS14) was purified to homogeneity, diluted in a range of decreasing concentrations, and immunoblotted with the RGS14 antibody. The antibody recognizes a prominent single band at 61 kDa and detects as little as 0.75 ng of pure TxH6-RGS14 (Fig. 1D). The doublet observed with 100 ng and 50 ng of pure TxH6-RGS14 reflects minor degradation products of the purified protein.
RGS14 mRNA and protein are upregulated during postnatal brain development
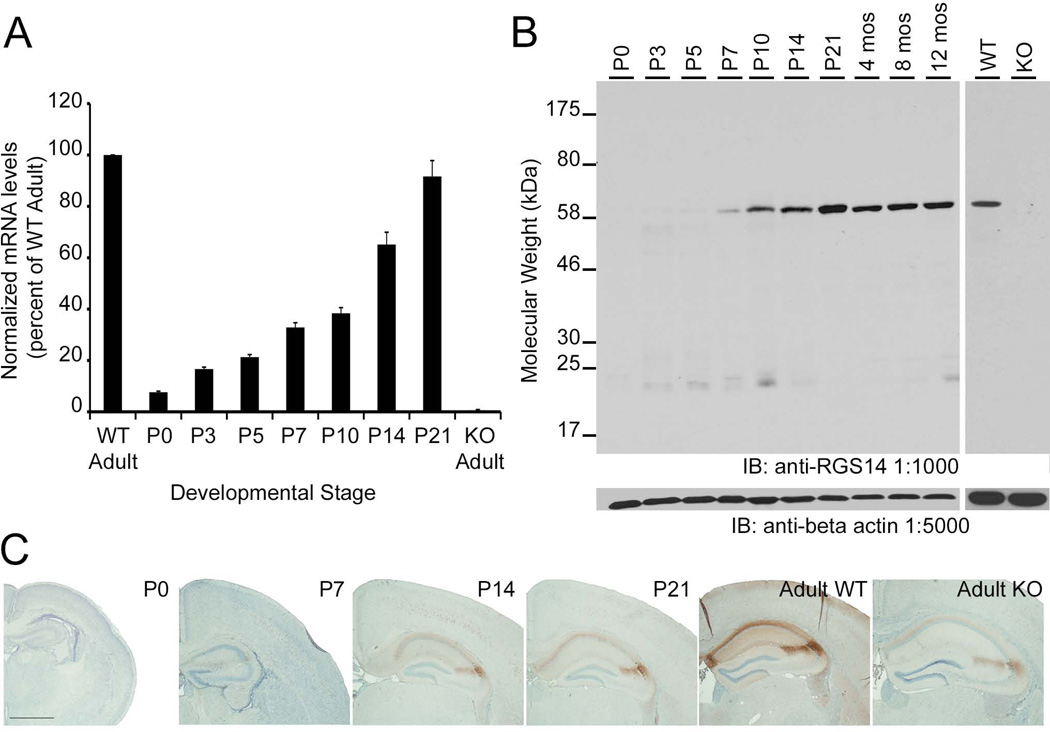
To quantify total levels of RGS14 mRNA throughout postnatal mouse brain development, mRNA was extracted from whole mouse brains at various postnatal stages and SYBR Green quantitative real-time PCR (qRT-PCR) reactions were performed. Quantitative analysis of mRNA levels reveals that RGS14 mRNA is found at very low levels in P0 brain and is gradually upregulated throughout postnatal development to reach the highest levels during adulthood (Fig. 2A). No RGS14 mRNA is amplified from adult RGS14-KO mouse brain, demonstrating the specificity of the oligononucleotide primers.
Figure 2.
RGS14 mRNA and protein are upregulated in mouse brain during postnatal development. A: Quantification of RGS14 mRNA levels determined by quantitative real-time PCR (qRT-PCR). Within each qPCR reaction, sample C(t) values were averaged and RGS14 levels were normalized to GAPDH using the ΔΔC(t) method (ΔΔC(t)=2−(RGS14-GAPDH)). ΔΔC(t) values from each experiment were averaged and expressed as percent of wild-type adult. Error bars represent the standard error of the mean. B: Left, Equal amounts of protein from wild-type mouse brain homogenates were subjected to SDS-PAGE and immunoblotting with an anti-RGS14 antibody (1:1,000). Right, Equal amounts of protein from adult RGS14 wild-type and knockout mouse brain homogenates were similarly subjected to SDS-PAGE and immunoblotting with an anti-RGS14 antibody (1:1,000). All samples were also probed with an anti-beta actin antibody (1:5,000) to demonstrate loading of equal protein amounts between samples. WT, wild-type; KO, RGS14 knockout. C: A series of low power micrographs of coronal mouse brain sections showing RGS14 immunoreactivity in hippocampus at different developmental stages. Scale bar = 1.0 mm.
To analyze the expression levels of RGS14 protein throughout postnatal development, equal amounts of protein from mouse brain homogenates collected from different postnatal stages were subjected to SDS-PAGE, immunoblotting, and probed with the RGS14 antibody. In agreement with the observed increasing mRNA levels, RGS14 protein is first detected by immunoblot as a prominent 61 kDa band at P7 and is gradually upregulated during postnatal development until it reaches the highest levels in adult mouse brain (Fig. 2B, left). Of note, RGS14 protein levels do not change after reaching adulthood as similar amounts of RGS14 are observed in mice aged up to one year. Immunoblots loaded with equal amounts of protein from adult wild-type and RGS14-KO brain homogenates were probed with the RGS14 monoclonal antibody to demonstrate specificity. The antibody recognizes a single 61 kDa band in WT brain, but no signal is detected in RGS14-KO brain, indicating that this antibody specifically recognizes RGS14 (Fig 2B, right). The same blots were probed with an anti-beta actin antibody to demonstrate equal protein loading for all samples.
To validate RGS14 protein levels observed by immunoblot experiments, mouse brain sections from P0, P7, P14, P21, adult WT, and adult RGS14-KO mice containing hippocampus were immunoperoxidase-labeled with the anti-RGS14 antibody and analyzed by light microscopy (LM, Fig. 2C). Consistent with immunoblot experiments, no immunoperoxidase labeling is observed in P0 hippocampus indicating absence of significant RGS14 protein expression. However, immunoperoxidase staining results in a dark brown deposit, which intensely labels CA2 pyramidal dendritic arbors and fasciola cinerea (FC) neurons in P14, P21, and adult WT hippocampus. Low levels of DAB labeling are observed in P7 hippocampus compared to older mice, consistent with less protein detected in immunoblot experiments at this age. Low-level background immunoreactivity is observed in the CA2 subfield of the adult RGS14-KO mouse brain. The nature of this staining is unclear, but may be non-specific background (see Discussion) as we find complete loss of the 61 kDa band corresponding full-length RGS14 protein by immunoblot with the same RGS14 antibody (Fig. 2B, right). Additionally, no RGS14 mRNA was detected in adult RGS14-KO mouse brain (Fig. 2A). Taken together, these studies demonstrate that RGS14 protein is not detectable until P7 after which time the protein is upregulated until adulthood where it continues to be expressed at the same level.
Overall distribution of RGS14 immunoperoxidase labeling
Detailed anatomical analysis of coronal brain sections at the LM level reveals that RGS14 immunoperoxidase labeling is not detectable in any region of P0 mouse brain (data not shown).However, neuronal RGS14 immunoperoxidase labeling is observed as a dark brown deposit as early as P7, which continues to increase thereafter until adulthood (summarized in Table 2). RGS14 immunolabeling is detected in the anterior olfactory nucleus (AON) and piriform cortex known as primary cortical areas for olfactory processing. Labeling is also present during development in specific layers of orbital and entorhinal cortices, with transient immunoreactivity in neocortex. Finally, RGS14 immunoperoxidase labeling is most prominent in hippocampal CA2 and FC with immunoreactivity increasing with age. After detailed examination for RGS14 immunoreactivity throughout the entire mouse brain, these areas are the only regions displaying RGS14 immunoreactivity throughout development and in adulthood. No immunoreactivity is observed in RGS14-KO mice (except minimal labeling in CA2 subfield-see Discussion), indicating specificity of immunoperoxidase labeling (data not shown).
TABLE 2.
Regional Brain Localization of RGS14
| Region | Developmental Stage | ||||
|---|---|---|---|---|---|
| P0 | P7 | P14 | P21 | Adult | |
| Anterior Olfactory Nucleus (AON) | − | + | + | ++ | +++ |
| Piriform Cortex (PIR) | − | + | + | ++ | +++ |
| Orbital Cortex (ORB) | − | + | +++ | + | + |
| Entorhinal Cortex (ENT) | − | + | +++ | +++ | + |
| Neocortex (Neo) | − | + | +++ | + | − |
| Hippocampus - CA2 | − | + | ++ | +++ | ++++ |
| Hippocampus - CA1 | − | − | ++ | +++ | + |
| Fasciola cinerea (FC) | − | + | ++ | ++ | ++++ |
Anterior olfactory nucleus (AON) and piriform cortex
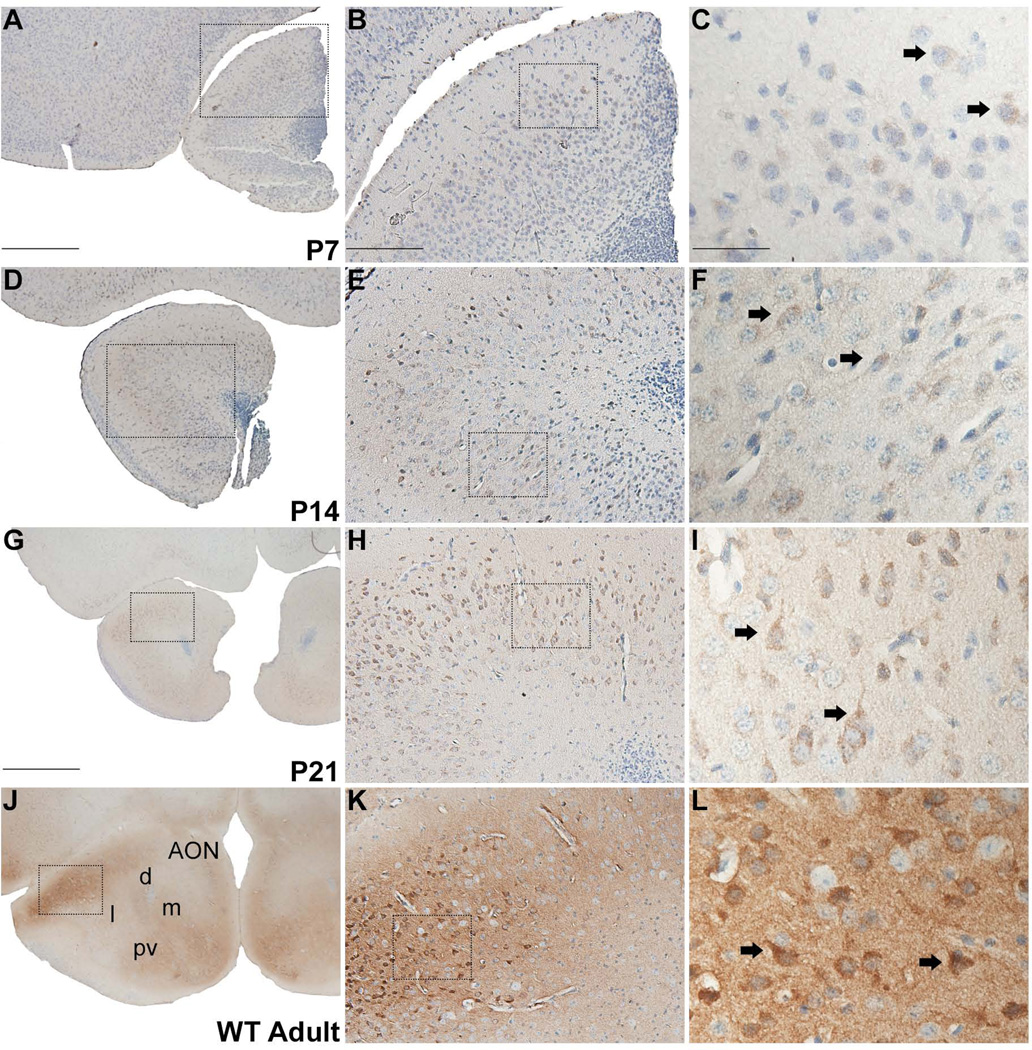
In the AON, RGS14 labeling is mainly concentrated in the soma and apical dendrites of neurons in P7, P14, P21 WT mice (Fig. 3A–I), while in the adult, strongly labeled neuronal cell bodies lay in a rich immunoreactive neuropil (Fig. 3J–L). The labeling is found throughout the whole extent of the AON in adolescent mice, but is particularly enriched in the dorsolateral portion of the structure in adult WT animals. At high magnification, some of the immunolabeled neurons display pyramidal morphology with a single, prominent apical dendrite extending from the soma (Fig 3I,L).
Figure 3.
RGS14 immunolabeling in anterior olfactory nucleus (AON) of postnatal mouse brain. Low, medium, and high magnification views of coronal hemisections at P7, P14, P21, and adult wild-type mouse brain labeled with immunoperoxidase using an anti-RGS14 monoclonal antibody (A–L). RGS14 immunoreactivity increases in the AON during postnatal development, and staining is restricted to soma and proximal dendrites of neurons, some of which have a pyramidal shaped cell body until P21 (F,I)). In adults, neuronal cell bodies are more heavily stained, and a significant neuropil immunostaining can be seen (J–L). Dashed boxes indicate region magnified in subsequent micrographs to the right. Arrows indicate immunolabeled neuronal perikarya. AON, anterior olfactory nucleus; d, dorsal; m, medial; l, lateral; pv, posteroventral. Scale bars = 500 µm in A (applies to D); 1.0 mm in G (applies to J); 200 µm in B (applies to E,H,K); 50 µm in C (applies to F,I,L).
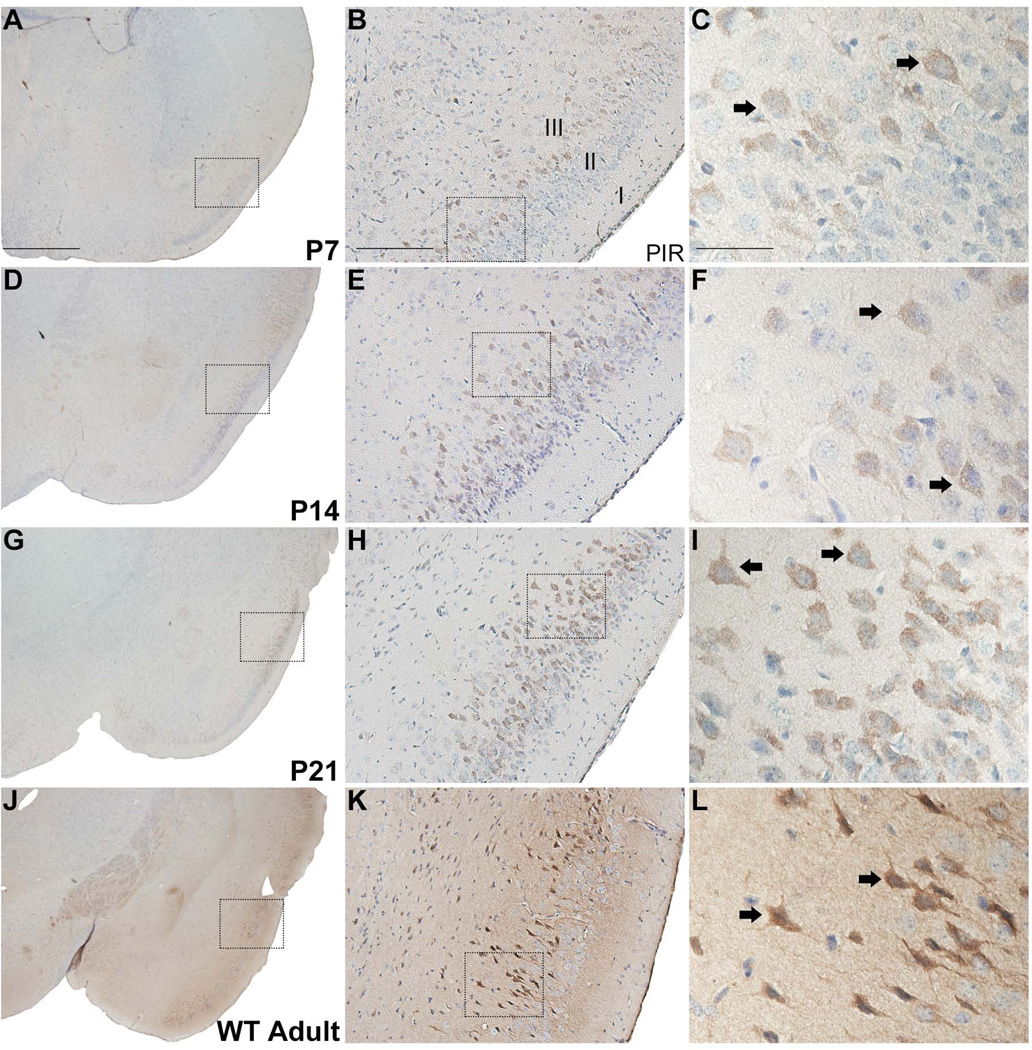
In the piriform cortex, RGS14 immunoreactivity is found mainly in layer II pyramidal neurons. Weak labeling is first observed at P7, but the immunolabeling intensity increases with age until reaching its highest level in adulthood (Fig. 4A–L). As found in AON, significant neuropil immunoreactivity and strong neuronal cell body and apical dendrite labeling are seen in the upper layers of the piriform cortex in adult animals (Fig. 4J–L).
Figure 4.
RGS14 immunoreactivity in postnatal mouse piriform cortex. Low, medium, and high magnification views of right coronal hemisections at P7, P14, P21, and adult wild-type mouse brain immunoperoxidase labeled with an anti-RGS14 antibody. RGS14 immunoreactivity increases in piriform cortex during postnatal development, and staining is localized to soma and proximal dendrites of neurons, some of which with a pyramidal shape (C,F,I,L). Dashed boxes indicate regions magnified in subsequent micrographs to the right. Arrows indicate immunolabeled neurons. PIR, piriform cortex. Scale bar = 1.0 mm in A (applies to D,G,J); 200 µm in B (applies to E,H,K); 50 µm in C (applies to F,I,L).
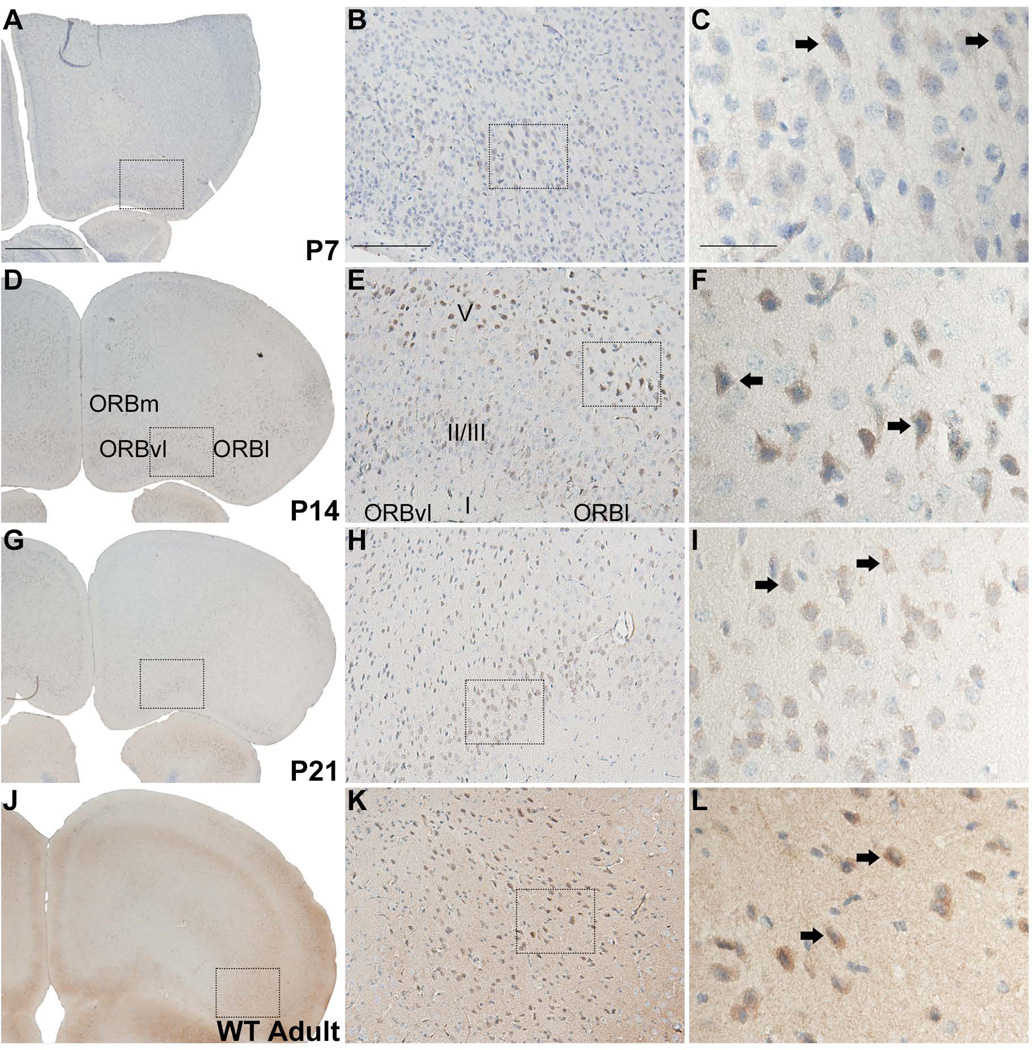
Orbital and entorhinal cortices
At the level of the AON, RGS14 labeling is found from P7 onward in layers II/III and V of the orbital cortex with highest immunoreactivity detectable at P14 and in adults (Fig. 5). At high magnification, labeling is mostly concentrated in neuronal somata, some of which have a pyramidal shape, in the orbital cortex (Fig. 5F). In adults, light neuropil staining is also found throughout the orbital cortex (Fig. 5J–L).
Figure 5.
RGS14 immunoreactivity in postnatal mouse orbital cortex. Low, medium, and high magnification views of right coronal hemisections at P7, P14, P21, and adult wild-type mouse brain immunoperoxidase labeled with an anti-RGS14 antibody. RGS14 immunoreactivity in the orbital cortex is highest at P14 and in adults, and staining is localized to soma and proximal dendrites of neurons, some with a pyramidal shape (F,L). Dashed boxes indicate regions magnified in subsequent micrographs to the right. Arrows indicate immunolabeled neurons.; ORB, orbital cortex; m, medial; vl, ventrolateral; l, lateral. Scale bars = 1.0 mm in A (applies to D,G,J); 200 µm in B (applies to E,H,K); 50 µm in C (applies to F,I,L).
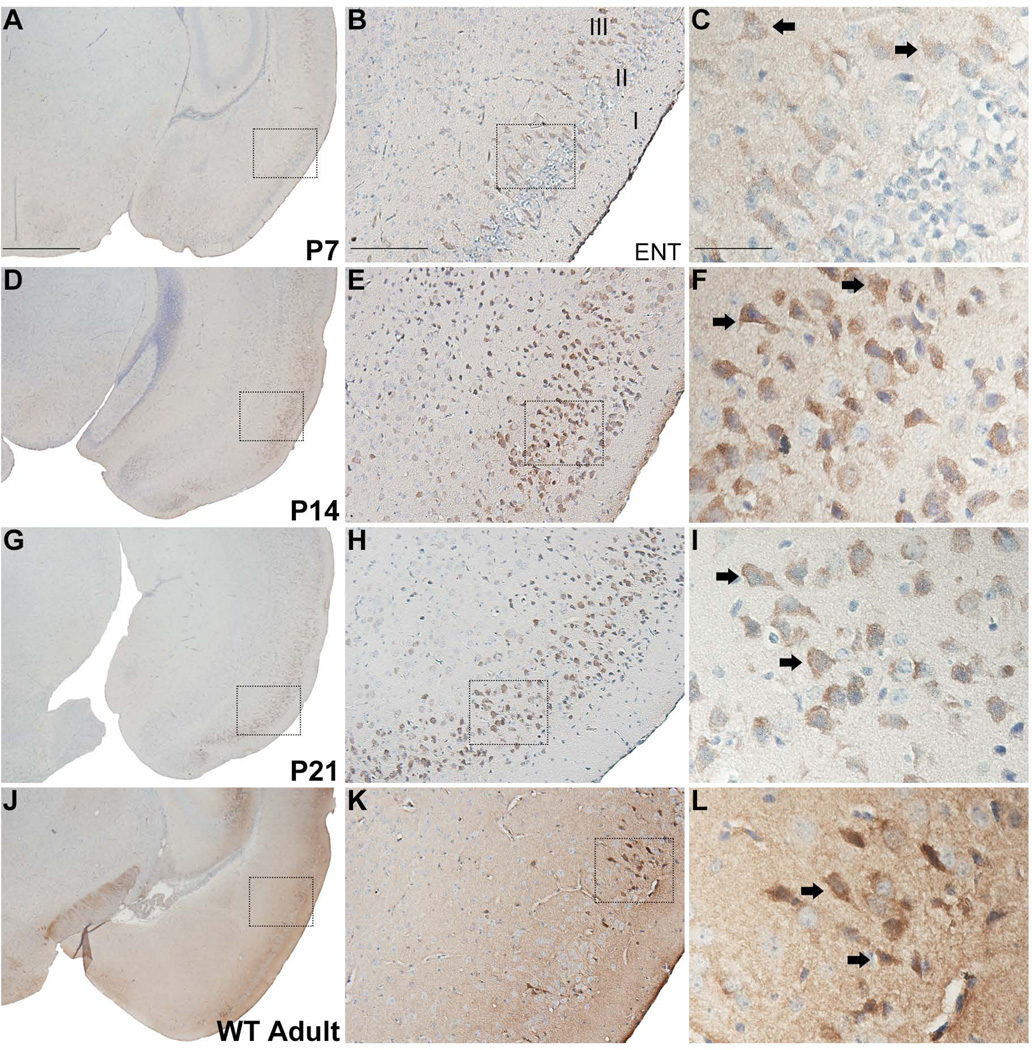
Similarly, weak immunoreactivity is first detected at P7 in layer II/III neurons of the entorhinal cortex (Fig. 6A–C). Immunolabeling is most prominent at P14 and in adults, with a slight decline in immunoreactivity at P21, (Fig. 6D–L). As in other cortical regions, some of the labeled cell bodies display a pyramidal shape appearance, and the neuropil labeling is most intense in adults (Fig. 6J–L).
Figure 6.
RGS14 immunoreactivity in postnatal mouse entorhinal cortex. Low, medium, and high magnification views of coronal hemisections from P7, P14, P21, and adult wild-type mouse brain immunoperoxidase labeled with an anti-RGS14 antibody. RGS14 immunoreactivity in the entorhinal cortex is highest at P14 and in adults, and the staining is localized to soma and apical dendrites of labeled neurons (F,I,L). In adults, a light immunoreactive neuropil can also be seen (J–L). Dashed boxes indicate regions magnified in subsequent micrographs to the right. Arrows indicate immunolabeled neurons. ENT, entorhinal cortex. Scale bars = 1.0 mm in A (applies to D,G,J); 200 µm in B (applies to E,H,K); 50 µm in C (applies to F,I,L).
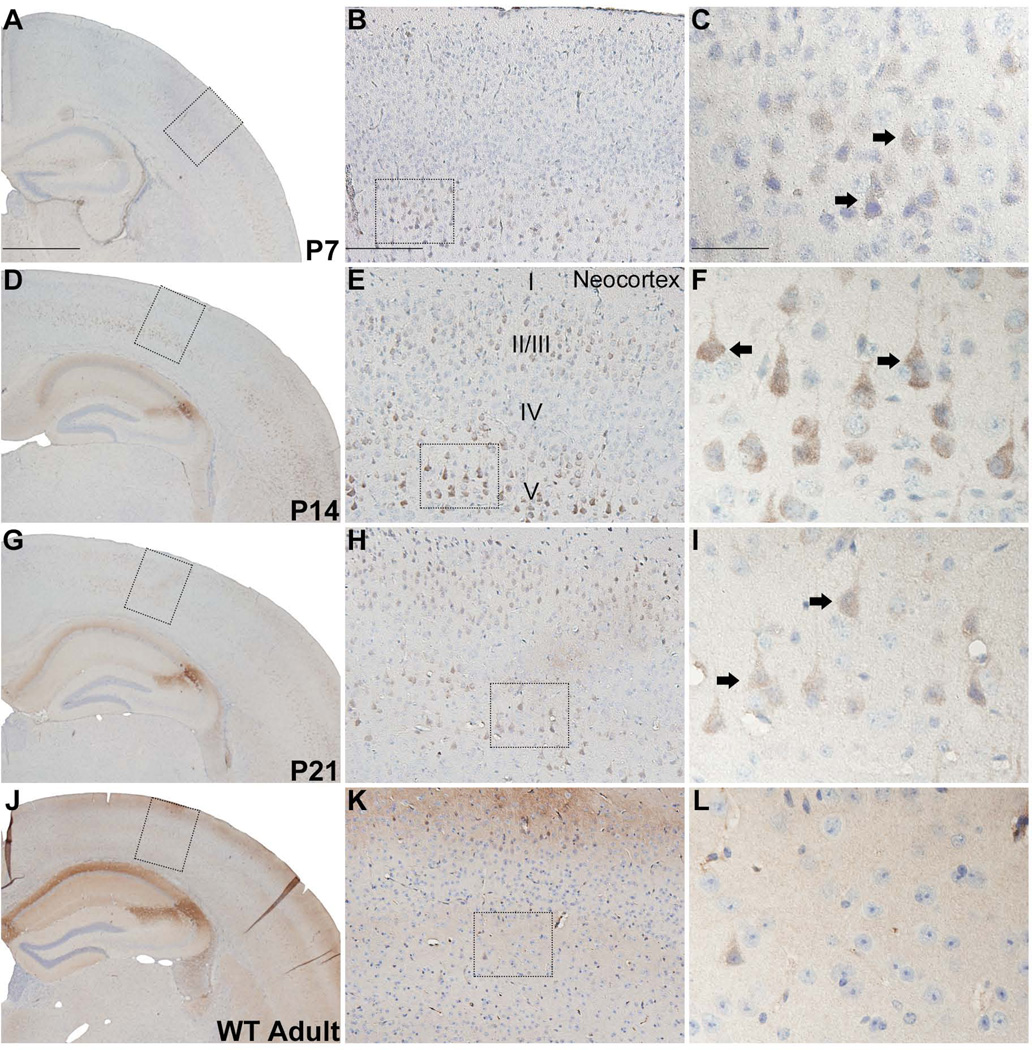
Transient expression of RGS14 in neocortex
Throughout the rostral-caudal axis of the adolescent mouse brain, light to moderate immunoreactivity for RGS14 is observed in layers II/III and V of pyramidal-shaped neocortical neurons between P7 and P21, while it is undetectable in adults (Fig. 7A–L).. A similar pattern of Immunolabeling is found across several regions of neocortex including orbital, somatomotor, somatosensory, auditory, and visual areas.
Figure 7.
RGS14 immunolabeling is transiently expressed in postnatal mouse neocortex. Low, medium, and high magnification views of coronal hemisections from P7, P14, P21, and adult wild-type mouse brain immunoperoxidase labeled with an anti-RGS14 antibody. RGS14 immunoreactivity is highest at P14 in neocortical layers II/III and V, and staining is localized to soma and apical dendrites of pyramidal neurons (F). Immunostaining is less intense at P21 (G–I) and is undetectable in adults (J–L). Dashed boxes indicate regions magnified in subsequent micrographs to the right. Arrows indicate immunolabeled neurons. Scale bars = 1.0 mm in A (applies to D,G,J); 200 µm in B (applies to E,H,K); 50 µm in C (applies to F,I,L).
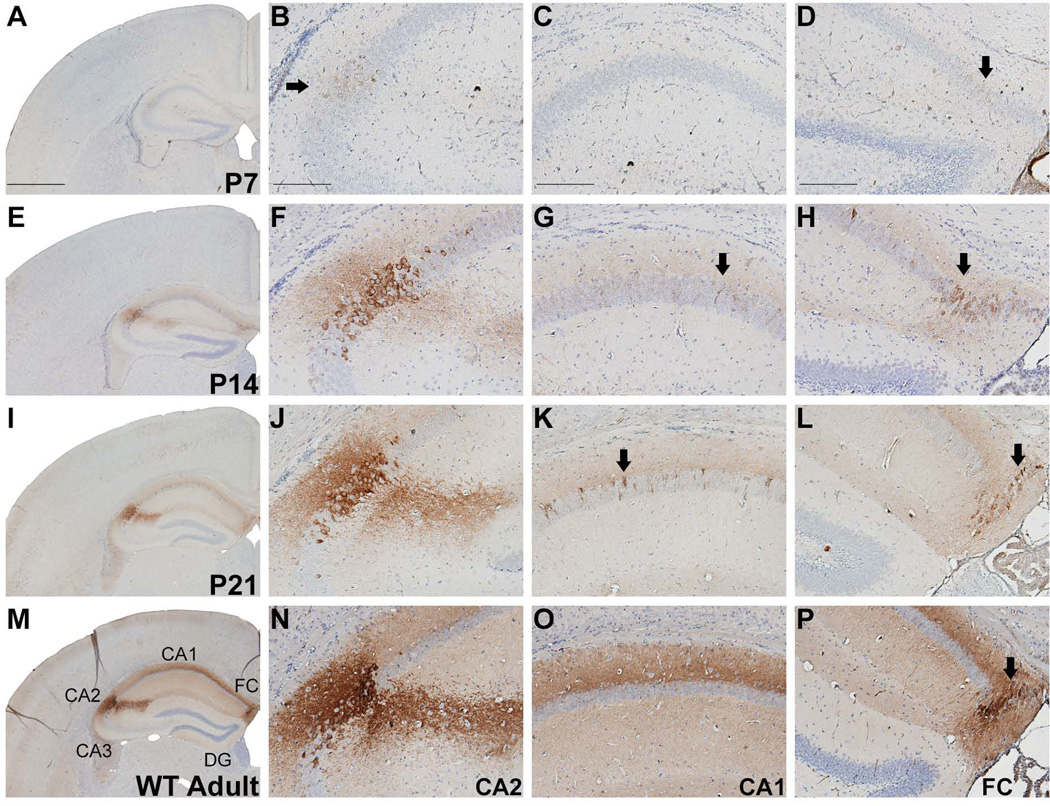
Hippocampal RGS14 immunoreactivity increases throughout development
In the hippocampus, weak RGS14 immunoperoxidase labeling, first observable at P7 in hippocampal CA2, is significantly upregulated until it reaches its highest levels in adult WT mouse brain (Fig. 8A–P). Staining is most prominent at the soma and dendritic arbors of CA2 pyramidal neurons as well as in fasciola cinerea (FC). Labeling is also occasionally observed in the soma and proximal dendrites of a very sparse population of CA1 neurons at P14 and P21 (Fig. 8G,K), while in adults the stratum lacunosum moleculare and stratum radiatum in CA1 region harbored significant neuropil immunoreactivity (8O). Of note, the CA1 neuropil immunoreactivity in adults is due to immunoperoxidase labeling of CA2 neurites, which extend through this plane of area CA1.
Figure 8.
RGS14 immunolabeling in postnatal mouse hippocampus. Low and medium magnification views of hemisections from P7, P14, P21, and adult wild-type mouse brains immunoperoxidase labeled with an anti-RGS14 antibody. RGS14 immunoreactivity increases in hippocampal CA2 during development, which results in an extensive labeling of the dendritic arbors of CA2 pyramidal neurons (F,J,N). RGS14 immunoreactivity in fasciola cinerea (FC) also increases with age, reaching its highest levels in the adult mouse brain (arrows in D,H,L,P). At P14 and P21, the cell bodies and proximal dendrites of a small population of CA1 pyramidal neurons display immunoreactivity, while in adults, a significant neuropil immunostaining of CA1 region can be seen (M,O). Arrows indicate immunolabeled neurons. FC, fasciola cinerea; DG, dentate gyrus. Scale bars = 1.0 mm in A (applies to E, I, M); 200 µm in B–D (applies to F–H, J–L, N–P).
DISCUSSION
Our results provide the first detailed anatomical analysis of the expression of RGS14 in the developing and adult mouse brain. In doing so, we also provide the first comprehensive characterization of a newly described anti-RGS14 monoclonal antibody, confirming its specificity and sensitivity for detecting RGS14 protein in mice. We show that both mRNA and protein levels are gradually upregulated throughout postnatal development, reaching their highest levels in the adult mouse brain, except for the neocortex, which displayed a lower level of immunostaining in adults than at earlier time points (i.e. P7–P21). As a complement to our previous report of RGS14 as a hippocampal CA2 and FC enriched gene (Lee et al., 2010), the current findings demonstrate that RGS14 protein is also significantly expressed in the anterior olfactory nucleus, piriform cortex, orbital cortex, entorhinal cortex, and neocortex in mice.
Contrary to previous immunohistochemical studies in monkey and rat brain (Lopez-Aranda et al., 2006) which reported broad RGS14 protein expression in both neurons and glia, our results show a much more restricted distribution of RGS14 protein expression limited to neuronal cell bodies in fewer brain regions. The differences in localization profiles between our study and these previous results could be due to species differences in RGS14 distribution or, more likely, from antibody specificity. In that regard, it is worthy noting that our findings are entirely consistent with independently reported adult mouse brain in situ hybridization data from the Allen Mouse Brain Atlas (http://mouse.brain-map.org) examining RGS14 mRNA expression and distribution patterns in the adult mouse brain.
Our findings are also consistent with independent microarray studies on adult human (http://human.brain-map.org/) and non-human primate (http://www.blueprintnhpatlas.org/) brain tissue, both reporting that RGS14 mRNA is most highly expressed in hippocampal CA2 and moderately expressed in CA1. Contrary to our findings in mice, these data also show high levels of RGS14 mRNA expression in the striatum (caudate nucleus and putamen), which could suggest a unique striatal function for RGS14 in primates relative to rodents. Germane to this, mRNA and protein variants of RGS14 have been reported in primates (see below), and it is possible that RGS14 variants could be differentially expressed in CA2 versus striatum in primates. However, this idea is speculative since the sequence(s) of the RGS14 transcript(s) detected in these microarray data sets are unknown. Further immunoperoxidase staining and in situ hybridization studies are required to characterize the localization of RGS14 protein and mRNA in the primate brain. A detailed characterization of the RGS14 mRNA/protein species found in primate brain and a comprehensive analysis of their distribution and subcellular localization could provide great insight into the roles of RGS14 in human physiology and disease.
Antibody characterization and specificity
Previous studies have shown that RGS14 protein is enriched in brain (Hollinger et al., 2001; Lopez-Aranda et al., 2006), but the lack of a fully characterized, specific anti-RGS14 antibody has limited immunohistochemical analysis of protein distribution in brain. Here we show that the anti-RGS14 mouse monoclonal antibody (Clone N133/21, NeuroMabs) used in our study is very specific and sensitive for RGS14, and that this antibody recognizes an epitope in the C-terminal region of the mouse and rat RGS14 protein. Furthermore, this antibody recognizes a single 61 kDa protein band in mouse brain corresponding to native, full-length RGS14 protein. Although whole-genome shotgun sequencing (Mural et al., 2002) has predicted lower molecular weight variants of RGS14 including the region of the protein containing the antibody epitope, this antibody did not detect these proteins by immunoblot. However, we cannot rule out the possibilities that these variants may be expressed at an undetectable level for immunoblot, or perhaps expressed outside of mouse brain.
We observed very light immunoperoxidase labeling with this antibody in the hippocampal CA2 subfield, but not in other regions of adult RGS14-KO mice (Figure 2C). While we cannot conclude that this staining necessarily represents non-specific background labeling, several lines of evidence suggest that this is the case. Previous studies showed that hippocampal CA2 exhibits background immunoreactivity to antibodies against proteins not present in this region (Holmseth et al., 2012) as well as a unique extracellular milieu surrounding these neurons that may be non-specifically labeled (Bruckner et al., 2003). Because this light background staining is not present in P0-P14 mice, it suggests that the CA2-specific antigen protein(s) the antibody cross-reacts with is also developmentally upregulated. Another possible explanation is that the low level of background labeling is caused by the general anti-mouse IgG secondary antibody, rather than the use of a IgG subclass-specific secondary antibody (Manning et al., 2012). However, no immunolabeling was observed in control experiments in which primary antibody was omitted. Alternatively, we cannot definitively exclude the possibility that the RGS14-KO mice used are not complete knock-outs. These mice were generated by deleting exons 2–7 of the RGS14 gene that may result in a low production of a smaller molecular weight variant from exons 8–11, which would contain the epitope recognized by the anti-RGS14 monoclonal antibody. Arguing against this possibility, however, are our findings (Figure 2A and 2B) showing that we could not detect either the full-length RGS14 protein or any lower molecular weight variants by immunoblot, or RGS14 mRNA by qRT-PCR. Taken together, our hypothesis is that this staining represents low-level non-specific staining as has been reported for other proteins (Holmseth et al., 2012).
Upregulation of RGS14 during postnatal development: implications for early learning and synaptic plasticity
We recently reported that RGS14 suppresses synaptic plasticity in CA2 neurons as well as hippocampal-based spatial and novel object recognition memory in adult mice (Lee et al., 2010). Other studies strongly suggest that CA2 plays a significant role in social behavior and temporal order for memories (DeVito et al., 2009), and initial experiments have shown that the social neuropeptides oxytocin and vasopressin induce synaptic potentiation in CA2 pyramidal neurons (Caruana et al., 2012). Therefore, the upregulation of RGS14 protein beginning at P7 may allow for a period of regionally enhanced plasticity and learning during early postnatal development to allow newborn pups to adapt to their environment and form strong social bonds, i.e. maternal attachment. For example, the increase in RGS14 protein after P7 may serve as a filter to allow hippocampal CA2 to encode episodic memories only under specific conditions. This may coincide with new synapse formation and/or pruning during early postnatal development, shaped by environmental inputs such as maternal bonding and other social interactions.
Olfaction plays a major role in guiding rodent behaviors including social recognition. In mammals, odorants are first processed by the olfactory bulb, which targets specific structures collectively referred to as primary olfactory cortex, including the anterior olfactory nucleus (AON) and piriform cortex (Wilson and Rennaker, 2010). The olfactory cortex is responsible for processing and associating odorants with specific events (Wilson and Sullivan, 2011). The expression of RGS14 in pyramidal neurons in both the AON and piriform cortex makes it well positioned to modulate primary olfactory inputs and thus guide olfactory and social learning. RGS14 is also expressed in orbital and entorhinal cortical neurons, which receives substantial input from primary olfactory cortical areas. This distribution pattern, therefore, suggests that RGS14 could play a pivotal role in modulating olfactory processing in different brain regions. Further studies are required to determine if RGS14 regulates olfaction in mice.
RGS14: a suppressor of plasticity in multiple neuronal populations?
Although we have found that RGS14 restricts CA2 synaptic plasticity, its role in the other brain areas remains to be determined. Of note, the subcellular immunoperoxidase labeling for RGS14 is distinct in CA2 compared with other regions. In hippocampal CA2, RGS14 staining is much more heavily concentrated in the apical and basal dendrites of pyramidal neurons while than in other areas where the labeling is lighter and often restricted to the soma and apical dendrites of pyramidal neurons, such as in CA1. Of note, RGS14 immunoreactivity in CA1 did not colocalize with any known markers of CA1 interneurons (Chris McBain personal communication), and we therefore deduce that RGS14 is likely expressed in a very small, sporadic subset of CA1 pyramidal neurons. This difference in localization suggests that RGS14 may play a distinct role in plasticity suppression for area CA2 rather than in other neuronal populations outside of the hippocampus. One possibility is that RGS14 modulates synaptogenesis and dendrite development as its expression is dramatically upregulated during the first two postnatal weeks, a time of extensive synapse formation and pruning in rodent brain (Fiala et al., 1998). We have recently shown that RGS14 coordinates Giα1 and H-Ras signaling to modulate neurite outgrowth in PC12 cells (Vellano et al., 2013). The transient expression of RGS14 in neocortex, which peaks at P14, suggests that it must have a distinct purpose at this time in early development.
Implications for RGS14 and area CA2 in human cognition and behavior
Humans and primates also express a roughly equivalent long isoform (approximately 63 kDa) of RGS14 as well as shorter splice variants that lack the N-terminal RGS domain (Lopez-Aranda et al., 2006). Our findings indicate that, at least in mouse brain, RGS14 is expressed as the full length 61 kDa isoform and that expression is largely limited to hippocampal area CA2 in adulthood. Hippocampal long-term potentiation (LTP) is believed to underlie certain key aspects of human learning, memory, and cognition, most notably spatial and contextual learning. While hippocampal LTP has been thoroughly characterized in the dentate gyrus (DG)-CA3-CA1 trisynaptic pathway, significantly less is known about the role of LTP-resistant area CA2 in overall hippocampal functions or behaviors (Scorza et al., 2011; Zhao et al., 2007). It has recently been shown that, in contrast to Schaffer collateral inputs, entorhinal cortex inputs to CA2 are capable of producing LTP, but the cellular mechanisms underlying this plasticity remain to be demonstrated (Chevaleyre and Siegelbaum, 2010). Moreover, a number of genes, including RGS14, are highly expressed in CA2, but not CA1 (Lein et al., 2005; Lein et al., 2007), suggesting that the cellular mechanisms regulating plasticity in CA2 may differ from the well-known pathways in CA1 neurons. Further experiments are required to understand the unique cellular machinery governing CA2 plasticity, and the role of CA2 in mediating overall hippocampal function. Our findings are consistent with the hypothesis of Caruana et al. (2012), suggesting that the relative synaptic stability of CA2 is designed to allow encoding of memories only under specific circumstances, such as in early postnatal development when RGS14 protein is not expressed.
The contribution of area CA2 to putatively linked behaviors is in the early stages of exploration, but initial experiments strongly suggest that CA2 plays a significant role in several forms of learning/memory including social, spatial, object recognition, and temporal order (Caruana et al., 2012). Area CA2, in particular, is associated with human pathologies including Alzheimer’s disease, schizophrenia, autism and bipolar spectrum disorders, as well as ischemia/epilepsy (Benes et al., 1998; Braak, 1980; DeVito et al., 2009; Kirino, 1982; Sadowski et al., 1999; Sloviter, 1991). RGS14 has also recently been identified as a candidate gene in a study of fear learning in mice (Parker et al., 2012), which could suggest a role for RGS14 in human post-traumatic stress disorder (PTSD). Thus, RGS14 could function to selectively allow encoding of CA2 under certain conditions, e.g. maternal attachment in the neonate, while filtering storage of memories that could be maladaptive, e.g. traumatic life events. Microarray studies reporting high levels of RGS14 expression in adult human and non-human primate CA2 suggest that RGS14’s role as a suppressor of plasticity and hippocampal-dependent learning in mice may also extend to primates. The unique expression of RGS14 in the primate striatum indicates that it could serve yet unknown functions specific to primates. Further studies are needed to elucidate roles for RGS14 and hippocampal region CA2 in human behavior and disease.
ACKNOWLEDGEMENTS
We thank Marla Gearing and Deborah Cooper for exceptional technical assistance and helpful discussion of data. This research project was supported in part by the histopathology core of the Emory Neuroscience NINDS Core Facilities grant, P30NS055077. We also thank Drs. Belvin Gong and Bianca Bautista of the NIH/NINDS-sponsored NeuroMabs facility at the University of California-Davis for their outstanding work in developing the anti-RGS14 monoclonal antibody used in these studies.
Financial Support: These studies were supported by grants NIH/NINDS 5R01 NS37112 and NIH/NINDS 1R21NS074975 both awarded to JRH. The work was supported by NIH T32GM008605 training grant awarded to Emory Graduate Program in Neuroscience. This work was also supported by the National Center for Research Resources P51RR000165 and the Office of Research Infrastructure Programs / OD P51OD011132 to the Yerkes National Primate Center.
ROLE OF AUTHORS
P.R.E., S.E.L., and J.R.H. designed research; P.R.E. and S.E.L. performed experiments; P.R.E. analyzed and interpreted data; P.R.E. wrote the manuscript; P.R.E., Y.S., and J.R.H critically revised the manuscript.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
LITERATURE CITED
- Benes FM, Kwok EW, Vincent SL, Todtenkopf MS. A reduction of nonpyramidal cells in sector CA2 of schizophrenics and manic depressives. Biological psychiatry. 1998;44(2):88–97. doi: 10.1016/s0006-3223(98)00138-3. [DOI] [PubMed] [Google Scholar]
- Bernstein LS, Ramineni S, Hague C, Cladman W, Chidiac P, Levey AI, Hepler JR. RGS2 binds directly and selectively to the M1 muscarinic acetylcholine receptor third intracellular loop to modulate Gq/11alpha signaling. The Journal of biological chemistry. 2004;279(20):21248–21256. doi: 10.1074/jbc.M312407200. [DOI] [PubMed] [Google Scholar]
- Braak H. Architectonics of the human telencephalic cortex. 1980 [Google Scholar]
- Bruckner G, Grosche J, Hartlage-Rubsamen M, Schmidt S, Schachner M. Region and lamina-specific distribution of extracellular matrix proteoglycans, hyaluronan, and tenascin-R in the mouse hippocampal formation. Journal of chemical neuroanatomy. 2003;26(1):37–50. doi: 10.1016/s0891-0618(03)00036-x. [DOI] [PubMed] [Google Scholar]
- Caruana DA, Alexander GM, Dudek SM. New insights into the regulation of synaptic plasticity from an unexpected place: hippocampal area CA2. Learn Mem. 2012;19(9):391–400. doi: 10.1101/lm.025304.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevaleyre V, Siegelbaum SA. Strong CA2 pyramidal neuron synapses define a powerful disynaptic cortico-hippocampal loop. Neuron. 2010;66(4):560–572. doi: 10.1016/j.neuron.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kozasa T, Takekoshi K, De Gunzburg J, Kehrl JH. RGS14, a GTPase-activating protein for Gi alpha, attenuates Gi alpha- and G13 alpha-mediated signaling pathways. Molecular pharmacology. 2000;58(3):569–576. doi: 10.1124/mol.58.3.569. [DOI] [PubMed] [Google Scholar]
- DeVito LM, Konigsberg R, Lykken C, Sauvage M, Young WS, 3rd, Eichenbaum H. Vasopressin 1b receptor knock-out impairs memory for temporal order. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29(9):2676–2683. doi: 10.1523/JNEUROSCI.5488-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC, Feinberg M, Popov V, Harris KM. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18(21):8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstein-Dunn E, Young KH, Cockett MI, Khawaja XZ. Regional distribution of regulators of G-protein signaling (RGS) 1, 2, 13, 14, 16, and GAIP messenger ribonucleic acids by in situ hybridization in rat brain. Molecular Brain Research. 2001;88(1–2):113–123. doi: 10.1016/s0169-328x(01)00038-9. [DOI] [PubMed] [Google Scholar]
- Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacological reviews. 2002;54(3):527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- Hollinger S, Taylor JB, Goldman EH, Hepler JR. RGS14 is a bifunctional regulator of G alpha(i/o) activity that exists in multiple populations in brain. Journal of neurochemistry. 2001;79(5):941–949. doi: 10.1046/j.1471-4159.2001.00629.x. [DOI] [PubMed] [Google Scholar]
- Holmseth S, Dehnes Y, Huang YH, Follin-Arbelet VV, Grutle NJ, Mylonakou MN, Plachez C, Zhou Y, Furness DN, Bergles DE, Lehre KP, Danbolt NC. The density of EAAC1 (EAAT3) glutamate transporters expressed by neurons in the mammalian CNS. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(17):6000–6013. doi: 10.1523/JNEUROSCI.5347-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel C, Wohlgemuth S, Rousseau F, Schymkowitz J, Ferkinghoff-Borg J, Wittinghofer F, Serrano L. Recognizing and defining true Ras binding domains II: in silico prediction based on homology modelling and energy calculations. Journal of molecular biology. 2005;348(3):759–775. doi: 10.1016/j.jmb.2005.02.046. [DOI] [PubMed] [Google Scholar]
- Kimple AJ, Bosch DE, Giguere PM, Siderovski DP. Regulators of G-protein signaling and their Galpha substrates: promises and challenges in their use as drug discovery targets. Pharmacological reviews. 2011;63(3):728–749. doi: 10.1124/pr.110.003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple RJ, De Vries L, Tronchere H, Behe CI, Morris RA, Farquhar MG, Siderovski DP. RGS12 and RGS14 GoLoco motifs are G alpha(i) interaction sites with guanine nucleotide dissociation inhibitor activity. Journal of Biological Chemistry. 2001;276(31):29275–29281. doi: 10.1074/jbc.M103208200. [DOI] [PubMed] [Google Scholar]
- Kirino T. Delayed neuronal death in the gerbil hippocampus following ischemia. Brain research. 1982;239(1):57–69. doi: 10.1016/0006-8993(82)90833-2. [DOI] [PubMed] [Google Scholar]
- Larminie C, Murdock P, Walhin JP, Duckworth M, Blumer KJ, Scheideler MA, Garnier M. Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Molecular Brain Research. 2004;122(1):24–34. doi: 10.1016/j.molbrainres.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Lee SE, Simons SB, Heldt SA, Zhao M, Schroeder JP, Vellano CP, Cowan DP, Ramineni S, Yates CK, Feng Y, Smith Y, Sweatt JD, Weinshenker D, Ressler KJ, Dudek SM, Hepler JR. RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc Natl Acad Sci U S A. 2010;107(39):16994–16998. doi: 10.1073/pnas.1005362107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Callaway EM, Albright TD, Gage FH. Redefining the boundaries of the hippocampal CA2 subfield in the mouse using gene expression and 3-dimensional reconstruction. The Journal of comparative neurology. 2005;485(1):1–10. doi: 10.1002/cne.20426. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lopez-Aranda MF, Acevedo MJ, Carballo FJ, Gutierrez A, Khan ZU. Localization of the GoLoco motif carrier regulator of G-protein signalling 12 and 14 proteins in monkey and rat brain. European Journal of Neuroscience. 2006;23(11):2971–2982. doi: 10.1111/j.1460-9568.2006.04838.x. [DOI] [PubMed] [Google Scholar]
- Manning CF, Bundros AM, Trimmer JS. Benefits and pitfalls of secondary antibodies: why choosing the right secondary is of primary importance. PloS one. 2012;7(6):e38313. doi: 10.1371/journal.pone.0038313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal V, Linder ME. The RGS14 GoLoco domain discriminates among G alpha(i) isoforms. Journal of Biological Chemistry. 2004;279(45):46772–46778. doi: 10.1074/jbc.M407409200. [DOI] [PubMed] [Google Scholar]
- Mural RJ, Adams MD, Myers EW, Smith HO, Miklos GL, Wides R, Halpern A, Li PW, Sutton GG, Nadeau J, Salzberg SL, Holt RA, Kodira CD, Lu F, Chen L, Deng Z, Evangelista CC, Gan W, Heiman TJ, Li J, Li Z, Merkulov GV, Milshina NV, Naik AK, Qi R, Shue BC, Wang A, Wang J, Wang X, Yan X, Ye J, Yooseph S, Zhao Q, Zheng L, Zhu SC, Biddick K, Bolanos R, Delcher AL, Dew IM, Fasulo D, Flanigan MJ, Huson DH, Kravitz SA, Miller JR, Mobarry CM, Reinert K, Remington KA, Zhang Q, Zheng XH, Nusskern DR, Lai Z, Lei Y, Zhong W, Yao A, Guan P, Ji RR, Gu Z, Wang ZY, Zhong F, Xiao C, Chiang CC, Yandell M, Wortman JR, Amanatides PG, Hladun SL, Pratts EC, Johnson JE, Dodson KL, Woodford KJ, Evans CA, Gropman B, Rusch DB, Venter E, Wang M, Smith TJ, Houck JT, Tompkins DE, Haynes C, Jacob D, Chin SH, Allen DR, Dahlke CE, Sanders R, Li K, Liu X, Levitsky AA, Majoros WH, Chen Q, Xia AC, Lopez JR, Donnelly MT, Newman MH, Glodek A, Kraft CL, Nodell M, Ali F, An HJ, Baldwin-Pitts D, Beeson KY, Cai S, Carnes M, Carver A, Caulk PM, Center A, Chen YH, Cheng ML, Coyne MD, Crowder M, Danaher S, Davenport LB, Desilets R, Dietz SM, Doup L, Dullaghan P, Ferriera S, Fosler CR, Gire HC, Gluecksmann A, Gocayne JD, Gray J, Hart B, Haynes J, Hoover J, Howland T, Ibegwam C, Jalali M, Johns D, Kline L, Ma DS, MacCawley S, Magoon A, Mann F, May D, McIntosh TC, Mehta S, Moy L, Moy MC, Murphy BJ, Murphy SD, Nelson KA, Nuri Z, Parker KA, Prudhomme AC, Puri VN, Qureshi H, Raley JC, Reardon MS, Regier MA, Rogers YH, Romblad DL, Schutz J, Scott JL, Scott R, Sitter CD, Smallwood M, Sprague AC, Stewart E, Strong RV, Suh E, Sylvester K, Thomas R, Tint NN, Tsonis C, Wang G, Wang G, Williams MS, Williams SM, Windsor SM, Wolfe K, Wu MM, Zaveri J, Chaturvedi K, Gabrielian AE, Ke Z, Sun J, Subramanian G, Venter JC, Pfannkoch CM, Barnstead M, Stephenson LD. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science. 2002;296(5573):1661–1671. doi: 10.1126/science.1069193. [DOI] [PubMed] [Google Scholar]
- Oner SS, Maher EM, Breton B, Bouvier M, Blumer JB. Receptor-regulated interaction of activator of G-protein signaling-4 and Galphai. The Journal of biological chemistry. 2010;285(27):20588–20594. doi: 10.1074/jbc.C109.088070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CC, Sokoloff G, Cheng R, Palmer AA. Genome-wide association for fear conditioning in an advanced intercross mouse line. Behavior genetics. 2012;42(3):437–448. doi: 10.1007/s10519-011-9524-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M, Wisniewski HM, Jakubowska-Sadowska K, Tarnawski M, Lazarewicz JW, Mossakowski MJ. Pattern of neuronal loss in the rat hippocampus following experimental cardiac arrest-induced ischemia. Journal of the neurological sciences. 1999;168(1):13–20. doi: 10.1016/s0022-510x(99)00159-8. [DOI] [PubMed] [Google Scholar]
- Scorza CA, Araujo BH, Leite LA, Torres LB, Otalora LF, Oliveira MS, Garrido-Sanabria ER, Cavalheiro EA. Morphological and electrophysiological properties of pyramidal-like neurons in the stratum oriens of Cornu ammonis 1 and Cornu ammonis 2 area of Proechimys. Neuroscience. 2011;177:252–268. doi: 10.1016/j.neuroscience.2010.12.054. [DOI] [PubMed] [Google Scholar]
- Shu FJ, Ramineni S, Amyot W, Hepler JR. Selective interactions between Gi alpha 1 and Gi alpha 3 and the GoLoco/GPR domain of RGS14 influence its dynamic subcellular localization. Cellular Signalling. 2007;19(1):163–176. doi: 10.1016/j.cellsig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Shu FJ, Ramineni S, Hepler JR. RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAPkinase signalling pathways. Cellular Signalling. 2010;22(3):366–376. doi: 10.1016/j.cellsig.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloviter RS. Permanently altered hippocampal structure, excitability, and inhibition after experimental status epilepticus in the rat: the"dormant basket cell" hypothesis and its possible relevance to temporal lobe epilepsy. Hippocampus. 1991;1(1):41–66. doi: 10.1002/hipo.450010106. [DOI] [PubMed] [Google Scholar]
- Snow BE, Antonio L, Suggs S, Gutstein HB, Siderovski DP. Molecular cloning and expression analysis of rat Rgs12 and Rgs14. Biochemical and biophysical research communications. 1997;233(3):770–777. doi: 10.1006/bbrc.1997.6537. [DOI] [PubMed] [Google Scholar]
- Traver S, Bidot C, Spassky N, Baltauss T, de Tand MF, Thomas JL, Zalc B, Janoueix-Lerosey I, de Gunzburg J. RGS14 is a novel Rap effector that preferentially regulates the GTPase activity of G alpha(o) Biochemical Journal. 2000;350:19–29. [PMC free article] [PubMed] [Google Scholar]
- Vellano CP, Brown NE, Blumer JB, Hepler JR. Assembly and Function of the Regulator of G protein Signaling 14 (RGS14){middle dot}H-Ras Signaling Complex in Live Cells Are Regulated by Galphai1 and Galphai-linked G Protein-coupled Receptors. The Journal of biological chemistry. 2013;288(5):3620–3631. doi: 10.1074/jbc.M112.440057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellano CP, Lee SE, Dudek SM, Hepler JR. RGS14 at the interface of hippocampal signaling and synaptic plasticity. Trends in pharmacological sciences. 2011;32(11):666–674. doi: 10.1016/j.tips.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DA, Rennaker RL. Cortical Activity Evoked by Odors. In: Menini A, editor. The Neurobiology of Olfaction. Boca Raton (FL): 2010. [PubMed] [Google Scholar]
- Wilson DA, Sullivan RM. Cortical processing of odor objects. Neuron. 2011;72(4):506–519. doi: 10.1016/j.neuron.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Choi Y-S, Obrietan K, Dudek SM. Synaptic Plasticity (and the Lack Thereof) in Hippocampal CA2 Neurons. The Journal of Neuroscience. 2007;27(44):12025–12032. doi: 10.1523/JNEUROSCI.4094-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]