Abstract
Objectives
Genome wide association studies have shown 32 loci to influence BMI in European-American adults but replication in other studies is inconsistent and may be attributed to gene-by-age effects. The aims of this study were to determine if the influence of the summed risk score of these 32 loci (GRS) on BMI differed across age from birth to 40 years, and to determine if additive genetic effects other than those in the GRS differed by age.
Design and Methods
Serial measures of BMI were calculated at 0, 1, 3, 6, 9, 12, 18, and 28 months, and 4, 7, 11, 15, 19, 23, 30, and 40 years for 1176 (605 females, 571 males) European-American participants in the Fels Longitudinal Study. SOLAR was used for genetic analyses.
Results
GRS was significant (p< 0.05) at ages: 6, 9 months, 4–15 years, and 23–40 years. Remaining additive genetic effects independently influenced BMI (p<5.3×10−5, 0.40<h2<0.76). Some genetic correlations between ages were not significant. Differential GRS effects did not retain significance after multiple comparisons adjustments.
Conclusions
While well-known BMI variants do not appear to have significant differential effects, other additive genes differ over the lifespan.
INTRODUCTION
Over 32 common loci associated with body mass index (BMI) have been identified through genome-wide association studies in the last five years (1–4). These studies and others, however, have yielded inconsistent associations between these single nucleotide polymorphisms (SNPs) and BMI at different ages across the lifespan. That is, while some studies show that some BMI variants (i.e., SNPs) influence BMI in infancy (5) and childhood (1, 6), others do not consistently find significant influences within a life stage (7–9). For example, Zhao et al. (9) found variants in FTO, MC4R, SEC16B and GNPDA2 to be significant at ages 6–10 years, but at ages 11–14 years, only variants in BDNF and FTO were significant. Because of the cross-sectional design of many of these studies, there are little data describing how genetic effects differ over age. While some longitudinal studies have examined changes in the influence of various SNPs across age, to our knowledge, few studies have examined genetic effects from infancy through childhood and adolescence to adulthood (10, 11). Understanding the effect of genes on BMI during childhood and adolescence is made more difficult because weight gain is a natural part of growth and development. Thus identification of possible gene-by-age interactions advances our understanding of growth and obesity, by distinguishing those genes that are influential at different times of the lifespan, including both adulthood and childhood. In turn, this may enable us to distinguish those biological pathways directly related to obesity, as opposed to those which jointly influence growth, development and maturation. By understanding which genes are influential over the whole lifespan or at different times, the metabolic pathways can be more specifically targeted for future therapy. The aims of this study were to examine whether the covariate effect of a genetic risk score (GRS) comprised of 32 well-replicated variants (SNPs) on BMI assessed longitudinally, differed across the life course (from birth to 40 years of age), and to determine whether the remaining additive genetic effects also differed by age.
METHODS
Study sample and measurements
The study sample consisted of 1176 white participants (605 females, 571 males) from the Fels Longitudinal Study, a study of human growth, development, and body composition conducted in the Dayton, Ohio area of the United States (12), that started in 1929. At the beginning of the study, participants were enrolled in utero, and family members such as parents and older siblings were also recruited. The current study sample consists of white participants who have at least one measurement of weight and height between birth and 44.9 years of age and have been genotyped for a high density panel of SNPs. Birth weight was obtained from birth records. Weight and recumbent length were measured, by trained anthropometrists according to standard references (13). Dates of measurement ranged from May 1934 to December 2011. For measurements of individuals over 24 months of age, standing height was used in lieu of recumbent length. BMI was calculated across 5 different life stages that included infancy, childhood, adolescence, early adulthood, and mid-adulthood. The corresponding ages within lifestage are listed in Table 1
Table 1.
Descriptive statistics of study sample.
| Variable | N | Mean | SD | Range | |
|---|---|---|---|---|---|
| BMI during infancy | |||||
| 0 (+ 0.03) months | 561 | 13.1 | 1.4 | 6.0 | 18.9 |
| 1 (± 0.5) month | 425 | 14.1 | 1.3 | 10.2 | 19.2 |
| 3 (± 1.5) months | 490 | 16.0 | 1.4 | 11.9 | 19.9 |
| 6 (± 1.5) months | 511 | 17.0 | 1.4 | 13.3 | 21.2 |
| 9 (± 1.5) months | 505 | 17.4 | 1.4 | 12.8 | 24.2 |
| 12 (± 3) months | 533 | 17.2 | 1.3 | 13.3 | 20.6 |
| 18 (± 3) months | 534 | 16.5 | 1.3 | 12.9 | 21.9 |
| 28 (± 7.5) months | 612 | 16.0 | 1.2 | 12.7 | 22.7 |
| BMI during childhood | |||||
| 4 (± 1) years | 654 | 15.6 | 1.2 | 12.8 | 22.1 |
| 7 (± 2) years | 779 | 15.9 | 1.9 | 12.1 | 28.7 |
| BMI during adolescence | |||||
| 11 (± 2) years | 776 | 18.0 | 3.1 | 12.4 | 34.7 |
| 15 (± 2) years | 744 | 20.9 | 3.5 | 14.0 | 38.2 |
| BMI during early adulthood | |||||
| 19 (± 2) years | 671 | 22.4 | 3.9 | 15.0 | 43.8 |
| 23 (± 2) years | 507 | 23.5 | 4.6 | 15.6 | 49.7 |
| 30 (± 5) years | 654 | 24.6 | 5.1 | 15.8 | 50.5 |
| BMI during mid-adulthood | |||||
| 40 (± 5) years | 590 | 26 | 5.3 | 16.3 | 59.4 |
| Year of birth | 1176 | 1964.2 | 21.5 | 1901 | 2007 |
| Genetic risk score (GRS) | 1176 | 28.5 | 3.5 | 18.1 | 39.9 |
| Sex (females)a | 1176 | 605 (51.4%) | |||
Number (%) listed under mean
Relative age at each of the time points was calculated as the age at measurement minus the exact target age and was included in all analyses as a covariate to account for individual variations in the exact timing of the measurements. Year of birth, sex, and year of birth-by-sex were also included as covariates. Year of birth was included to account for potential era or cohort effects. Because not all participants were alive and measured at the same time, the sample sizes for the different target ages ranged from 425 to 779. Means and ranges of the covariates used for analyses are listed by target age in Table 1.
Genetic data
For all participants in this study, DNA was extracted from whole blood collected via venipuncture using standard procedures and stored at −80 degrees Celsius. Individuals were genotyped using the genome-wide Illumina Human 610-Quad Bead-chip array (Illumina, USA) at the Texas Biomedical Research Institute. SimWalk2 mistyping analysis (14–16) was used to determine genotypes that had a high probability of being incorrectly called, and these Mendelian errors were removed by blanking these genotypes. HapMap 2 SNP genotypes were imputed using MaCH1 (17, 18), and were further cleaned using SimWalk2 (14–16). Merlin (19) was used to impute missing genotypes using familial data. The squared correlation between imputed and true genotypes (R2) for these 32 SNPs used in the study ranged from 0.89 to 1.0.
Table 2 lists the 32 SNPs significantly related to BMI (1–3) that were reported in Table 1 from Speliotes et al (4) and were used to calculate the BMI GRS for each individual using the HapMap 2 SNP data. For each SNP, the allele that was reported by Speliotes et al. (4) to be associated with greater BMI was considered to be the risk allele, and given a score of 1. Thus, for each SNP, an individual could have scores ranging from 0 to 2.
Table 2.
SNP and corresponding risk alleles used to calculate genetic risk score (GRS) based on Speltiotes et al, 2010
| SNP | Nearest gene | Risk allele | Frequency of risk allele in Fels Longitudinal Study |
|---|---|---|---|
| rs10150332 | NRXN3 | C | 0.22 |
| rs10767664 | BDNF | A | 0.81 |
| rs10938397 | GNPDA2 | G | 0.41 |
| rs10968576 | LRRN6C | G | 0.31 |
| rs11847697 | PRKD1 | T | 0.03 |
| rs12444979 | GPRC5B | C | 0.85 |
| rs13078807 | CADM2 | G | 0.19 |
| rs13107325 | SLC39A8 | T | 0.05 |
| rs1514175 | TNNI3K | A | 0.43 |
| rs1555543 | PTBP2 | C | 0.61 |
| rs1558902 | FTO | A | 0.39 |
| rs206936 | NUDT3 | G | 0.22 |
| rs2112347 | FLJ35779 | T | 0.64 |
| rs2241423 | MAP2K5 | G | 0.77 |
| rs2287019 | QPCTL | C | 0.80 |
| rs2815752 | NEGR1 | A | 0.63 |
| rs2867125 | TMEM18 | C | 0.84 |
| rs2890652 | LRP1B | C | 0.14 |
| rs29941 | KCTD15 | G | 0.68 |
| rs3810291 | TMEM160 | A | 0.68 |
| rs3817334 | MTCH2 | T | 0.38 |
| rs4771122 | MTIF3 | G | 0.25 |
| rs4836133 | ZNF608 | A | 0.45 |
| rs4929949 | RPL27A | C | 0.51 |
| rs543874 | SEC16B | G | 0.18 |
| rs571312 | MC4R | A | 0.25 |
| rs713586 | RBJ | C | 0.48 |
| rs7138803 | FAIM2 | A | 0.35 |
| rs7359397 | SH2B1 | T | 0.40 |
| rs887912 | FANCL | T | 0.29 |
| rs9816226 | ETV5 | T | 0.79 |
| rs987237 | TFAP2B | G | 0.18 |
Because these individual SNPs often have different effects at different ages, and also describe only a small proportion of the phenotypic variance (20), we, as have others (6, 11, 21), decided to investigate the influence of the combined effects of the BMI variants on BMI at different ages. Recognizing that not all studies combine the same gene variants, we elected to follow the exact variants used by Speliotes et al. (4), as they had identified most of the individual variants that influenced BMI. The number of risk alleles per individual was summed across the 32 BMI variants.
Quantitative genetic analysis
BMI at each age were first adjusted for year of birth, sex, year of birth-by-sex, and the exact age difference using linear regression models, and the residuals were then inverse-normalized to handle residual kurtosis. Effectively then, BMI at each age had a mean of 0, and a standard deviation of 1. Because mean BMI tends to increase with age, this method has the added benefit of standardizing all effect sizes to the same scale. Thus, beta coefficients from subsequent genetic analyses are interpreted as changes in a standard deviation unit (SDU) of BMI and do not coincide with the size of mean BMI.
Maximum-likelihood variance components methods in SOLAR (22) were used to estimate narrow-sense heritabilities (h2) at each age, adjusting for the GRS and population stratification (23) using the first 4 principal components (PCs). Briefly, PCs were calculated using 27, 966 cleaned Illumina SNPs that had minimal linkage disequilibrium (r < 0.1) across a 2Mb sliding window, and that had a relatively high minor allele frequency (15.5%) for our entire sample of SNP genotyped participants. From these participants, 452 unrelated participants were identified using the unrelate command in PEDSYS (24). Principal components analysis was performed on the 27,966 genotype scores of the 452 unrelated participants using prcomp in R (http://www.r-project.org). PC scores for all genotyped participants were calculated from the loadings using predict in R. Narrow sense heritability, defined as the proportion of the phenotypic variance attributable to additive genetic effects, was estimated as h2 = σ2G / σ2P, where σ2G is the additive genetic variance and σ2P is the total phenotypic variance.
Likelihood ratio tests (LRT) were used to test if models allowing h2 to be estimated were different from models where h2 was not parameterized or was fixed to zero. Similarly, GRS effects were tested for significance by comparing models where GRS was estimated in the model against models where GRS was fixed to zero. To determine the proportion of the variation explained by the GRS, at each age, we used the same models described above and subtracted the proportion of the variance explained by the covariates in the restricted model (where GRS was not estimated) from the full model that included the GRS.
Bivariate extensions to the univariate model were used to search for genotype-by-age effects. To simplify the analysis, we restricted our pair-wise analyses to include one age from each of the 5 life stages. For each life stage, we chose the age with the greatest total amount of variation explained (i.e., h2 + variation explained by GRS). For life stages with more than two ages, we also included the ages at minimum total variation explained. In SOLAR, the covariance is given by a 2 × 2 covariance matrix with elements defined as Ωab = 2ΦρGσgaσgb + IρEσeaσeb, where 2Φ is the kinship matrix that structures σga and σgb, the variance due to the additive effects of genes for time points a and b; I is an identity matrix that structures σea and σeb, the variance due to unmeasured, non-genetic factors in a and b; ρG is the additive genetic correlation between time points a and b, and ρE is the unmeasured environmental correlation between the two time points.
In our study for each pair of ages modeled, we first investigated gene-by-age effects among the additive genes other than the BMI variants by examining whether the genetic correlation (ρG), a measure of shared genetic influences (pleiotropy), between BMI measured at two different ages was significantly different from one (complete pleiotropy), or whether the genetic variances (σG) of BMI measured at two different ages were significantly different at the two ages (25). For each pair of ages modeled, LRT were used to test for complete pleiotropy, which occurs when the same gene (or set of genes) that influences BMI at one age also influences BMI at another age. LRT were also used to test for situations of no pleiotropy, where ρG is not significantly different from zero, and the genes that influence BMI at one age are wholly different from the genes that influence BMI at the other age. Incomplete pleiotropy occurs when ρG is different from both zero and one. In this case, some but not all of the genes that influence BMI at one age also influence BMI at other ages. Instances of complete and incomplete pleiotropy are indications of gene-by-age interactions for additive genes other than the BMI variants.
We also tested for differences in h2 by age by examining differences in the decomposed variances of BMI at each age. We tested whether genetic variances (σG) or environmental variances (σE) were significantly different at different ages. By modeling parameters of σG and σE, differences in h2 can be decomposed into changes due to polygenes (σG), residual environmental effects (σE), or both. LRT were used to test for differences in the genetic variances estimated at different ages, which can be interpreted as an indicator of differences in the expression of polygenes, other than BMI variants, and are also indicative of gene-by-age interaction effects.
Differences in GRS effect across age
When modeling bivariate parameters for each pair of ages, essentially the number of parameters from each univariate model for each age is summed and two additional parameters, ρG and ρE, are estimated. Thus for each age-pair, there are two βGRS estimates, one for each age. We can test if the influence of the GRS on BMI across age-pairs is the same (i.e., there are no GRS-by-age effects) by restricting the individual βGRS covariate estimates for each age to be equal. As with other parameter estimates, we used LRT to test for differences in the effects of GRS on BMI at different ages.
RESULTS
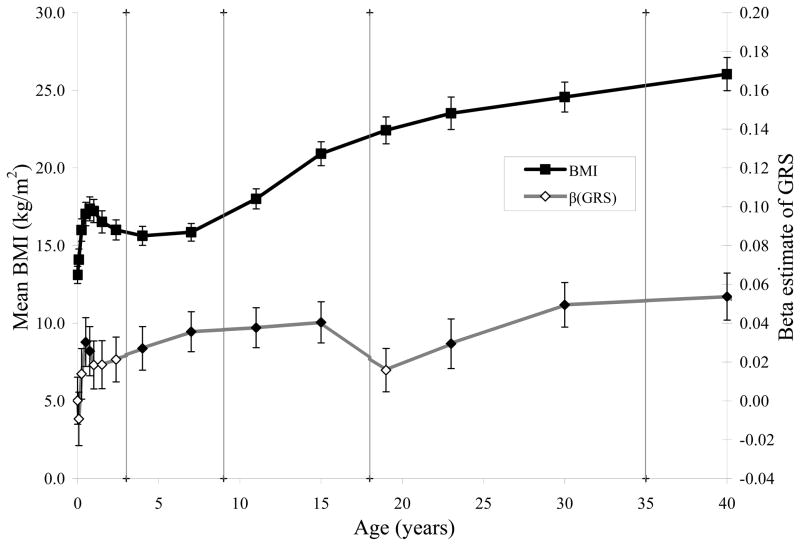
Raw mean BMI across age (solid squares, primary axis) and the estimates of the covariate effect of the GRS (diamonds, secondary axis) are shown in Figure 1. LRT indicate that βGRS was significant (solid diamonds) at 6 and 9 months during infancy and during all of childhood and adolescence. During adulthood, βGRS was significant at 23, 30, and 40 years of age.
FIGURE 1.
Mean BMI (solid squares, left axis) and influence of BMI variants (diamonds, right axis) across age at birth, infancy (1, 3, 6, 9, 12, 18, and 28 months) childhood (4 and 7 years), adolescence (11 and 15 years), adulthood (19, 23, and 30 years) and mid-adulthood (40 years). βGRS is significantly different from zero (p < 0.05) at 6 and 9 months, all of childhood and adolescence, and at ages 23, 30 and 40 years (solid diamonds).
Heritability of BMI across age
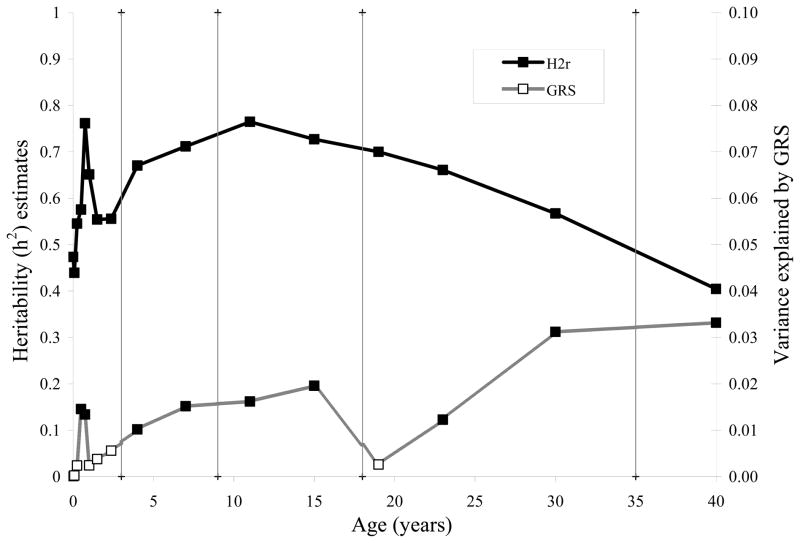
Additive genetic effects (h2) after adjusting for GRS were significant at all ages across the lifespan (p < 5.3x10−5) and explained from 40 to 76% of the variance in BMI (Figure 2, solid squares, Supplemental Table S1). Heritability estimates of BMI during infancy were moderate at birth and 1 month (h2 = 0.47 and 0.44 respectively), and peaked at 9 months (h2 = 0.76). Peak heritability during childhood was at 7 years of age (h2 = 0.71), and during adolescence was at 11 years of age (h2 = 0.76). During early adulthood, heritabilities peaked at 19 years of age (h2 = 0.70), and declined until 30 years of age (h2 = 0.49). In mid-adulthood, the heritability continued to decline until 40 years of age (h2 = 0.40).
FIGURE 2.
Proportion of the variance explained by BMI variants (open squares, right axis) and additive genetic effects (h2) adjusting for BMI variants (solid squares, left axis), across age at birth, infancy (1, 3, 6, 9, 12, 18, and 28 months) childhood (4 and 7 years), adolescence (11 and 15 years), adulthood (19, 23, and 30 years) and mid-adulthood (40 years). All heritability estimates were significant (p < 5.3×10−5).
The proportion of the variation explained by the GRS (Figure 2, open squares, Supplemental Table S1) shows a similar pattern to the GRS parameter estimates (βGRS, Figure 1, diamonds). Significant GRS effects explain from 1.0 to 3.3% of the variation in BMI across age, and show a pattern that is slightly different from the residual additive genetic effects. The variation explained by additive genetic effects and GRS generally appear to be parallel in childhood and adolescence, however, in adulthood, they appear to have opposite trends, with the additive genetic effects decreasing, and the GRS increasing. Note, however, that the magnitudes of the additive genetic effects are much larger than the GRS.
Gene-by-age differences in additive genetic effects and βGRS
Differences in the heritability of BMI at different ages were tested at birth, 9 months, 7, 11, 19, 30, and 40 years. Table 3 shows that the genetic correlations (ρG ± SE) for BMI age-pairs range from −0.07 to 0.99. After adjusting for multiple testing, the correlations at birth and infancy, with adolescence and adulthood (i.e., first 2 columns, last four rows) were not significantly different from zero. Thus, additive genetic effects (excluding GRS effects) that influence BMI during infancy and at birth are entirely different from those that influence BMI during adolescence and adulthood. The ρG between BMI at ages 7 and 40, and BMI at ages 11 and 40 were also not statistically different from zero. All other ρG were significantly greater than zero (adjusted p < 0.005) and significantly less than one (adjusted p < 0.008), except for the ρG between birth and infancy, BMI at age 19 and age 30, and BMI at age 30 and age 40. These results indicate that the genes that influence BMI earlier in life tend to differ from those influencing BMI later in life. These results indicate that gene-by-age interaction effects, excluding GRS variants, are present for BMI across the lifespan.
Table 3.
Genetic correlations (ρG ± SE) and sample size between BMI measured at different times in the lifespan.
| Birth | Infancy | Childhood | Adolescence | Adult (19 yrs) | Adult (30 yrs) | |
|---|---|---|---|---|---|---|
| Infancy (9 mos) | 0.81 ± 0.29a (340) | |||||
| Childhood (7 yrs) | 0.49 ± 0.15b (485) | 0.59 ± 0.09b (488) | ||||
| Adolescence (11 yrs) | 0.20 ± 0.15NS (479) | 0.36 ± 0.12bc (467) | 0.96 ± 0.02b (715) | |||
| Young adulthood (19 yrs) | 0.19 ± 0.18NS (410) | 0.25 ± 0.13NS (400) | 0.80 ± 0.06b (593) | 0.84 ± 0.05b (611) | ||
| Young adulthood (30 yrs) | 0.68 ± 0.32ac (290) | 0.04 ± 0.18NS (321) | 0.62 ± 0.09b (474) | 0.78 ± 0.08b (472) | 0.99 ± 0.05a (452) | |
| Mid-Adulthood (40 yrs) | 0.45 ± 0.30NS (209) | −0.07 ± 0.20NS (308) | 0.34 ± 0.15bc (385) | 0.49 ± 0.14bc (379) | 0.64 ± 0.12b (359) | 0.96 ± 0.04a (441) |
Not significantly different from zero, indicating that all genes influencing BMI at one age are wholly different from those influencing BMI at the other age (i.e., there are gene-by-age effects)
Not significantly different from one, but significantly different from zero, indicating that all of the genes influencing BMI at one age are the same genes influencing BMI at another age.
Significantly different from zero, and significantly different from one, indicating that some by not all the genes influencing BMI at one age are the same genes influencing BMI at another age (i.e., there are gene-by-age effects)
Not significantly different from zero, after adjusting using Holm-Bonferroni.
Tests for differences in the genetic variance (σG), residual environmental variance (σE) shown in Table 4 indicate that at each age, the heritabilities of BMI from birth to 40 years of age are generally stable (Table 4, first two columns) as the σG and σE tend to have opposite trends. While there were some differences in the age-pairs (indicated by different letters) for σG, and σE, after adjusting for multiple testing using Holm-Bonferroni, all pair-wise comparisons were not significant.
Table 4.
Comparison of the GRS effect (βGRS), genetic variance (σG), and environmental variance (σE) across various life stages.
| σE | σG | βGRS | |
|---|---|---|---|
| Birth | 0.71 (0.07)ac | 0.67 (0.08)a | 0.0001 (0.012)a |
| Infancy (9 mos) | 0.48 (0.10)b | 0.86 (0.08)b | 0.026 (0.013)acd |
| Childhood (7 yrs) | 0.53 (0.07)ab | 0.84 (0.07)ab | 0.036 (0.010)cd |
| Adolescence (11 yrs) | 0.48 (0.07)b | 0.87 (0.06)bc | 0.038 (0.010)cd |
| Young adulthood (19 yrs) | 0.54 (0.07)bc | 0.83 (0.07)abc | 0.016 (0.011)abc |
| Young adulthood (30 yrs) | 0.64 (0.06)abc | 0.73 (0.06)abc | 0.049 (0.011)d |
| Mid-Adulthood (40 yrs) | 0.75 (0.06)c | 0.61 (0.08)ac | 0.054 (0.012)ad |
Estimates with different letters are significantly (p < 0.05) different from each other. All comparisons are not significant after adjusting for multiple testing using Holm-Bonferroni.
The results for GRS-by-age effects are shown in Table 4, last column. Again, while there were some differences in the age-pairs (indicated by different letters) for βGRS, all pair-wise comparisons were not significant after adjusting for multiple testing. That is, there were no GRS-by-age effects.
DISCUSSION
While there are a few studies that examine how heritabilities of BMI change over childhood and adulthood (6, 11, 26–32), not all of them comprehensively examine changes in heritability across the entire lifespan, from birth through adulthood. Our results show that heritability estimates start out moderately at birth, spike at ~ 9 months, immediately drop for the rest of infancy, and then increase again during childhood. At 11 years of age, the heritabilities begin to decline on through adulthood.
As mentioned previously, some studies have shown that the variants associated with BMI in adulthood also influence BMI in infancy (5) and childhood (1, 21), while others have had mixed results (7–9). For example, the effect of the FTO gene has been shown to be significant from age 4 years to 11 years (1, 28, 33), but show no effect at birth. Hardy et al. (10), also show FTO effects at 11, 15, 20, 26, and 36 years, but not earlier or later.
In our study, we found combined BMI variant effects at 6 and 9 months during infancy, in all of childhood and adolescence, and in all adulthood, except early adulthood. A recent publication using GRS based on 11 SNPs (11) indicates that the influence of obesity risk scores are significant at birth, and reach the 2nd highest effect at 11 years of age, dips slightly at 15 years of age, and then peaks at 20 years of age. In our study, our first peak occurred at 15 years, dipped at 20 years, and had the highest effect occurring in late to mid adulthood. Using the same subset of 11 SNPs (11), we found that the combined 11 variants showed a similar pattern to our results with 32 SNPs, but showed a lesser magnitude of association, with fewer ages at which BMI was influenced by the GRS of 11 SNPs (not shown).
We wanted to know if the pattern of differential genetic influence on BMI at cross-sectional ages were representative of gene-by-age effects, including both the BMI variants and other additive genetic effects. In our formal test of the combined effect of BMI variants on BMI at different ages, we were unable to identify gene-by-age interactions, after adjustments for multiple testing. That is, the effects of BMI variants at different ages were not statistically different from each other. To date, only a few studies have formally tested if the influence of BMI variants changes across age (7, 11, 34, 35), with most of them using mixed modeling methods to evaluate BMI variant(s)-by-age interactions. In a combined examination of BMI from participants in Project Heart Beat! and the Bogalusa study (34), the FTO-by-age interaction was only significant for the Bogalusa participants, while in a British cohort born in 1946 (35), the LIN28B-by-age2 interaction was significant for women only. A GRS comprised of 11 SNPs (11) was also found to have a significant GRS-by-age2 interaction, where the effects were positive from 2–11 years, while it was negative from 11 to 53 years.
With respect to the remaining additive genetic effects, the genetic variances in our study are not significantly different from each other and agree with other studies that have shown the heritabilities of BMI to be stable (31). The examination of the genetic correlations, however, indicates that there are gene-by-age effects, where the putative genes (other than the BMI variants) that influence BMI early in life differ from those later in life. In a study examining differences in BMI across 28 years (36), the estimated genetic correlation for BMI at ages of 20 and 48 years was 0.60 (incomplete pleiotropy). Our 19–40 years genetic correlation of 0.64, which approximates the 28 year lifespan analysed in the aforementioned study, is comparable, while our 11–30 years genetic correlation of 0.78 is slightly higher.
There are some limitations to our study, one of which is our sample size. It is possible that we do not have sufficient power to determine genetic effects, particularly differences between the genetic variances and GRS. Using a mean (across the 32 SNPs) MAF of 0.281 (Table 2), a mean h2 of 0.61 (across all traits, Supplemental Table S1) and a sample size of 590 (approximate mean sample size for all traits), post hoc tests of power indicate that we have 74% power to detect a SNP with an effect size of 1.14% (the mean %GRS across all traits). Regardless, the GRS was significant at some ages, and these significant results did not necessarily occur at ages with the largest sample sizes.
The data used for analysis in our study offers several advantages over previous studies. For instance, the measurements of weight and height used to calculate BMI were not self-reported. This study is also one of a few to report genetic correlations for BMI at different ages across the lifespan from birth throughout childhood and adolescence into adulthood. We also rigorously tested for statistical differences in the association of both BMI variants and other additive genetic effects. Additionally, our study is one of a few to purposely separate out the influences of known BMI variants (4) from remaining additive genetics in order to determine when the influence of genes may differ.
In summary, our study shows that the influence of 32 identified BMI SNP variants on BMI is not significant at birth and in infancy after 9 months, but is significant during childhood, adolescence, and adulthood after age 19 years. Other additive genetic effects are also significant at all ages, from birth, through infancy, childhood, adolescence and mid-adulthood. The small fluctuations in additive genetic effects do not appear to be due to changes in the underlying genetic variances, while the small changes in the effects of the BMI variants on BMI at different ages are also negligible. However, the results of the gene-by-age interaction analyses suggest that different genes influence BMI at different ages.
Supplementary Material
Acknowledgments
This work was supported by NIH grants (R01HD053685, R01HD012252). We thank the participants of the Fels Longitudinal Study for their continued participation and the research staff at the Texas Biomedical Research Institute and the Lifespan Health Research Center.
Footnotes
DISCLOSURES
The authors have no conflicts of interest to declare.
The authors have no competing interests.
Author contributions were are follows. Design and concept of study: ACC, SAC, ML, EWD, WOJ; data analysis and interpretation: ACC, ML, SAC, JWK, VPD, JB; data quality control and cleaning: JEC, TDD, CB, JB, JWK, ACC, SAC, BT, RMS; acquisition of funding: SAC, EWD, BT, RMS. All authors were involved in writing the paper and had final approval of the submitted and published versions.
LITERATURE CITED
- 1.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 3.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Bermejo A, Petry CJ, Diaz M, et al. The association between the FTO gene and fat mass in humans develops by the postnatal age of two weeks. J Clin Endocrinol Metab. 2008;93:1501–5. doi: 10.1210/jc.2007-2343. [DOI] [PubMed] [Google Scholar]
- 6.Elks CE, Loos RJ, Sharp SJ, et al. Genetic markers of adult obesity risk are associated with greater early infancy weight gain and growth. PLoS Med. 2010;7:e1000284. doi: 10.1371/journal.pmed.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mei H, Chen W, Jiang F, et al. Longitudinal replication studies of GWAS risk SNPs influencing body mass index over the course of childhood and adulthood. PLoS One. 2012;7:e31470. doi: 10.1371/journal.pone.0031470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mei H, Chen W, Srinivasan SR, et al. FTO influences on longitudinal BMI over childhood and adulthood and modulation on relationship between birth weight and longitudinal BMI. Hum Genet. 2010;128:589–96. doi: 10.1007/s00439-010-0883-7. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J, Bradfield JP, Li M, et al. The role of obesity-associated loci identified in genome-wide association studies in the determination of pediatric BMI. Obesity. 2009;17:2254–7. doi: 10.1038/oby.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardy R, Wills AK, Wong A, et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet. 2010;19:545–52. doi: 10.1093/hmg/ddp504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elks CE, Loos RJ, Hardy R, et al. Adult obesity susceptibility variants are associated with greater childhood weight gain and a faster tempo of growth: the 1946 British Birth Cohort Study. Am J Clin Nutr. 2012;95:1150–6. doi: 10.3945/ajcn.111.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roche AF. Growth, maturation, and body composition : the Fels Longitudinal Study, 1929–1991. Cambridge University Press; Cambridge; New York, NY: 1992. p. xiii.p. 282. [Google Scholar]
- 13.Lohman TG, Roche AF, Martorell R, editors. Anthropometric Standardization Reference Manual. Human Kinetics Publishers, Inc; Champaign: 1988. [Google Scholar]
- 14.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–37. [PMC free article] [PubMed] [Google Scholar]
- 15.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobel E, Sengul H, Weeks DE. Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum Hered. 2001;52:121–31. doi: 10.1159/000053366. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 20.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.den Hoed M, Ekelund U, Brage S, et al. Genetic susceptibility to obesity and related traits in childhood and adolescence: influence of loci identified by genome-wide association studies. Diabetes. 2010;59:2980–8. doi: 10.2337/db10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 24.Dyke B. PGL Technical Report No 2. Southwest Foundation for Biomedical Research; San Antonio, Texas: 1992. Pedsys: A Pedigree Data Management System. [Google Scholar]
- 25.Towne B, Siervogel RM, Blangero J. Effects of genotype-by-sex interaction on quantitative trait linkage analysis. Genet Epidemiol. 1997;14:1053–8. doi: 10.1002/(SICI)1098-2272(1997)14:6<1053::AID-GEPI82>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 26.Elks CE, den Hoed M, Zhao JH, et al. Variability in the heritability of body mass index: a systematic review and meta-regression. Front Endocrinol (Lausanne) 2012;3:29. doi: 10.3389/fendo.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubois L, Ohm Kyvik K, Girard M, et al. Genetic and environmental contributions to weight, height, and BMI from birth to 19 years of age: an international study of over 12,000 twin pairs. PLoS One. 2012;7:e30153. doi: 10.1371/journal.pone.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haworth CM, Carnell S, Meaburn EL, Davis OS, Plomin R, Wardle J. Increasing heritability of BMI and stronger associations with the FTO gene over childhood. Obesity. 2008;16:2663–8. doi: 10.1038/oby.2008.434. [DOI] [PubMed] [Google Scholar]
- 29.Silventoinen K, Pietilainen KH, Tynelius P, Sorensen TI, Kaprio J, Rasmussen F. Genetic and environmental factors in relative weight from birth to age 18: the Swedish young male twins study. Int J Obes. 2007;31:615–21. doi: 10.1038/sj.ijo.0803577. [DOI] [PubMed] [Google Scholar]
- 30.Coady SA, Jaquish CE, Fabsitz RR, Larson MG, Cupples LA, Myers RH. Genetic variability of adult body mass index: a longitudinal assessment in Framingham families. Obes Res. 2002;10:675–81. doi: 10.1038/oby.2002.91. [DOI] [PubMed] [Google Scholar]
- 31.Haberstick BC, Lessem JM, McQueen MB, et al. Stable genes and changing environments: body mass index across adolescence and young adulthood. Behav Genet. 2010;40:495–504. doi: 10.1007/s10519-009-9327-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornes BK, Zhu G, Martin NG. Sex differences in genetic variation in weight: a longitudinal study of body mass index in adolescent twins. Behav Genet. 2007;37:648–60. doi: 10.1007/s10519-007-9165-0. [DOI] [PubMed] [Google Scholar]
- 33.Jess T, Zimmermann E, Kring SI, et al. Impact on weight dynamics and general growth of the common FTO rs9939609: a longitudinal Danish cohort study. Int J Obes. 2008;32:1388–94. doi: 10.1038/ijo.2008.110. [DOI] [PubMed] [Google Scholar]
- 34.Hallman DM, Friedel VC, Eissa MA, et al. The association of variants in the FTO gene with longitudinal body mass index profiles in non-Hispanic white children and adolescents. Int J Obes. 2012;36:61–8. doi: 10.1038/ijo.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ong KK, Elks CE, Wills AK, et al. Associations between the pubertal timing-related variant in LIN28B and BMI vary across the life course. J Clin Endocrinol Metab. 2011;96:E125–9. doi: 10.1210/jc.2010-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franz CE, Grant MD, Jacobson KC, et al. Genetics of body mass stability and risk for chronic disease: a 28-year longitudinal study. Twin Res Hum Genet. 2007;10:537–45. doi: 10.1375/twin.10.4.537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.