Abstract
Objectives
Caries process comprises acidogenic and aciduric bacteria that are responsible for lowering the pH and subsequent destruction of hydroxyapatite matrix in enamel and dentine. The aim of this study was to identify the correlation between the pH gradient of a carious lesion and proportion and distribution of four bacterial genera; lactobacilli, streptococci, prevotellae, and fusobacteria with regard to total load of bacteria.
Materials and methods
A total of 25 teeth with extensive dentinal caries were sampled in sequential layers. Using quantitative real-time PCR of 16S rRNA gene, we quantified the total load of bacteria as well as the proportion of the abovementioned genera following pH measurement of each sample with a fine microelectrode.
Results
We demonstrated the presence of a pH gradient across the lesion with a strong association between the quantity of lactobacilli and the lowest pH range (pH 4.5–5.0; p = 0.003). Streptococci had a tendency to occupy the most superficial aspect of the carious lesion but showed no correlation to any pH value. Prevotellae showed clear preference for the pH range 5.5–6.0 (p = 0.042). The total representation of these four genera did not reach more than one quarter of the total bacterial load in most carious samples.
Conclusion
We revealed differential colonization behavior of bacteria with respect to pH gradient and a lower than expected abundance of lactobacilli and streptococci in established carious lesions. The data indicate the numerical importance of relatively unexplored taxa within the lesion of dentinal caries.
Clinical relevance
The gradient nature of pH in the lesion as well as colonization difference of examined bacterial taxa with reference to pH provides a new insight in regard to conservative caries management.
Keywords: Caries, Real-time PCR, Gram-positive bacteria, Gram-negative bacteria, pH gradient, 16S rRNA
Introduction
Dental caries is a continuum in which acidogenic and aciduric bacteria, availability of fermentable carbohydrate, salivary function, and fluoride concentration will define the direction for de- and remineralization [1]. The large body of literature on microbiology of caries emphasizes the role of streptococci and lactobacilli, suggesting that these should comprise the majority of the complex and diverse total load of bacteria in established dentinal lesions. A particular focus has been the capacity of species within these genera to produce and tolerate low pH environments sufficient to demineralize hydroxyapatite matrices [2]. Moreover, gram-negative species including prevotellae and fusobacteria have also been detected frequently in carious dentine [3, 4]. In an earlier study, members of Prevotella comprised 15 % of the species among 75 bacterial taxa identified in 10 carious lesions [3]. On the other hand, strains of Prevotella intermedia and Fusobacterium nucleatum are shown to increase their metabolic activity under acidic conditions [5]. However, quantitative insight into the contribution of these genera to the total load of bacteria is not available. How these organisms colonize and invade dentine is largely unknown. An ecological plaque hypothesis has been proposed to explain the succession of species leading to a mature, climax community, in which nutrient availability, oxygen tension, and environmental concentration of hydrogen ions have a major influence on bacterial composition of the lesion [6, 7]. The profiles of pH and organic acids within carious dentine have been of interest to researchers in the field, since Stephan's description of caries as a pH-mediated disease [8, 9]. Macgregor studied the presence of acid in dentine and enamel caries using methyl red to confirm low pH values in dentine lesions [10]. While acid and base production by bacteria and their pH lowering potential and tolerance has been widely studied in vitro [5, 11–15], the pH and organic acid composition of dentinal caries was not examined, until Hojo et al. demonstrated ex vivo the organic acid profile and pH of carious lesions of dentine [16]. Nevertheless, to date, the relationship between the bacterial component of the lesion and its pH value has not been studied in a quantitative manner.
The aim of this study was to assess pH gradient of a dentine caries lesion and to identify any correlation between the pH of the lesion and the quantity of four bacterial genera, including two widely proclaimed acidogenic and/or aciduric gram-positive genera, lactobacilli and streptococci, as well as two gram-negative genera, Prevotella and Fusobacterium, which are highly associated with caries. The total load of bacteria was also measured in order to have a unified scale. Findings from this study provide further knowledge to improve conservative restorative approaches including atraumatic restorative treatment of caries.
Materials and methods
Sampling and DNA isolation
Permanent molar or premolar teeth with occlusal or proximal caries (n = 25) comprising open cavities and with a diagnosis of irreversible pulpitis were extracted, with informed consent from adult patients, following a discussion on possible treatment options in regard to the affected tooth according to the guidelines of the ethics committee (NSW Health, protocol number X07-0261). Lesions in the sampled teeth presented with relative characteristics of an active lesion, including soft to leathery dentine texture. However, since further classification of dentine caries (powdery, leathery, soft, semihard, or hard) is not universally standardized and can be subjective to some extent, we did not attempt to categorize these lesions according to their presenting dentine texture. We did not include any recurrent or arrested (hard, darkly stained) lesion in this study. Subjects had not taken antibiotics within 4 weeks prior to extraction, and no carbohydrate intake was noted for at least 2 h prior to extraction. Using a sterile slow speed handpiece and sterile size one round bur, four or five sequential layers of 1-mm-thick dentine samples were taken from the tooth, and as accurately as the technique allowed, starting circumferentially from the carious lesion and progressing into sound dentine as the last layer. Minimal cross-contamination between layers was unavoidable, although all care was taken to prevent this. All samples were taken by a single calibrated dentist. A total of 111 samples with a minimum wet weight of 7 mg were suspended in 0.9 % NaCl to a concentration of 1 mg in 4 µl of saline. Calibration trials confirmed the stability of hydrogen ion concentration of the suspension over the limited time period of sampling. Following immediate pH measurement at ambient temperature, 4 µl of the suspension was taken for DNA isolation. Purification and extraction of DNA was performed using the QIAamp DNA Mini Kit (Qiagen, Australia), as described previously [3, 17, 18]. DNA concentration (A260) and purity (A260/A280) were determined using a NanoPhotometer (Implen, Munich, Germany).
Primer and probe sets for real-time qPCR
Over 340 complete sequences of 16S rRNA genes of species and strains of oral bacteria were collected from the National Collection for Biotechnology Information database (http://www.ncbi.nlm.nih.gov), including 91 Lactobacillus spp., 79 Streptococcus spp., 16 Fusobacterium spp., 37 Prevotella spp., and 122 species or strains of known human bacterial flora (see “Supplementary data”). Alignment of these 16S rRNA gene sequences using ClustalW2 multiple sequence alignment software [19] enabled the identification of optimal conserved and hypervariable regions for the design of quantitative real-time PCR (qPCR) primers and fluorophore–quencher probe sets for the streptococcal, prevotellae, and fusobacteria genera (Table 1, column A). The possibility of cross-hybridization to other bacterial genes was also excluded by BLAST search [20]. Primers and probes were synthetized by Integrated DNA Technologies, Inc. (Coralville, Iowa) and validated for specificity against a number of representative species (Table 1, column B; Fig. 1.). Lactobacilli SYBR® Green primer set was as designed and validated previously [17]. Total load of bacteria in each sample was also measured using the fluorophore–quencher TaqMan® universal primer and probe set as designed and tested previously [21].
Table 1.
Column A: universal and genera-specific primer and probe sets employed for qPCR. See Supplementary data for details on DNA library preparation and real time PCR conditions. Column B: extracted DNA of bacteria used for PCR test of specificity of designed primer and probe sets
| Primer and probe sets for selected bacteria | List of species/strains primer sets were PCR tested against | ||
|---|---|---|---|
| Sequence | |||
| 5′→3′ | (A) | Tma | (B) |
| Universalb (Amplicon size = 466 bp) | Lactobacillus rhamnosus ATCC 7469 | ||
| Forward: | |||
| TCCTACGGGAGGCAGCAGT | 60.8 | Lactobacillus salivarius ATCC 11741 | |
| Reverse: | Lactobacillus casei ATCC 393 | ||
| GGACTACCAGGGTATCTAATCCTGTT | 57.2 | Lactobacillus plantarum ATCC 14917 | |
| Probec: | Lactobacillus gasseri ATCC 33323 | ||
| CGTATTACCGCGGCTGCTGGCAC | 64.8 | Streptococcus mutans NCTC 10449 | |
| Streptococcus oralis NCTC 11427 | |||
| Lactobacillusd (SYBR® Green)(Amplicon size = 223 bp) | Streptococcus mitis NCTC 12261 | ||
| Forward: | |||
| TGGAAACAGRTGCTAATACCG | 52.4 – 54.8 | Streptococcus parasanguinis ATCC 15912 | |
| Reverse: | Streptococcus gordonii ATCC 10558 | ||
| GTCCATTGTGGAAGATTCCC | 53.4 | Streptococcus thermophilus ATCC 19258 | |
| Streptococcus vestibularis ATCC 49124 | |||
| Streptococcus (Amplicon size = 86 bp) | Fusobacterium nucleatum ATCC 25586 | ||
| Forward: | |||
| CGGATCGTAAAGCTCTGTTGTAAG | 55.5 | Fusobacterium necrophorum ATCC 25286 | |
| Reverse: | Bifidobacterium breve ATCC 15700 | ||
| GCCGTCCCTTTCTGGTAAG | 55.6 | Actinomyces israelii ATCC 12102 | |
| Probe: | Neisseria meningitidis ATCC 13077 | ||
| GTGTGAGAGTGGAAAGTTCACACWGTGACGGT | 64.1 | Neisseria gonorrhoeae ATCC 19424 | |
| Neisseria elongata ATCC 25295 | |||
| Prevotella (Amplicon size = 116 bp) | Pseudomounas aeruginosa ATCC 10145 | ||
| Forward: | |||
| GAAGGTGCGGGTATCGAAC | 55.8 | Porphyromonas gingivalis W83 | |
| Reverse: | Haemophilus influenzae ATCC 33391 | ||
| GGG ATGCTTAATGCTTTCGCTT | 56.2 | Haemophilus parainfluenzae ATCC 33392 | |
| Probe: | Prevotella intermedia ATCC 25611 | ||
| CCGCACAGTAAACGATGGATGCCCGC | 65.7 | Prevotella denticola ATCC 35308 | |
| Prevotella bivia ATCC 29303 | |||
| Fusobacterium (Amplicon size = 87 bp) | Prevotella melaninogenica ATCC 25845 | ||
| Forward: | |||
| GCG GAACTACAAGTGTAGAGGT | 56.4 | Alloprevotella tannerae ATCC 51259 | |
| Reverse: | Bacteroides uniformis ATCC 8492 | ||
| AGCGTCAGTATCTGTCCAGT | 55.4 | Bacteroides eggerthii ATCC 27754 | |
| Probe: | Tannerella forsythia ATCC 43037 | ||
| AGCTGGCTTCCCCATCGGCATTCCTACAA | 66.5 | Parvimonas micra ATCC 33270 | |
| Escherichia coli JM109 | |||
Melting temperature (Tm) of oligonucleotide determined by Integrated DNA Technologies (Coralville, Iowa) www.idtdna.com/scitools
From Nadkarni et al. [18]
Oligonucleotide probes were labeled with 6-carboxyfluorescein (FAM) fluorescent reporter at the 5′ end and Iowa Black® FQ dark quencher at the 3′ end
From Byun et al. [14]
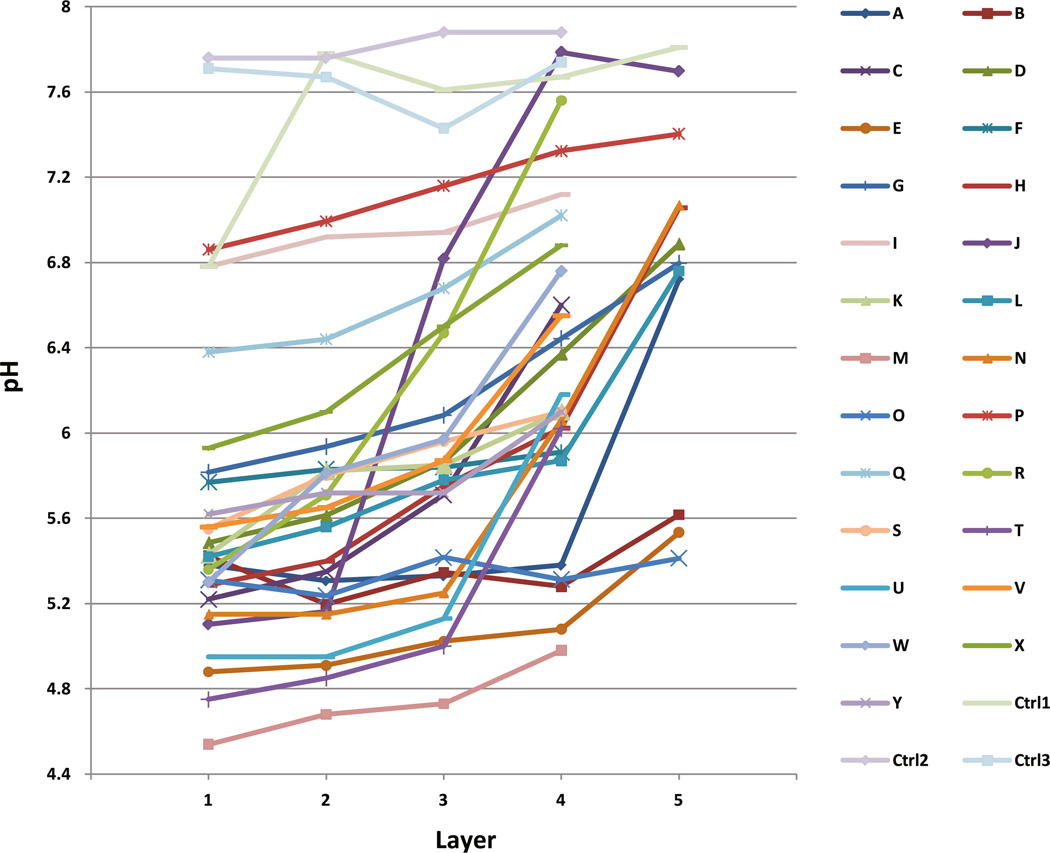
Fig. 1.
pH gradients of dentine samples at different depth in 25 carious teeth as well as in three noncarious controls. The most superficial layer of the carious dentine is Layer 1, progressing deeper at 1 mm intervals into sound dentine. The last layer of each tooth was within sound dentine. There is significant correlation between increasing pH and increasing depth of the dentin sample (R = 0.497; p < 0.001). Controls were three sound third molar teeth extracted for noncaries reasons
Quantitative real-time PCR and enumeration of bacterial cell numbers
qPCR is a relatively accurate and sensitive method in quantification of small number of bacteria [22]. In a bacterial community, where dead and alive mixture of bacteria may present, the nuclease activity of bacteria will decompose the remnants of DNA, and hence, false overdetection and overquantification of dead bacteria is inherently minimized [23]. We carried out qPCR in triplicate in 25 µl volume reactions using Platinum® quantitative PCR SuperMix-UDG (Invitrogen™) for TaqMan primer and probe sets and Platinum® SYBR® Green qPCR SuperMix-UDG (Invitrogen™) for the lactobacilli primer set, containing 200 µM of forward and reverse primers and 100 pg to 1 µg of genomic DNA template, as well as 100 µM of fluorogenic probe for TaqMan primer and probe sets. The real-time PCR conditions were set at 95 °C for 2 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s for the universal primer and probe set. The PCR condition for streptococci, Prevotella spp., and Fusobacterium spp. was set at 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, and 58 °C for 30 s. The lactobacilli SYBR Green PCR condition was set at 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, and 62 °C for 1 min and a third step of one cycle of 95 °C for 1 min and 55 °C for 30 s and 95 °C for 30 s, as these conditions were optimized according to annealing temperatures for oligonucleotide primer and probe sets as well as instrument documentation (Stratagene Mx3005P, Agilent Technologies, Inc., Santa Clara, CA). To determine the theoretical cell numbers with qPCR, a standard curve using extracted DNA from Streptococcus mutans against the streptococcal probe and primer set was employed to represent streptococcal numbers. The quantities, based on the molecular weight of the DNA standard and number of 16 s rRNA gene copies [24], were converted to bacterial numbers, using the known genome size of S. mutans NCTC 10449 and adjusted for 5.39 copies of 16S rRNA gene in an average streptococcal genome. Similarly, P. intermedia ATCC 25611 was used as standard for the Prevotella genus, with three copies of 16S rRNA gene per genome, F. nucleatum ATCC 25586 was used as the standard for the Fusobacterium genus, with five copies of 16S rRNA gene per genome, and Lactobacillus salivarius ATCC 11741 was used as the standard for the Lactobacillus genus, with 5.58 copies of 16S rRNA gene per genome. S. mutans NCTC 10449 DNA was used as the universal qPCR standard, with 5.39 average number of copies of 16S rRNA gene.
pH measurements
We performed pH measurements using a minimum of 30 µl suspension of dentine samples at 1 mg per 4 µl of saline, to give a stable suspension [16]. To measure pH, we used a palladium Touch Microelectrode (Beetrode® NMPH3,World Precision Instruments Ltd., UK) with a 100 µm sensor tip. We measured pH of the samples within 15 min following suspension preparation. Calibration trials confirmed the stability of hydrogen ion concentration of the suspension over the limited time period of sampling. The time frame from tooth extraction to dentine removal and pH measurement was about 45 min. However, pH reads were consistent and reliable at ambient temperature for 15 min from the time of suspension preparation. Three noncarious control teeth were also included in the pH measurement.
Statistical analysis
Nonparametric Mann–Whitney U (Wilcoxon rank-sum test) as well as median test was performed to compare the concentration of bacteria within different pH groups. Correlation coefficient and bivariate regression analysis were used to assess pH gradient trend within layers. Pearson two-tailed correlation analysis was used to assess association between bacterial genera at different layers. Multiple regression analysis was performed to find if there was a statistically meaningful predictive factor for pH as the dependent variable. Holm–Bonferroni correction of significance level was applied to avoid overestimated interpretation of sequential group comparisons [25]. All statistical analyses of data were performed using IBM® SPSS® Statistics 19.0 GradPack software.
Results
pH gradient within a carious tooth
The pH gradients of carious dentine were found to range from 4.54 to 7.79, with the most superficial dentine layer invariably the most acidic (Fig. 1). Correlation coefficient from bivariate regression analysis showed a significant linear trend (R = 0.497; p < 0.001) of increasing pH from the superficial layer of carious dentine to the deeper sound dentine. In general, pH difference between superficial dentine to the deepest dentine sample within a carious tooth averaged 1.132 pH units. By comparison, three control healthy teeth showed dentinal pH values of between 6.8 and 7.9 but without graded changes according to depth of sample.
Colonization of carious dentine by selected bacterial genera
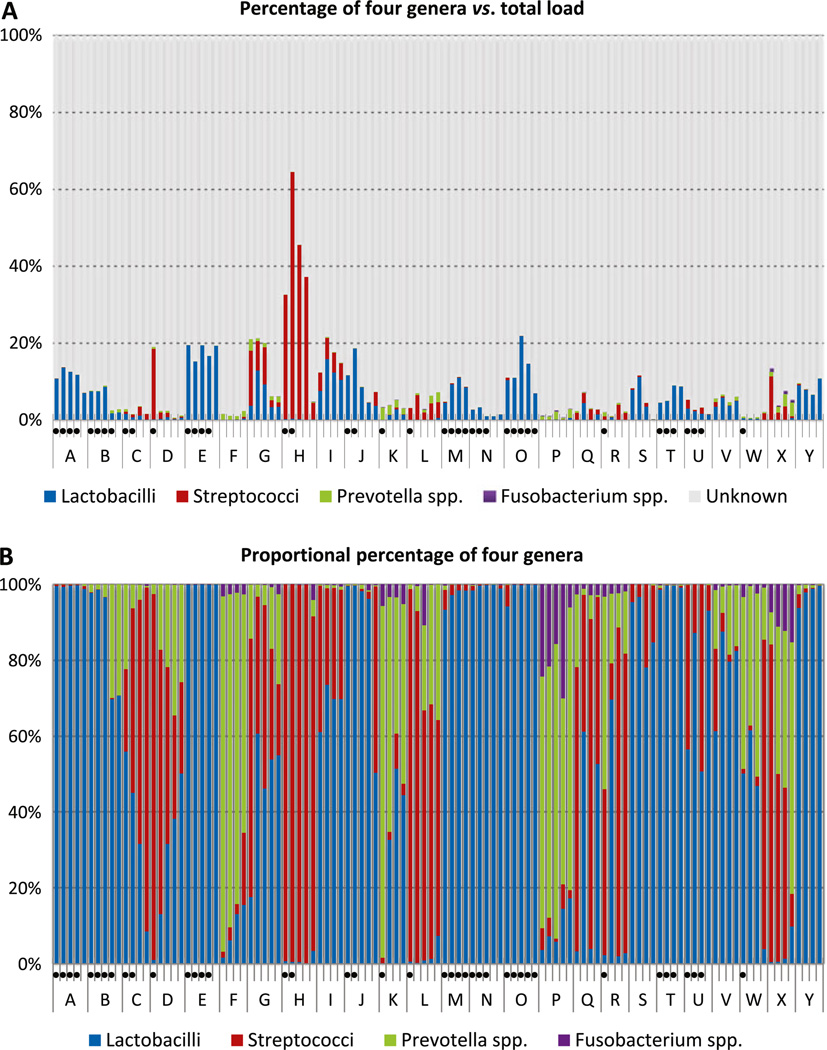
Total load of bacteria as quantified by a universal primer/probe set showed a range of 1.379 × 103 to 1.153 × 108 bacteria per milligram of dentine. As expected, total bacterial load decreased with the depth of dentine (p = 0.002). Streptococci were detected in the range of 1 to 5.506 × 106 bacteria/mg. Fusobacteria ranged from undetectable to 1.605 × 104 bacteria/ mg, prevotellae from undetectable to 2.605 × 105 bacteria/mg, and lactobacilli ranging from 1 to 8.706 × 106 bacteria/mg dentine (Table 2.). The cumulative percentage contributed by the four genera reached only 22 % of total bacterial load, except for one tooth, where streptococci contributed up to 64 % of total bacterial load (Fig. 2a). As a proportion of total bacterial load, only streptococci were significantly more abundant in the most superficial layer of dentine (p = 0.048), while the proportion of other genera studied did not show a correlation with the depth of the lesion. The maximum percentage contribution of streptococci, lactobacilli, prevotellae, and fusobacteria to total bacteria load was 64, 22, 3.5, and 0.9 %, respectively. The mean and median rank of lactobacilli was generally higher than for other taxa, followed by streptococci, prevotellae, and fusobacteria. However, streptococci comprised a negligible proportion in 15 teeth, and lactobacilli were proportionally dominant in 11 of these (teeth A, B, E, J, M, N, O, S, T, V, and Y; Fig. 2b). Similarly, in 8 teeth (C, F, H, L, P, R, W, and X), lactobacilli made up less than 1 % of the total bacterial flora, yet streptococci proportionally dominated the lesion in three of these teeth (H, L, and X). Overall, prevotellae and fusobacteria contributed only a small percentage of the total bacterial population in most samples, but there was a consistent correlation (p = 0.002) between these two groups of bacteria overall and also in all layers of carious dentine, as determined by two-tailed Pearson correlation analysis. Details of bacterial concentration as well as pH values are presented in the “Supplementary table”.
Table 2.
Descriptive concentration (bacteria/mg) of individual genera and total load of bacteria in sample
| Descriptive Statistics | ||||
|---|---|---|---|---|
| Bacteria | Minimum (cell/mg) | Maximum (cell/mg) | Mean (cell/mg) | Std. Deviation |
| Lactobacilli | 3 | 8,708,604 | 209,469 | 910,650 |
| Streptococci | 1 | 5,506,524 | 67,036 | 525,144 |
| Prevotellae | 0 | 260,318 | 10,369 | 36,060 |
| Fusobacteria | 0 | 16,057 | 1,040 | 2,872 |
| Total load of bacteria | 1,380 | 115,334,797 | 3,273,796 | 11,850,195 |
Fig. 2.
Contribution of four genera in carious samples. Samples from the same tooth are labeled under the same letter with the most superficial layer on the left and progressing to the deepest layer on the right. Dots below a sample indicate a pH below hydroxyapatite dissolution pH of 5.5. a Percentage of four genera versus total load of bacteria. b Proportional percentage of the four genera
Correlation between dentinal pH and colonization of specific genera
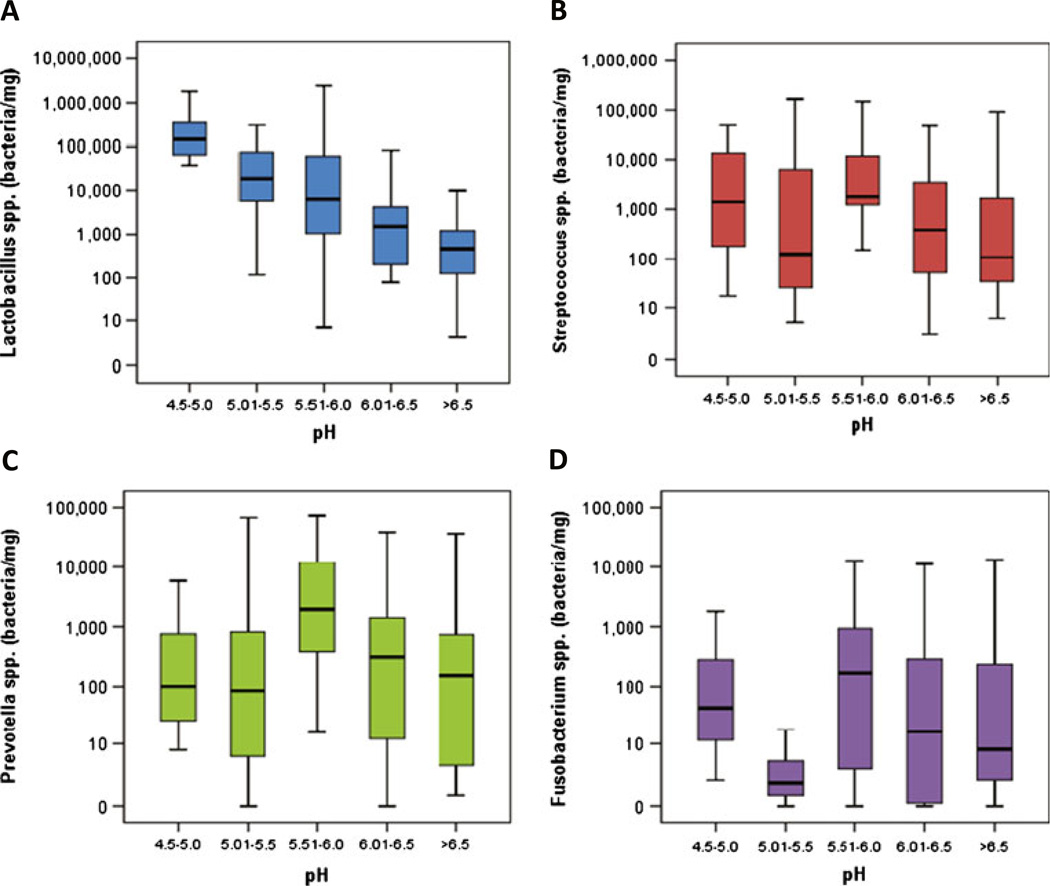
By grouping dentine samples into five pH intervals of 0.5 units, a number of colonization patterns could be detected. Lactobacilli showed a clear propensity for a low pH environment, being most abundant in the pH range 4.5–5.0 (p = 0.003) (Table 3; Fig. 3). Within eight teeth showing predominant dentinal pH below 5.5 (a critical pH, below which dissolution of hydroxyapatite crystals occurs), six showed relative dominance of lactobacilli over the other three genera (Fig. 2b). On the other hand, prevotellae were observed to favor the pH range of 5.5–6.0 (p = 0.042). No statistically significant correlation was found between the colonization of streptococci or fusobacteria with dentinal pH levels. Multiple regression analysis for four genera as well as total load as predictive factors for pH did not indicate statistically significant trends.
Table 3.
Percentile distribution of bacteria at pH ranges
| pH range | Percentile concentration (cell/mg) | |||||||
|---|---|---|---|---|---|---|---|---|
| P5 | P10 | P25 | P50 | P75 | P90 | P95 | ||
| Lactobacilli | 4.5–5 | 37,734 | 37,986 | 48,773 | 144,437 | 427,739 | 1,570,823 | – |
| 5.0–5.5 | 64 | 184 | 3,132 | 17,073 | 76,295 | 137,577 | 215,897 | |
| 5.5–6.0 | 7 | 23 | 1,010 | 6,209 | 79,560 | 859,200 | 2,356,856 | |
| 6.0–6.5 | 1 | 46 | 143 | 1,443 | 4,109 | 43,098 | – | |
| >6.5 | 3 | 22 | 125 | 556 | 1,622 | 858,798 | 6,641,739 | |
| Streptococci | 4.5–5 | 18 | 28 | 82 | 1,380 | 26,971 | 48,401 | – |
| 5.0–5.5 | 5 | 9 | 25 | 205 | 7,046 | 21,114 | 145,797 | |
| 5.5–6.0 | 1 | 159 | 1,188 | 1,761 | 12,435 | 85,956 | 134,346 | |
| 6.0–6.5 | 2 | 17 | 55 | 532 | 4,246 | 42,171 | – | |
| >6.5 | 6 | 7 | 36 | 186 | 3,004 | 306,770 | 4,043,407 | |
| Prevotellae | 4.5–5 | 8 | 8 | 14 | 101 | 1,554 | 5,054 | – |
| 5.0–5.5 | 0 | 1 | 6 | 88 | 1,063 | 15,926 | 155,019 | |
| 5.5–6.0 | 1 | 17 | 499 | 2,402 | 16,317 | 60,926 | 151,229 | |
| 6.0–6.5 | 0 | 1 | 2 | 225 | 2,746 | 21,288 | – | |
| >6.5 | 1 | 2 | 6 | 355 | 2,011 | 28,576 | 41,926 | |
| Fusobacteria | 4.5–5 | 2 | 2 | 3 | 43 | 598 | 1,616 | – |
| 5.0–5.5 | 0 | 0 | 1 | 2 | 8 | 1,013 | 10,040 | |
| 5.5–6.0 | 0 | 1 | 3 | 170 | 983 | 5,887 | 10,426 | |
| 6.0–6.5 | 0 | 0 | 0 | 17 | 481 | 5,283 | – | |
| >6.5 | 0 | 1 | 2 | 17 | 546 | 5,656 | 12,162 | |
| Total load of bacteria | 4.5–5 | 435,153 | 496,692 | 884,003 | 1,651,535 | 5,876,933 | 18,043,634 | – |
| 5.0–5.5 | 50,513 | 80,862 | 264,972 | 598,880 | 780,630 | 2,837,841 | 12,010,584 | |
| 5.5–6.0 | 10,985 | 30,779 | 112,218 | 1,292,971 | 4,522,828 | 16,140,983 | 27,998,151 | |
| 6.0–6.5 | 1,434 | 16,203 | 43,337 | 161,326 | 429,581 | 2,285,414 | – | |
| >6.5 | 2,033 | 4,752 | 17,766 | 48,980 | 344,247 | 7,354,811 | 84,180,190 | |
Fig. 3.
Concentration (bacteria/mg) of four genera at each pH range with standard deviation and median. a Lactobacillus spp. show a robust propensity (p = 0.003) for the lowest pH range (4.5–5.0). b Streptococcus spp. have variable concentrations at different pH ranges, and no correlation to any pH group was observed. c Prevotella spp. revealed a strong tendency (p = 0.042) for pH range (5.5–6.0). d Fusobacterium spp. showed no propensity for any pH group. They could be considered outliers with very low concentration levels
Discussion
pH of dentine caries
Although there have been numerous studies that measured the pH of the superficial plaque biofilm [26–34], there are only a limited number of pH studies on dentinal carious lesion in the literature. The first study on hydrogen ion concentration on the tooth surface dates back to Stephen's report in 1940 [28]. Further, using finer antimony electrodes, a positive correlation between late stages of caries and the fall in pH value on the surface of the tooth was concluded [29]. Methyl red was used later and showed pH of below 5 in carious dentine [10]. The use of an antimony pH electrode was reported as a useful tool in plaque pH studies [27]. Dirksen et al. used 2 mm tip micro-antimony electrodes to study pH of the carious dentine and showed relatively higher acidities compared to the present study and earlier pH studies on carious lesions [35]. Values obtained for pH in the present study concur with previous analysis of acid gradient in caries, where a single sample was taken from each tooth according to the estimated active or arrested status of the lesion [16]. However, by assessing sequential samples in our study, we report that it is the most superficial part of the lesion that is most acidic. This contrasts with an earlier report that acidity increases in the depth of the lesion. Early pH study of dentine caries examined lesions in two layers only and used a 2 mm tip pH electrode [35]. In comparison, we used a 20 times finer pH microelectrode with a 100 µm tip diameter. Multiple sequential samples were taken from each lesion in our study, which enabled us to provide a more realistic pH gradient analysis of a dentine lesion. Progress in understanding is restricted by available technology. In the present study, a minimum of 30 µl saline suspension was needed to measure pH of dentine samples with the fine probe, and therefore at least 7 mg of dentine was required to prepare a 1 mg in 4 µl suspension. Accordingly, samples could not be separated at a finer level than 7 mg. Furthermore, the method used multiple sterile slow speed round burs, so that sampling through macroscopically unaffected sound dentine was possible. Hence, a degree of cross contamination between different dentine samples on each tooth was unavoidable. Nevertheless, the technique was reproducible and the findings informative.
pH and bacterial colonization
Previous analysis of pH and organic acid profile in highly acidic lesions (mean pH 4.9) showed high levels of lactate as a major product of acidogenic lactobacilli and streptococci [16, 36]. Lesions with less acidic pH (mean pH 5.7) and more neutral pH were associated with acetic and propionic acids. Acetate and propionate have been shown to be produced by gram-negative bacteria such as prevotellae [37]. These findings explain our observation that prevotellae showed tendency for pH range 5.5–6.0 (p = 0.42), while lactobacilli were associated with the more acidic regions of carious lesions (p = 0.003). Some prevotellae and fusobacteria are capable of amino acid fermentation [38, 39]. The production of lactic acid by lactobacilli has been proposed to be significant in generating and maintaining a low pH environment [2].
The tendency for streptococci to occupy more superficial layers of the lesion suggests a preference of this genus for open access to exogenous substrates. It was noteworthy that numbers of streptococci did not correlate to dentinal pH, as members of this genus are implicated in caries initiation [40]. A possible explanation is that not all oral streptococci are aciduric, despite their in vitro pH lowering potential and acid tolerance [11, 12]. Functional coassociation between a number of acid sensitive Streptococcus spp. with Veillonella spp. has been reported, due to the utilization of lactate by Veillonella species to create a less acidic local environment [41, 42]. Therefore, a number of different streptococcal species may colonize at different pH values. There remain large numbers of unidentified bacteria within these samples that may also contribute to the generation and maintenance of the pH gradient or alternatively, preferentially, colonize these acidic microenvironments within the carious lesion.
It has been proposed that S. mutans and lactobacilli can dominate a lesion at pH 5 or lower, according to controlled chemostat, and batch culture studies showing that they are more competitive under highly acidic conditions [7, 40, 43]. Our combined pH and bacterial study showed that in vivo, no constant domination of a lesion by these more aciduric and acidogenic genera could be found. Following enamel destruction, dentine provides a different environmental niche for bacteria involved in caries progression. It contains a higher proportion of organic matrix and lower inorganic component than enamel. The hydroxyapatite crystals of dentine are reported to be calcium poor and carbonate rich in comparison to pure hydroxyapatite [44], and dentine dissolution can occur at higher pH levels compared to enamel [45]. Hence, this may allow colonization of diverse bacterial taxa that are not as acidogenic as required for initial enamel destruction. Acid-intolerant members of other bacterial taxa including gram-negative species, which are numerous in the oral cavity, may colonize the less acidic regions of a lesion and give rise to a more complex bacterial consortium. Nevertheless, pH of the environment and bacterial colonization are closely related and can have dynamic influence on each other [7]. Further, the availability of metabolic substrate at different part of a lesion and oxygen tension can also have an impact on this microbial selection [46, 47].
Proportion of examined bacterial genera
Despite emphasis on mutans and non-mutans streptococci as well as Lactobacillus spp. in the carious process [11, 36, 48–51], a lower than expected contribution of these genera to total microbial load was observed in this study. The role of streptococci in caries initiation is not excluded by our results; however, the role of S. mutans has been argued extensively from combined longitudinal and cross-sectional bacterial profile analysis of caries [52]. Our results indicate that progression of the lesion is probably not dependent on members of this genus. Recent molecular study by cloning and sequencing showed that 10 % of teeth with rampant caries did not have detectable levels of S. mutans [53]. Gross et al. grouped carious lesions as either S. mutans dominant, Lactobacillus spp. dominant, S. mutans and Lactobacillus spp. dominant, or a group with poor representation of both S. mutans and lactobacilli [54]. Cross-sectional analysis in the present study detected streptococci in most lesions, although in 15/25 of teeth, streptococci comprised less than 1 % of total bacterial population. In 8/25 of teeth, the contribution of lactobacilli was less than 1 % of total load. We found Prevotella spp. at proportionally lower levels but higher than Fusobacterium spp., findings compatible with previous investigations of advanced caries [18, 48]. Fusobacterium spp. showed significant correlation with concentration of Prevotella spp. in caries, possibly reflecting microhabitat influences [55].
Although results from this work provided information regarding the quantity of lactobacilli, streptococci, prevotellae, and fusobacteria in carious lesions, there were restrictions imposed by the methods employed. In designing an optimum qPCR primer and probe set, many factors should be addressed; namely, length of the amplicon, position of the probe, GC content, optimum melting temperature, standard curve and correct cycle threshold detection level, as well as sensitivity and specificity of the primers [22, 56]. All attempts were made to address the criteria required for an optimum qPCR assay; however, real-time PCR requires sequence data of the target genes, and the detection level is limited to known bacteria, which have previously been sequenced [22]. Hence, coverage of the target bacterial genera in the present study may have been restricted by the availability of sequence data for the species within each genus. Also, what is ultimately achieved from the quantification of bacteria in a qPCR-based study is relative to the processed standard curve, and any unanticipated error through standard curve construction can adversely affect the interpretation of results [57, 58]. Also, it would impose a considerable cost and time to individually assess all samples for more than a few bacteria by real-time PCR. In order to curb qPCR limitations and also to achieve a breadth of knowledge in regard to caries microbiota, pyrosequencing analysis may provide a comprehensive knowledge in the field.
Potential clinical implication of findings
The presence of a pH gradient in a lesion may have particular significance in minimal intervention dentistry with particular reference to atraumatic restorative treatment (ART) of caries. ART removes the most active site of the lesion, known as infected dentine, presumably excluding the most acidic part containing the majority of acidogenic and aciduric bacteria, and sealing the lesion with a restorative material. This has been a relatively successful method in minimal intervention dentistry [59]. A wide range of materials have been used to seal the lesion, including the placement of composite resin following chemomechanical removal of caries. The outcome achieved is 46 % success rate over a 2-year period [60]. However, comparison of ART approaches to conventional restorative treatment of caries has indicated little difference between the two methods [61]. Compomer and glass ionomer (GI) materials were compared at 1-year review and found to be equivalent, although the authors noted the follow-up time was not long enough to extrapolate a conclusive success rate [62]. Resin-modified glass ionomer materials have shown promising longer-term success rates of over 70 % in both primary and permanent dentitions [63, 64]. A 3-year study of ART using amalgam and glass ionomer materials in single-and multi-surface restorations showed over 80% and less than 50 % survival rate, respectively; however, there was no statistically significant difference in longevity between the two materials [65]. In contrast, lower survival rates were observed in a 5-year follow-up study of ART procedure on first permanent molars with composite resin and glass ionomer, with less than 14 % survival rate for both materials [66]. Overall, using high viscosity glass ionomer materials is known as a long lasting approach in ART, especially in single-surface restorations [59, 65–67]. Nevertheless, there remain high failure rates due to both mechanical defects and caries development under such restorations [67].
Interestingly, although ART aims to remove the active caries site that is associated with greater acidity, none of the materials used in the ART procedure demonstrate a proven buffering capacity. In fact, a number of acids are used in glass ionomer material to optimize the workability, setting time, and also to improve physical properties of the GI materials in an acid–base reaction, which leads to an initial acidic phase [68]. While clinical macroscopic evaluation of a lesion in guiding removal of the active parts of the lesion can be relatively acceptable, the presence of an acidic pH gradient in the depth of the lesion can provide an acceptable environment for aciduric and acidogenic anaerobic bacteria under a sealed restoration. To date, no pre or posttreatment of the lesion with a high-pH conditioner has been examined to evaluate the effect on the ART approach. Findings from this study indicate that chemical treatment of a lesion to influence pH may improve the success rate of the ART procedure in minimal intervention dentistry. Acid conditioning of dentine widens dentine tubular system to provide a better chemomechanical interlocking retention [69]. To minimize failure due to secondary caries formation, it appears reasonable to pretreat the prepared cavity with a high-pH conditioner.
Supplementary Material
Acknowledgments
The authors acknowledge funding from NIH/NIDCR (grant no. R01 DEO15272-07), Australian National Health and Medical Research Council (NHMRC grant no. 512524.3), Australian Dental Research Foundation (grant no. 91–2011]), and New South Wales Dental Board (grant no. 2011).We thank Dr Derek Harty and Dr Mangala Nadkarni (Institute of Dental Research, Westmead, NSW, Australia) for provision of some DNA materials. We also thank Mr. Mitchell Brown and Mr. Terry Flood (Center for Infectious Disease and Microbiology, Institute for Clinical Pathology and Medical Research, Westmead, Australia) who provided us with valuable assistance with culture and magnetic DNA extraction of some bacteria.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00784-013-1009-0) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare no potential conflicts of interest in regard to authorship and/or publication of this article.
Contributor Information
Nima Kianoush, Email: nima.kianoush@sydney.edu.au, Institute of Dental Research, Westmead Center for Oral Health and Westmead Millennium Institute, Sydney, Australia; Department of Oral Biology, Faculty of Dentistry, University of Sydney, Sydney, Australia; Department of Oral Biology, Institute of Dental Research, Level 2, Westmead Center for Oral Health C24, Wentworthville, NSW 2145, Australia.
Ky-Anh T. Nguyen, Institute of Dental Research, Westmead Center for Oral Health and Westmead Millennium Institute, Sydney, Australia Department of Oral Biology, Faculty of Dentistry, University of Sydney, Sydney, Australia.
Gina V. Browne, Institute of Dental Research, Westmead Center for Oral Health and Westmead Millennium Institute, Sydney, Australia
Mary Simonian, Institute of Dental Research, Westmead Center for Oral Health and Westmead Millennium Institute, Sydney, Australia.
Neil Hunter, Institute of Dental Research, Westmead Center for Oral Health and Westmead Millennium Institute, Sydney, Australia; Department of Oral Biology, Faculty of Dentistry, University of Sydney, Sydney, Australia.
References
- 1.Featherstone JD. The continuum of dental caries–evidence for a dynamic disease process. J Dent Res. 2004;83(Spec No C):C39–C42. doi: 10.1177/154405910408301s08. [DOI] [PubMed] [Google Scholar]
- 2.van Houte J. Role of microorganisms in caries etiology. J Dent Res. 1994;73(3):672–681. doi: 10.1177/00220345940730031301. [DOI] [PubMed] [Google Scholar]
- 3.Chhour KL, Nadkarni MA, Byun R, Martin FE, Jacques NA, Hunter N. Molecular analysis of microbial diversity in advanced caries. J Clin Microbiol. 2005;43(2):843–849. doi: 10.1128/JCM.43.2.843-849.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nadkarni MA, Caldon CE, Chhour KL, Fisher IP, Martin FE, Jacques NA, Hunter N. Carious dentine provides a habitat for a complex array of novel Prevotella-like bacteria. J Clin Microbiol. 2004;42(11):5238–5244. doi: 10.1128/JCM.42.11.5238-5244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi N, Saito K, Schachtele CF, Yamada T. Acid tolerance and acid-neutralizing activity of Porphyromonas gingivalis, Prevotella intermedia, and Fusobacterium nucleatum. Oral Microbiol Immunol. 1997;12(6):323–328. doi: 10.1111/j.1399-302x.1997.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 6.Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8(2):263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90(3):294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 8.Stephan RM. Two factors of possible importance in relation to the etiology and treatment of dental caries and other dental diseases. Science. 1940;92(2399):578–579. doi: 10.1126/science.92.2399.578. [DOI] [PubMed] [Google Scholar]
- 9.Featherstone JD, Rodgers BE. Effect of acetic, lactic, and other organic acids on the formation of artificial carious lesions. Caries Res. 1981;15(5):377–385. doi: 10.1159/000260541. [DOI] [PubMed] [Google Scholar]
- 10.Macgregor AB. The position and extent of acid in the carious process. Archives of oral biology. 1961;4:86–91. doi: 10.1016/0003-9969(61)90084-x. [DOI] [PubMed] [Google Scholar]
- 11.Van Houte J, Sansone C, Joshipura K, Kent R. Mutans streptococci and non-mutans streptococci acidogenic at low pH and in vitro acidogenic potential of dental plaque in two different areas of the human dentition. J Dent Res. 1991;70(12):1503–1507. doi: 10.1177/00220345910700120601. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi N, Yamada T. Acid-induced acid tolerance and acid-ogenicity of non-mutans streptococci. Oral Microbiol Immunol. 1999;14(1):43–48. doi: 10.1034/j.1399-302x.1999.140105.x. [DOI] [PubMed] [Google Scholar]
- 13.Iwami Y, Abbe K, Takahashi-Abbe S, Yamada T. Acid production by streptococci growing at low pH in a chemostat under anaerobic conditions. Oral Microbiol Immunol. 1992;7(5):304–308. doi: 10.1111/j.1399-302x.1992.tb00593.x. [DOI] [PubMed] [Google Scholar]
- 14.Badet MC, Richard B, Dorignac G. An in vitro study of the pH-lowering potential of salivary lactobacilli associated with dental caries. J Appl Microbiol. 2001;90(6):1015–1018. doi: 10.1046/j.1365-2672.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- 15.McLean JS, Fansler SJ, Majors PD, McAteer K, Allen LZ, Shirtliff ME, Lux R, Shi W. Identifying low pH active and lactate-utilizing taxa within oral microbiome communities from healthy children using stable isotope probing techniques. PLoS One. 2012;7(3):e32219. doi: 10.1371/journal.pone.0032219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hojo S, Komatsu M, Okuda R, Takahashi N, Yamada T. Acid profiles and pH of carious dentin in active and arrested lesions. J Dent Res. 1994;73(12):1853–1857. doi: 10.1177/00220345940730121001. [DOI] [PubMed] [Google Scholar]
- 17.Byun R, Nadkarni MA, Chhour KL, Martin FE, Jacques NA, Hunter N. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J Clin Microbiol. 2004;42(7):3128–3136. doi: 10.1128/JCM.42.7.3128-3136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin FE, Nadkarni MA, Jacques NA, Hunter N. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J Clin Microbiol. 2002;40(5):1698–1704. doi: 10.1128/JCM.40.5.1698-1704.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148(Pt 1):257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 22.Smith CJ, Osborn AM. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol Ecol. 2009;67(1):6–20. doi: 10.1111/j.1574-6941.2008.00629.x. [DOI] [PubMed] [Google Scholar]
- 23.Nadkarni MA, Martin FE, Hunter N, Jacques NA. Methods for optimizing DNA extraction before quantifying oral bacterial numbers by real-time PCR. FEMS Microbiol Lett. 2009;296(1):45–51. doi: 10.1111/j.1574-6968.2009.01629.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee ZM, Bussema C, 3rd, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37:D489–D493. doi: 10.1093/nar/gkn689. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm S. A simple sequentially rejective multiple test procedure. Scand J Statist. 1979;6:65–70. [Google Scholar]
- 26.Schachtele CF, Jensen ME. Comparison of methods for monitoring changes in the pH of human dental plaque. J Dent Res. 1982;61(10):1117–1125. doi: 10.1177/00220345820610100201. [DOI] [PubMed] [Google Scholar]
- 27.Kleinberg I, Jenkins GN, Chatterjee R, Wijeyeweera L. The antimony pH electrode and its role in the assessment and interpretation of dental plaque pH. J Dent Res. 1982;61(10):1139–1147. doi: 10.1177/00220345820610100601. [DOI] [PubMed] [Google Scholar]
- 28.Stephen RM. Changes in hydrogen ion concentration on tooth surfaces and in carious lesions. J Am Dent Assoc. 1940;27:718–723. [Google Scholar]
- 29.Bentley KD, Haldi J, Law ML, Ramsey DA, Wynn W. Dental caries in relation to pH on tooth surfaces. I. pH and lactate concentration in relation to the extent of the lesions in rats' teeth. J Nutr. 1956;60(3):427–435. doi: 10.1093/jn/60.3.427. [DOI] [PubMed] [Google Scholar]
- 30.Igarashi K, Kamiyama K, Yamada T. Measurement of pH in human dental plaque in vivo with an ion-sensitive transistor electrode. Archives of oral biology. 1981;26(3):203–207. doi: 10.1016/0003-9969(81)90131-x. [DOI] [PubMed] [Google Scholar]
- 31.Hassell TM. Construction of micro-antimony electrodes for use in radio telemetry of plaque pH. Helvetica odontologica acta. 1971;15(1):50–51. [PubMed] [Google Scholar]
- 32.Tahmassebi JF, Duggal MS. The effect of different methods of drinking on the pH of dental plaque in vivo. Intern J Paediatr Dent Br Paedodontic Soc Intern Assoc Dent Children. 1997;7(4):249–254. doi: 10.1046/j.1365-263x.1997.00054.x. [DOI] [PubMed] [Google Scholar]
- 33.Roos EH, Donly KJ. In vivo dental plaque pH variation with regular and diet soft drinks. Pediatr Dent. 2002;24(4):350–353. [PubMed] [Google Scholar]
- 34.Jawale BA, Bendgude V, Mahuli AV, Dave B, Kulkarni H, Mittal S. Dental plaque pH variation with regular soft drink, diet soft drink, and high energy drink: an in vivo study. J Contemp Dent Pract. 2012;13(2):201–204. doi: 10.5005/jp-journals-10024-1121. [DOI] [PubMed] [Google Scholar]
- 35.Dirksen TR, Little MF, Bibby BG. The pH of carious cavities-II. The pH at different depths in isolated cavities. Archives of oral biology. 1963;8:91–97. doi: 10.1016/0003-9969(63)90046-3. [DOI] [PubMed] [Google Scholar]
- 36.Hojo S, Takahashi N, Yamada T. Acid profile in carious dentin. J Dent Res. 1991;70(3):182–186. doi: 10.1177/00220345910700030501. [DOI] [PubMed] [Google Scholar]
- 37.Hopkins MJ, Englyst HN, Macfarlane S, Furrie E, Macfarlane GT, McBain AJ. Degradation of cross-linked and non-cross-linked arabinoxylans by the intestinal microbiota in children. Appl Environ Microbiol. 2003;69(11):6354–6360. doi: 10.1128/AEM.69.11.6354-6360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jansen HJ, van der Hoeven JS. Protein degradation by Prevotella intermedia and Actinomyces meyeri supports the growth of nonprotein-cleaving oral bacteria in serum. J Clin Periodontol. 1997;24(5):346–353. doi: 10.1111/j.1600-051x.1997.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi N. Acid-neutralizing activity during amino acid fermentation by Porphyromonas gingivalis, Prevotella intermedia, and Fusobacterium nucleatum. Oral Microbiol Immunol. 2003;18(2):109–113. doi: 10.1034/j.1399-302x.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 40.Bradshaw DJ, Marsh PD. Analysis of pH-driven disruption of oral microbial communities in vitro. Caries Res. 1998;32(6):456–462. doi: 10.1159/000016487. [DOI] [PubMed] [Google Scholar]
- 41.Periasamy S, Kolenbrander PE. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J Bacteriol. 2010;192(12):2965–2972. doi: 10.1128/JB.01631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chalmers NI, Palmer RJ, Jr, Cisar JO, Kolenbrander PE. Characterization of a Streptococcus sp.–Veillonella sp. community micromanipulated from dental plaque. J Bacteriol. 2008;190(24):8145–8154. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horiuchi M, Washio J, Mayanagi H, Takahashi N. Transient acid-impairment of growth ability of oral Streptococcus, Actinomyces, and Lactobacillus: a possible ecological determinant in dental plaque. Oral Microbiol Immunol. 2009;24(4):319–324. doi: 10.1111/j.1399-302X.2009.00517.x. [DOI] [PubMed] [Google Scholar]
- 44.Berkovitz BKB, Holland GR, Moxham BJ. Oral anatomy, histology and embryology. 3rd edn. Mosby; 2002. [Google Scholar]
- 45.Vanuspong W, Eisenburger M, Addy M. Cervical tooth wear and sensitivity: erosion, softening, and rehardening of dentine: effects of pH, time, and ultrasonication. J Clin Periodontol. 2002;29(4):351–357. doi: 10.1034/j.1600-051x.2002.290411.x. [DOI] [PubMed] [Google Scholar]
- 46.Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149(2):279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 47.Bradshaw DJ, Marsh PD, Allison C, Schilling KM. Effect of oxygen, inoculum composition, and flow rate on development of mixed-culture oral biofilms. Microbiology. 1996;142(3):623–629. doi: 10.1099/13500872-142-3-623. [DOI] [PubMed] [Google Scholar]
- 48.Munson MA, Banerjee A, Watson TF, Wade WG. Molecular analysis of the microflora associated with dental caries. J ClinMicrobiol. 2004;42(7):3023–3029. doi: 10.1128/JCM.42.7.3023-3029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Houte J, Lopman J, Kent R. The predominant cultivable flora of sound and carious human root surfaces. J Dent Res. 1994;73(11):1727–1734. doi: 10.1177/00220345940730110801. [DOI] [PubMed] [Google Scholar]
- 50.Sansone C, Van Houte J, Joshipura K, Kent R, Margolis HC. The association of mutans streptococci and non-mutans streptococci capable of acidogenesis at a low pH with dental caries on enamel and root surfaces. J Dent Res. 1993;72(2):508–516. doi: 10.1177/00220345930720020701. [DOI] [PubMed] [Google Scholar]
- 51.Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50(4):353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One. 2012;7(10):e47722. doi: 10.1371/journal.pone.0047722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46(4):1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, Asnani K, Griffen AL. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. 2010;48(11):4121–4128. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jenkinson HF. Beyond the oral microbiome. Environ Microbiol. 2011;13(12):3077–3087. doi: 10.1111/j.1462-2920.2011.02573.x. [DOI] [PubMed] [Google Scholar]
- 56.Klein D. Quantification using real-time PCR technology: applications and limitations. Trends Mol Med. 2002;8(6):257–260. doi: 10.1016/s1471-4914(02)02355-9. [DOI] [PubMed] [Google Scholar]
- 57.Smith CJ, Nedwell DB, Dong LF, Osborn AM. Evaluation of quantitative polymerase chain reaction-based approaches for determining gene copy and gene transcript numbers in environmental samples. Environ Microbiol. 2006;8(5):804–815. doi: 10.1111/j.1462-2920.2005.00963.x. [DOI] [PubMed] [Google Scholar]
- 58.Love JL, Scholes P, Gilpin B, Savill M, Lin S, Samuel L. Evaluation of uncertainty in quantitative real-time PCR. J Microbiol Methods. 2006;67(2):349–356. doi: 10.1016/j.mimet.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 59.Frencken JE, Leal SC, Navarro MF. Twenty-five-year atraumatic restorative treatment (ART) approach: a comprehensive overview. Clinical oral investigations. 2012;16(5):1337–1346. doi: 10.1007/s00784-012-0783-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Topaloglu-Ak A, Eden E, Frencken JE, Oncag O. Two-year survival rate of class II composite resin restorations prepared by ART with and without a chemomechanical caries removal gel in primary molars. Clinical oral investigations. 2009;13(3):325–332. doi: 10.1007/s00784-008-0241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eden E, Topaloglu-Ak A, Frencken JE, van't Hof M. Survival of self-etch adhesive Class II composite restorations using ART and conventional cavity preparations in primary molars. Am J Dent. 2006;19(6):359–363. [PubMed] [Google Scholar]
- 62.Louw AJ, Sarvan I, Chikte UM, Honkala E. One-year evaluation of atraumatic restorative treatment and minimum intervention techniques on primary teeth. SADJ: J S Afr Dental Assoc=tydskrif van die Suid-Afrikaanse Tandheelkundige Vereniging. 2002;57(9):366–371. [PubMed] [Google Scholar]
- 63.Cefaly DF, Barata TJ, Bresciani E, Fagundes TC, Lauris JR, Navarro MF. Clinical evaluation of multiple-surface ART restorations 12-month follow-up. J Dent Child (Chic) 2007;74(3):203–208. [PubMed] [Google Scholar]
- 64.Faccin ES, Ferreira SH, Kramer PF, Ardenghi TM, Feldens CA. Clinical performance of ART restorations in primary teeth: a survival analysis. J Clin Pediatr Dent. 2009;33(4):295–298. doi: 10.17796/jcpd.33.4.001283522r230h70. [DOI] [PubMed] [Google Scholar]
- 65.Taifour D, Frencken JE, Beiruti N, van 't Hof MA, Truin GJ. Effectiveness of glass-ionomer (ART) and amalgam restorations in the deciduous dentition: results after 3 years. Caries Res. 2002;36(6):437–444. doi: 10.1159/000066531. [DOI] [PubMed] [Google Scholar]
- 66.Beiruti N, Frencken JE, van't Hof MA, Taifour D, van Palenstein Helderman WH. Caries-preventive effect of a one-time application of composite resin and glass ionomer sealants after 5 years. Caries Res. 2006;40(1):52–59. doi: 10.1159/000088907. [DOI] [PubMed] [Google Scholar]
- 67.Zanata RL, Fagundes TC, Freitas MC, Lauris JR, Navarro MF. Ten-year survival of ART restorations in permanent posterior teeth. Clin Oral Inv. 2011;15(2):265–271. doi: 10.1007/s00784-009-0378-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prentice LH, Tyas M, Burrow MF. The effect of oxalic acid incorporation on the setting time and strength of glass-ionomer cement. Acta biomaterialia. 2006;2(1):109–112. doi: 10.1016/j.actbio.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 69.McInnes-Ledoux PM, Weinberg R, Grogono A. Bonding glass-ionomer cements to chemomechanically-prepared dentin. Dent Mater Off Publ Acad Dent Mater. 1989;5(3):189–193. doi: 10.1016/0109-5641(89)90012-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.