Abstract
Purpose
Lung cancer is a leading cause of cancer deaths and efforts are underway to identify novel therapies to treat these tumors. Diacylglycerol kinase η (DGKη), an enzyme that phosphorylates diacylglycerol to form phosphatidic acid, has been shown to modulate MAPK signaling downstream of EGFR, which is an oncogenic driver in some lung cancers. Since mutations in EGFR and K-Ras are common in lung cancer, we hypothesized that limiting the function of DGKη would attenuate oncogenic properties of lung cancer cells.
Methods
We determined the expression levels of DGKη in a mouse models of mutant EGFR and K-Ras lung cancer and in human lung cancer cell lines with activating mutations in either EGFR or K-Ras. We also tested the effects of shRNA-mediated depletion of DGKη in lung cancer cells and tested if DGKη depletion augmented the effects of afatinib, a new generation EGFR inhibitor.
Results
DGKη was expressed in malignant epithelium from mice with mutant EGFR or K-Ras lung cancer. It was also expressed in human lung cancer cell lines with EGFR or K-Ras mutations. Depleting DGKη in lung cancer cell lines, harboring mutant EGFR, reduced their growth on plastic and in soft agar and also augmented the effects of afatinib, an EGFR inhibitor. DGKη depletion also reduced growth of one of two lung cancer cell lines that harbored mutant K-Ras.
Conclusions
Our data indicate that DGKη is a potential therapeutic target in lung cancers, especially those harboring EGFR mutations. Our findings warrant further studies to examine the effects of limiting its function in vivo.
Keywords: Diacylglycerol kinase, Epidermal growth factor receptor, Lipid signaling, Diacylglycerol, Lung cancer
Introduction
Lung cancer is the leading cause of cancer deaths worldwide. Much of this disease is due to tobacco smoking, but globally up to 15 % of lung cancers in men and almost half of them in women are not attributable to smoking [1]. Often, lung cancers in never smokers harbor specific mutations in genes that are important for tumor maintenance and growth. One gene that is mutated in almost 60 % of lung cancers in never smokers is EGFR [2]. These mutations in EGFR, such as the L858R mutation, increase its kinase activity, leading to a state of oncogene addiction. As such, about 75 % of tumors harboring mutant EGFR respond to small molecule EGFR kinase inhibitors [2], which are superior to chemotherapy as initial treatment for these tumors [3–7]. Unfortunately, most patients eventually develop resistance to them by a variety of mechanisms [8–10]. This has led to additional efforts to find other signaling pathways that are important for tumorigenesis promoted by mutant EGFR.
Diacylglycerol kinases, which phosphorylate the lipid second messenger diacylglycerol to form phosphatidic acid, have been shown to mediate signaling downstream of EGFR. DGKδ, for example, modulates EGFR abundance and degradation by mediating its de-ubiquitination [11]. DGKη also modulates signaling downstream of EGFR by facilitating heterodimerization of B-Raf and C-Raf [12]. Both of these Raf isoforms bind to DGKη, which promotes their recruitment to the plasma membrane where they activate the MEK-ERK signaling cascade. RNAi-mediated knockdown of DGKη in HeLa cells limited proliferation by about 30 % and reduced EGF-mediated phosphorylation of both MEK and ERK [12]. Collectively, these data indicated that DGKη might be a novel cancer target downstream of EGFR or K-Ras, so we set out to determine the effects of limiting its function in lung cancer cell lines that harbor activating mutations in EGFR or K-Ras.
Materials and methods
Cell lines, expression plasmids, cell culture, and transfection
All cell lines (H441, H460, H1650, and H1975) were from American Type Culture Collection and propagated in RPMI 1640 (Invitrogen) with 10 % FBS and antibiotics. The type of lung cancers from which these cells were derived as well as their EGFR and K-Ras mutation status are indicated in Fig. 1a. DGKη RNAi was performed using Oligofectamine (Invitrogen) and the following siRNA duplexes: 5′-GGAUCUAGAUUCCGUAGAUTT-3′ and 5′-AUCUACGGAAUCUAGAUCCGGTT-3′. Scrambled siRNA duplexes were used as controls.
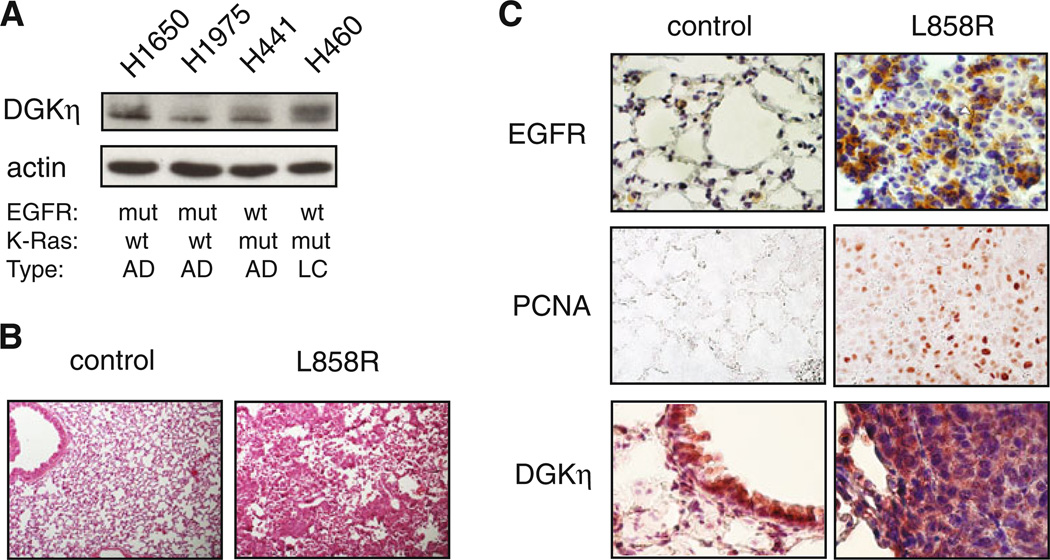
Fig. 1.
Expression of DGKη in lung cancer cell lines and in a mouse model of mutant EGFR lung cancer. a Lung cancer cell lines were grown to confluence, harvested, and then the levels of DGKη and actin were detected in the cell lysates by Western blotting. The mutation status of EGFR or K-Ras in each cell line is shown below the blot (mut mutation, wt wild-type). The type of lung carcinoma from which each line was obtained is also indicated (AD adenocarcinoma, LC large cell carcinoma). b Lungs from rtTA(−) × EGFR-L858R (control) or rtTA(+) × EGFR-L858R (L858R) mice treated with doxycycline for 8 weeks were harvested, fixed, sectioned, and stained with eosin. c Lung sections from control or L858R mice were immunostained to detect EGFR, PCNA, or DGKη
Generation of stable DGKη knockdown cell lines and proliferation, migration, and soft agar assays
To generate stable DGKη knockdown H1650 and H1975 cell lines, Dharmacon SMARTvector 2.0 lentiviral particles (#SK-006716-02) were used according to the instructions. Non-targeting particles (#S-005000-01) were used for a control cell line. Polyclonal stable cell lines were isolated under puromycin (2 µg/mL) selection. Cell proliferation was assessed by plating 10,000 cells in a 10-cm diameter plate, growing them in 1 % FBS with antibiotics for 5–7 days, and then counting the number of cells. Growth in soft agar was performed as described [13].
Reverse transcription polymerase chain reaction
Total RNA was prepared from H1650 stable cell lines using Trizol reagent (Invitrogen) and reverse transcribed as described [14]. RT-PCR using 10 ng of total cDNA was performed with the following primers: DGKη: 5′-GG ACCTCCAGAAGCATCTG-3′ (forward) and 5′-AACGTC ATCCCCAAGCTGC-3′; actin: 5′-AGGCACCAGGGCG TGAT-3′ (forward) and 5′-TCGTCCCAGTTGGTGACG AT-3′.
Western blotting
Western blotting was performed according to instructions provided by the suppliers. Anti-EGFR (#2232) antibodies were from Cell Signaling Technologies. Anti-actin (#691001) was from MP Biomedicals. Anti-DGKη (#13873-1-AP) was from Proteintech Group. Anti-PCNA (#2714-1) was from Epitomics.
Mice
All mouse experiments were reviewed and approved by the University of Utah Institutional Review Board. Transgenic mice expressing the reverse tetracycline-controlled transactivator (rtTA) protein under the control of the rat clara cell secretory protein (CCSP) gene promoter were obtained from The Jackson Laboratory (stock#006232). L858REGFR transgenic mice were obtained from the NCI mouse repository (strain#01XEA). Bi-transgenic CCSP-rtTA x L858R-EGFR or control L858R-EGFR mice were given doxycycline (1gm/L) in their water for 8 weeks. Mouse lungs harboring mutant K-Ras (LSL-KrasG12D/+) treated with or without adenoviral Cre as described [15] and harvested 12 weeks later were provided by Trudy Oliver (University of Utah).
Histological analysis
Lungs harvested from control or L858R-EGFR mice were fixed in 10 % neutral buffered formalin for 30–48 h, paraffin embedded, sectioned (5 lm), and stained with hematoxylin and eosin according to standard protocol. Immunostaining was performed using the ABC reagent (Vector Laboratories) and anti-EGFR (#1902-1) or anti-PCNA (#2714-1) from Epitomics or anti-DGKη (#13873-1-AP) from Proteintech Group according to instructions provided by the suppliers.
Results
DGKη is expressed in human lung cancer cell lines and a mouse model of mutant EGFR lung cancer
To determine if DGKη was expressed in human lung cancers, we surveyed a panel of human non-small cell lung cancer cell lines that had activating mutations in either EGFR (H1650 and H1975) or K-Ras (H441 or H460). We found that DGKη was expressed in all of them and that the levels of DGKη did not correlate with EGFR or K-Ras mutation status (Fig. 1a). This was consistent with prior data demonstrating that the levels of DGKη mRNA were not affected by activation of EGFR [16]. To further assess the expression levels of DGKη in mutant EGFR lung tumors, we obtained bi-transgenic mice in which doxycycline induces expression of an L858R-EGFR transgene in type II lung pneumocytes [17]. In preliminary experiments, we found that doxycycline induced significant tumor load after 8 weeks of treatment (Fig. 1b). As expected, EGFR was highly expressed in the tumors, but not in lungs of mice that were treated with doxycycline but did not carry the rtTA transgene necessary to induce expression of the L858R-EGFR transgene (Fig. 1c). Proliferating cell nuclear antigen (PCNA), a measure of cell proliferation, was also highly expressed in tumor-bearing lungs but was not detected in normal lungs (Fig. 1c). We found that DGKη was expressed in bronchial epithelium of control mice (Fig. 1c) and was highly expressed in malignant epithelium of L858R-EGFR mice (Fig. 1c). Collectively, these data indicate that DGKη is expressed in human lung cancer cell lines as well as in a mouse model of EGFR mutant lung cancer. Together with its known role in modulating EGFR signaling, these data suggest that DGKη might promote tumorigenesis in EGFR or K-Ras mutant lung cancer.
Depletion of DGKη reduces oncogenic properties of lung cancer cells harboring mutant EGFR
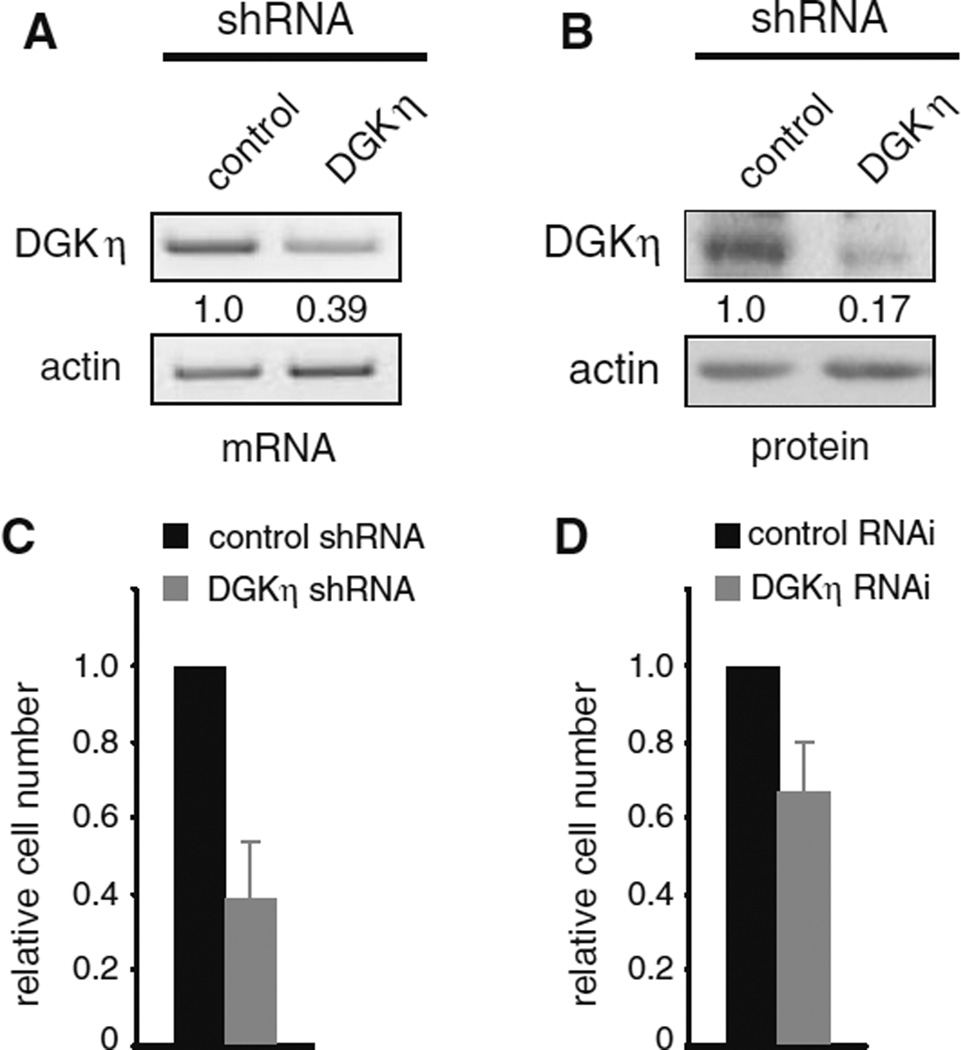
To further investigate the potentially oncogenic role of DGKη, we knocked down DGKη in human H1650 lung cancer cells. These cells express a gain-of-function mutation (delE746-A750) in the gene encoding EGFR. Using lentivirus we generated a polyclonal H1650 cell line stably expressing shRNA targeting DGKη that resulted in a ~60 % reduction in DGKη mRNA (Fig. 2a) and an ~80 % reduction in DGKη protein (Fig. 2b). To evaluate the effects of DGKη depletion on cell proliferation, we used cell counting assays and found 5 days after plating the cells in 1 % serum significantly reduced cell numbers of DGKη-deficient cells compared to control cells (Fig. 2c). Transient depletion of DGKη using siRNA oligonucleotides targeting another region of DGKη in H1650 cells also led to a *30 % reduction in cell number 3–4 days after performing RNAi (Fig. 2d). Together with prior results showing reduced cell proliferation of DGKη-deficient HeLa cells [12], these data indicate that DGKη depletion leads to reduced cell number in low serum conditions.
Fig. 2.
DGKη depletion reduces cell proliferation. H1650 cells were used to generate control or DGKη shRNA polyclonal cell lines. The cells were grown to confluence, harvested, and then DGKη and actin were detected using semi-quantitative RT-PCR (a) or Western blotting (b). The relative levels of DGKη normalized to actin are shown below the DGKη images. c Equal numbers of control or DGKη shRNA H1650 cells were grown for 5 days in medium with 1 % serum and then counted. The difference in cell numbers between control and DGKη shRNA cells was statistically significant (n = 3; p < 0.02). d Control or siRNA oligonucleotides were used for transient depletion of DGKη in H1650 cells grown in 1 % serum. Cell numbers were determined 3–4 days later. The difference in cell number between control and DGKη RNAi cells was statistically significant (n = 3; p < 0.05)
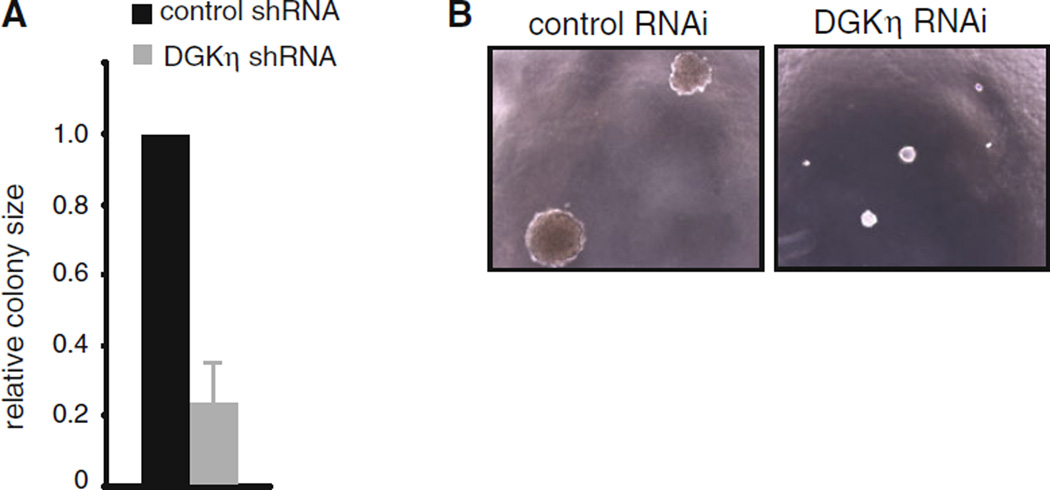
To assess the effects of DGKη deficiency on additional oncogenic properties, we tested the growth of the stable H1650 cells in conditions of anchorage independence by growing them in soft agar and found significantly reduced colony volumes in DGKη-deficient cells compared to control H1650 cells (Fig. 3a, b). Together with the above observations, these data indicated that depleting DGKη reduces the oncogenic potential of H1650 cells and led us to test the effects of its depletion in other EGFR mutant cell lines and to assess the potential therapeutic value of disrupting DGKη function in combination with an EGFR inhibitor.
Fig. 3.
DGKη depletion reduces growth in soft agar. a Control or DGKη shRNA H1650 cells were grown in soft agar for 14 days and then colony size was evaluated. The difference in colony size between control and DGKη shRNA cells was statistically significant (n > 50 colonies; p < 0.02). b Representative photos of colonies evaluated in panel a
Depletion of DGKη augments the inhibitory effects of EGFR inhibitors
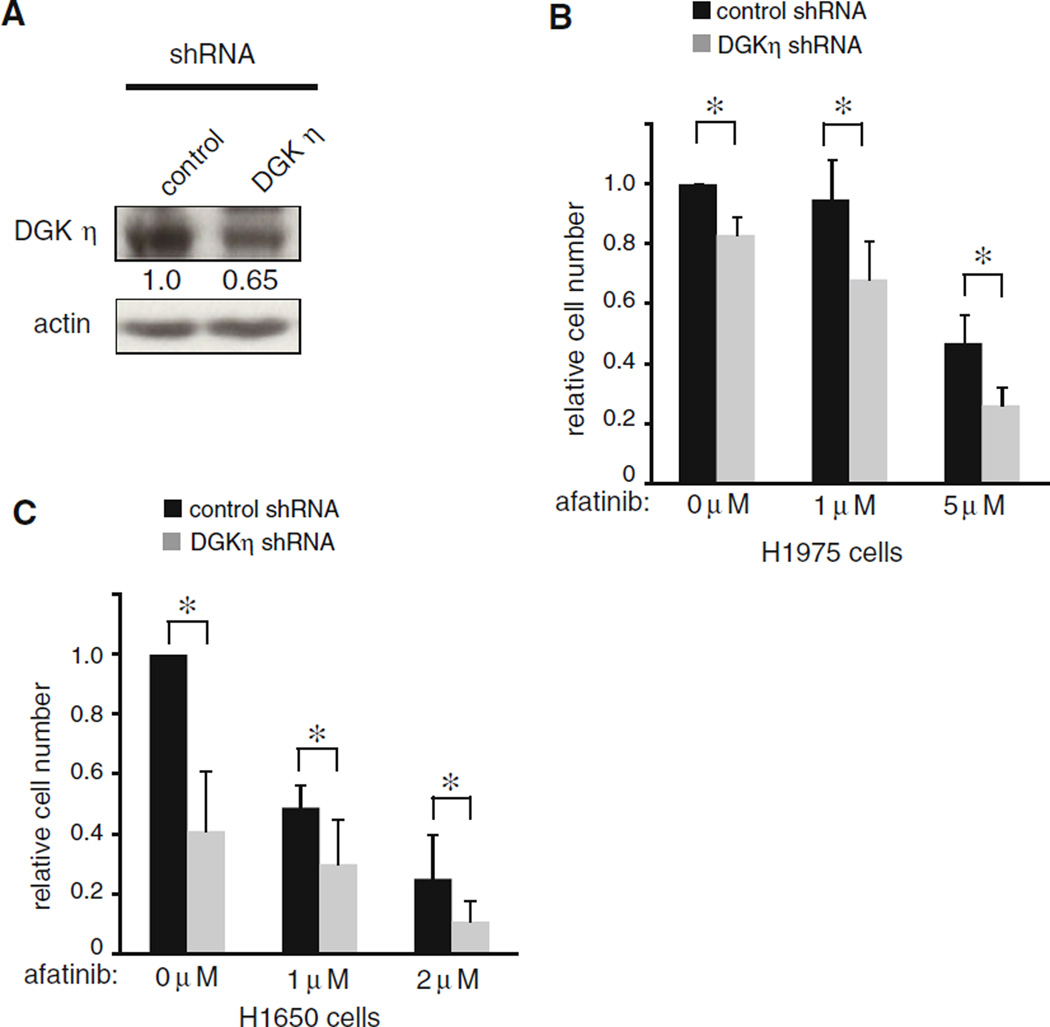
EGFR mutations in lung cancer can be targeted by several small molecule inhibitors that are either in clinical use or in late stage clinical trials. One of these inhibitors, afatinib (BIBW-2992), showed promising results in EGFR mutant lung cancer [18]. Since DGKη depletion reduced growth of cells harboring EGFR mutations, we tested the effects of combining afatinib with DGKη depletion. For these experiments, we generated a second cell line to test using H1975 cells, which harbor another activating EGFR mutation (L858R), to make stable DGKη knockdown cells (Fig. 4a). In this cell line, we were only able to reduce DGKη protein by about 35 %, which was less than the *80 % reduction that we achieved in the H1650 cells.
Fig. 4.
Depletion of DGKη augments the afatinib-induced growth inhibition. a H1975 cells were used to generate control or DGKη shRNA polyclonal cell lines. The cells were grown to confluence, harvested, and then DGKη and actin were detected in the cell lysates by Western blotting. The relative expression levels of DGKη normalized to actin are shown below the DGKη blot. b Control or DGKη shRNA H1975 cells were grown for 5 days in medium with 1 % serum in the absence or presence of escalating concentrations of afatinib and then counted. The asterisk indicates that the changes were statistically significant (n = 3; p < 0.05). c Control or DGKη shRNA H1650 cells were grown 5 days in medium with 1 % serum in the absence or presence of escalating concentrations of afatinib and then counted. The asterisk indicates that the changes were statistically significant (n = 3; p < 0.05)
We first tested growth of the control cell lines in the presence or absence of afatinib and in preliminary experiments we found that afatinib, alone, reduced growth of control H1650 cells more substantially than it reduced growth of control H1975 cells. This finding is consistent with the known T790 M mutation that is also harbored by H1975 cells, which reduces the affinity of EGFR for small molecule inhibitors. We then compared growth of control and DGKη-depleted cells and found in the absence of afatinib that DGKη-deficient H1975 cells proliferated more slowly than control H1975 cells (Fig. 4b), but the reduction in proliferation caused by DGKη depletion was not as substantial as in the H1650 cells. The reduced growth inhibition in the H1975 cells might reflect the limited knockdown of DGKη that we were able to achieve compared to the H1650 cells. In both cell lines, we found that DGKη depletion combined with afatinib led to more pronounced reductions in cell counts compared to afatinib alone (Fig. 4b, c). All differences between control and DGKη knockdown cells in the presence or absence of afatinib were statistically significant. Together, these data demonstrated that depleting DGKη reduced growth of another EGFR mutant cell line and also suggested that inhibitors of DGKη might be useful as adjuvant therapy combined with EGFR inhibitors in lung cancers harboring EGFR mutations.
DGKη depletion reduces growth of some lung cancer cell lines harboring activating mutations in K-Ras
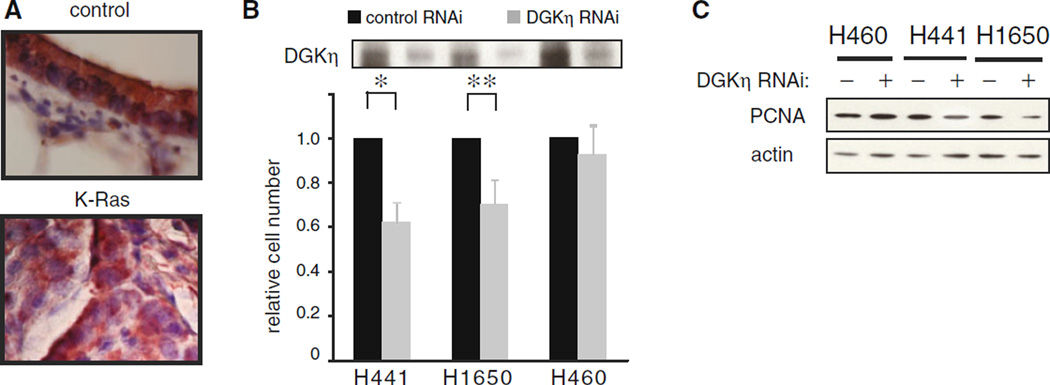
The reduced growth of DGKη deficient cell lines was consistent with the known effects of reducing expression of DGKη in HeLa cells [12], but the reduction of cell number was significantly greater in H1650 cells compared to the approximately 30 % reduction that was observed in HeLa cells [12]. Since HeLa cells express wild-type EGFR, the more pronounced reduction in the H1650 cells compared to HeLa cells was possibly due to the activating mutation in EGFR that the H1650 cells harbor. To further investigate the effects of DGKη deficiency on cell growth and determine if they are more pronounced in EGFR mutant cell lines, we set out to test the effects of depleting DGKη in lung cancer cell lines harboring activating mutations in K-Ras, which is mutated in about 16 % of lung cancers according to the catalog of somatic mutations in cancer (COSMIC) database. We first examined the expression levels of DGKη in a mouse model of mutant K-Ras lung cancer [15]. Similar to our observations in mutant EGFR lung tumors in mice, we found high levels of DGKη in mutant K-Ras lung tumors (Fig. 5a), indicating that DGKη might also be important in K-Ras-driven lung tumors.
Fig. 5.
DGKη is expressed in K-Ras mutant lung tumors and its depletion reduces proliferation of some K-Ras mutant cell lines. a DGKη was detected by immunostaining in lungs from control (sham-treated) or LSL-KrasG12D/+ mice. b Control or DGKη depleted lung cancer cell lines grown in 1 % serum were counted 3–4 days after RNAi. The asterisk indicates p < 0.04 (n = 3) and the double asterisk indicates p < 0.02 (n = 3). DGKη was detected by immunoblotting in each cell line and shown above the graph. c Levels of PCNA and actin in control or DGKη depleted cells grown in 1 % serum and harvested 3 days after RNAi were detected by immunoblotting
To determine if DGKη contributed to cell growth in lung tumor cell lines harboring activating mutations in K-Ras, we used transient siRNA-mediated depletion of DGKη and compared H460 and H441 cell lines, both of which harbor activated K-Ras, to H1650 cells. We found 3–4 days after performing RNAi and growing the cells in 1 % serum that depleting DGKη in H441 cells reduced cell number to the same extent as H1650 cells (Fig. 5b). But DGKη depletion caused only a small, non-significant reduction in H460 cell counts (Fig. 5b). These data indicated that the effects of DGKη depletion are not limited to cells harboring EGFR mutations.
Under the conditions of these growth assays, we could not measure consistent differences in the apoptosis markers cleaved PARP or caspase-3 in control or DGKη-depleted cells (data not shown), indicating that the reduced DGKη-deficient cell numbers that we observed were likely due to changes in the levels of proliferation. To assess the level of proliferation, we assayed the abundance of PCNA and found reduced levels in DGKη-deficient H441 and H1650 cells, but not in H460 cells (Fig. 5c). This lack of effect of DGKη depletion on PCNA levels in H460 cells was consistent with the lack of change in cell number caused by DGKη depletion. Together, these data indicate that DGKη has a prominent role in proliferation of EGFR mutant cell lines and can reduce proliferation of some cells harboring mutant K-Ras.
Discussion
Our data suggest that DGKη modulates oncogenic properties in lung cancer cells harboring EGFR mutations and in some lung cancer cells harboring K-Ras mutations. These findings are consistent with prior studies showing that DGKη depletion also reduced HeLa cell growth [12], indicating that DGKη modulates oncogenic properties in other types of cancer cell lines that do not harbor EGFR or K-Ras mutations. The broad effects of DGKη depletion on oncogenic behavior are not surprising given its role in facilitating Raf heterodimerization and membrane recruitment downstream of EGFR [12]. Since functional activation of EGFR is found in a majority of human epithelial cancers [19] and activating K-Ras mutations are common as well, our data indicate that DGKη is a novel cancer target and that inhibiting its function might affect a broad array of cancers.
Given the role of EGFR in lung cancer [2, 19], we focused our efforts on understanding the effects of depleting DGKη in lung cancer cell lines. We found evidence that DGKη promotes oncogenic properties of lung cancer cells. Consistent with this conclusion, DGKη was recently identified as part of a seventeen gene microarray signature that predicted likelihood of death from non-small cell lung cancer (NSCLC) [20]. Based on the cohort used in that study, low levels of DGKη mRNA correlated with longer survival in patients with NSCLC (Ref. [20] and S. Philipsen personal communication). The favorability of reduced DGKη expression in that study is in concert with our findings that DGKη depletion reduced oncogenic properties of lung cancer cells. Given its possible role in human lung cancer, we additionally tested growth of our stable control or DGKη-deficient H1650 or H1975 cell lines as xenografts in nude mice, but we could not maintain knockdown of DGKη over the 3 weeks of the experiments. Thus, it will take a more robust knockdown system or genetic models to further study the anti-oncogenic properties of depleting DGKη in vivo.
Targeted therapies directed against EGFR have been developed and used clinically during the past few decades. They work best when used to treat tumors “addicted” to EGFR signaling. Unfortunately, most tumors that initially respond to the inhibitors eventually develop resistance by a variety of mechanisms [8–10]. This acquired resistance to EGFR inhibitors and the inherent EGFR inhibitor resistance of some tumors has spawned efforts to find other ways of blocking the EGFR pathway. Our data indicate that inhibiting the function of DGKη in conjunction with an EGFR inhibitor might be a viable approach to treat tumors. Indeed, we discovered that when combined with afatinib, DGKη depletion further reduced growth of both H1650 and H1975 cell lines. Collectively, our data suggest that inhibitors of DGKη might have clinical efficacy either when used alone or when combined with EGFR inhibitors.
DGKη is a type II DGK that is structurally similar to DGKδ. We discovered that DGKδ also modulates EGFR signaling [11, 21] and that its depletion significantly reduced proliferation of H1650 cells in both two and three dimensions (M.K.T. manuscript in preparation). As such, targeting both DGKs δ and η might have additive or synergistic effects and could be a very effective therapeutic approach to limit EGFR signaling. Type II DGKs are unlike other DGK isoforms because they have a split catalytic domain [22]. This structural difference suggests that it might be possible to design inhibitors that specifically target both DGKs δ and g. This possibility and the antineoplastic effects of depleting DGKη warrant further studies to examine its role in human cancer, to study the effects of depleting DGKη in animal models of lung cancer and other cancers, and to develop strategies to inhibit its tumorigenic functions.
Acknowledgments
This work was supported by the Huntsman Cancer Foundation, the R. Harold Burton Foundation, the National Institutes of Health Grants R01-CA95463 (to M.K.T.) and by P30-CA042014.
Abbreviations
- EGFR
Epidermal growth factor receptor
- DGK
Diacylglycerol kinase
- DMEM
Dulbecco’s modified Eagle’s medium
- rtTA
Reverse tetracycline-controlled transactivator
- CCSP
Clara cell secretory protein
- PCNA
Proliferating cell nuclear antigen
- NSCLC
Non-small cell lung cancer
Footnotes
Conflict of interest The authors declare no conflicts of interest.
Contributor Information
T. Nakano, Huntsman Cancer Institute, University of Utah, 2000 East Circle of Hope, Salt Lake City, UT 84112-5550, USA
A. Iravani, Huntsman Cancer Institute, University of Utah, 2000 East Circle of Hope, Salt Lake City, UT 84112-5550, USA Department of Internal Medicine, University of Utah, Salt Lake City, UT 84112, USA.
M. Kim, Huntsman Cancer Institute, University of Utah, 2000 East Circle of Hope, Salt Lake City, UT 84112-5550, USA Department of Oncological Sciences, University of Utah, Salt Lake City, UT 84112, USA.
Y. Hozumi, Huntsman Cancer Institute, University of Utah, 2000 East Circle of Hope, Salt Lake City, UT 84112-5550, USA
M. Lohse, Huntsman Cancer Institute, University of Utah, 2000 East Circle of Hope, Salt Lake City, UT 84112-5550, USA
E. Reichert, Huntsman Cancer Institute, University of Utah, 2000 East Circle of Hope, Salt Lake City, UT 84112-5550, USA
T. M. Crotty, Huntsman Cancer Institute, University of Utah, 2000 East Circle of Hope, Salt Lake City, UT 84112-5550, USA
D. M. Stafforini, Huntsman Cancer Institute, University of Utah, 2000 East Circle of Hope, Salt Lake City, UT 84112-5550, USA Department of Internal Medicine, University of Utah, Salt Lake City, UT 84112, USA.
M. K. Topham, Email: matt.topham@hci.utah.edu, Huntsman Cancer Institute, University of Utah, 2000 East Circle of Hope, Salt Lake City, UT 84112-5550, USA; Department of Internal Medicine, University of Utah, Salt Lake City, UT 84112, USA; Department of Oncological Sciences, University of Utah, Salt Lake City, UT 84112, USA.
References
- 1.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. New Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 4.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 5.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. New Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 6.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 7.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutationpositive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 8.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. New Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 10.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai J, Crotty TM, Reichert E, Carraway KL, Stafforini DM, Topham MK. Diacylglycerol kinase delta and protein kinase C(alpha) modulate epidermal growth factor receptor abundance and degradation through ubiquitin-specific protease 8. J Biol Chem. 2010;285:6952–6959. doi: 10.1074/jbc.M109.055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasuda S, Kai M, Imai S, Takeishi K, Taketomi A, Toyota M, et al. Diacylglycerol kinase eta augments C-Raf activity and B-Raf/C-Raf heterodimerization. J Biol Chem. 2009;284:29559–29570. doi: 10.1074/jbc.M109.043604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Salihi MA, Ulmer SC, Doan T, Nelson CD, Crotty T, Prescott SM, et al. Cyclooxygenase-2 transactivates the epidermal growth factor receptor through specific E-prostanoid receptors and tumor necrosis factor-alpha converting enzyme. Cell Signal. 2007;19:1956–1963. doi: 10.1016/j.cellsig.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L, Traer E, McIntyre TM, Zimmerman GA, Prescott SM. The cloning and characterization of a novel human diacylglycerol kinase DGKi. J Biol Chem. 1998;273:32746–32752. doi: 10.1074/jbc.273.49.32746. [DOI] [PubMed] [Google Scholar]
- 15.Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24:837–852. doi: 10.1101/gad.1897010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakami T, Sakani F, Imai S, Houkin K, Kanoh H. Identification and characterization of two splice variants of diacylglycerol kinase η. J Biol Chem. 2003;278:34364–34372. doi: 10.1074/jbc.M301542200. [DOI] [PubMed] [Google Scholar]
- 17.Politi K, Zakowski MF, Fan P, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang JC, Shih JY, Su WC, Hsia TC, Tsai CM, Ou SH, et al. Afatinib for patients with lung adenocarcinoma and epidermal growth factor receptor mutations (LUX-Lung 2): a phase 2 trial. Lancet Oncol. 2012;13:539–548. doi: 10.1016/S1470-2045(12)70086-4. [DOI] [PubMed] [Google Scholar]
- 19.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. New Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 20.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crotty T, Cai J, Sakane F, Taketomi A, Prescott SM, Topham MK. Diacylglycerol kinase delta regulates protein kinase C and epidermal growth factor receptor signaling. Proc Natl Acad Sci USA. 2006;103:15485–15490. doi: 10.1073/pnas.0604104103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shulga YV, Topham MK, Epand RM. Regulation and functions of diacylglycerol kinases. Chem Rev. 2011;111:6186–6208. doi: 10.1021/cr1004106. [DOI] [PubMed] [Google Scholar]