Abstract
Hormone-sensitive prostate cancer typically progresses to castration resistant prostate cancer (CRPC) after the androgen deprivation therapy. We investigated the impact of microRNAs (miRs) in the transition of prostate cancer to CRPC. MiR-221/-222 was highly expressed in bone metastatic CRPC tumor specimens. We previously demonstrated that transient overexpression of miR-221/-222 in LNCaP promoted the development of the CRPC phenotype. In current study, we show that stably overexpressing miR-221 confers androgen independent (AI) cell growth in LNCaP by rescuing LNCaP cells from growth arrest at G1 phase due to the lack of androgen. Overexpressing of miR-221 in LNCaP reduced the transcription of a subgroup of androgen-responsive genes without affecting the androgen receptor (AR) or AR-androgen integrity. By performing systematic biochemical and bioinformatical analyses, we identified two miR-221 targets, HECTD2 and RAB1A, which could mediate the development of CRPC phenotype in multiple prostate cancer cell lines. Downregulation of HECTD2 significantly affected the androgen-induced and AR-mediated transcription, and downregulation of HECTD2 or RAB1A enhances AI cell growth. As a result of the elevated expression of miR-221, expression of many cell cycle genes was altered and pathways promoting epithelial to mesenchymal transition/tumor metastasis were activated. We hypothesize that a major biological consequence of upregulation of miR-221 is reprogramming of AR signaling, which in turn may mediate the transition to the CRPC phenotype.
Keywords: microRNA, miR-221, castration resistant prostate cancer, androgen receptor signaling
INTRODUCTION
Prostate cancer (CaP) is the most frequently diagnosed non-skin cancer in American men.1 Androgen receptor (AR) signaling has a critical role in the development and differentiation of normal prostate and CaP cells.2 Androgen deprivation therapy represents the most effective therapy for patients with hormone-sensitive CaP. However, patients invariably relapse with more aggressive castration resistant prostate cancer (CRPC).3 The mechanisms by which androgen signaling is modified in CRPC include AR mutations and amplification, intratumoral production of androgens, altered expressions of coactivators/corepressors, AR splice variants, AR reprogramming, ligand independent/AR-dependent mechanisms, as well as AR independent mechanisms.4–6 We found that microRNAs (miRs) have a crucial role in regulating AR function during the CRPC development.7,8
MiRs are groups of naturally occurring single-stranded small non-coding RNAs of 19–25 nt and negatively regulate gene expression.9 The mature miR is associated with a protein complex called the miRISC (miR-induced silencing complex), which typically contains an argonaute protein (AGO), and carries out the silencing of the miR target genes.10 Altered levels of miRs or dysfunction of miR pathways affect divergent cellular processes, thus leading to human diseases including cancer.11
Several reports have implicated specific miR expression patterns associated with tumorigenesis and the tumor grade in human CaP.12,13 We have been investigating molecular determinants involved in the transformation of hormone-sensitive CaP to CRPC and identified miR signatures in human CRPC.7,8 By undertaking a comprehensive miR expression profiling analysis in the androgen-dependent LNCaP cell line and the LNCaP-derived androgen independent (AI) LNCaP-Abl cell line, we identified a set of seven miRs that were differentially expressed.7 Among these miRs, the expression of miR-221/-222 was the most dramatically upregulated in LNCaP-Abl. Further functional studies demonstrated that transiently increased miR-221/-222 expression in LNCaP reduced the dihydrotestosterone (DHT)-induced cell growth and AR-mediated PSA expression.7 The miR-221/-222 expression was also significantly upregulated in human bone metastatic CRPC tumor specimens.8 We hypothesized that miR-221/-222 expression may have a crucial role in influencing AR signaling machinery and reprogramming AR-mediated transcription, as a result contributing to AI phenotype.
Upregulation of miR-221/-222 has been observed in a variety of cancer cells14,15 and appeared to be associated with the TRAIL-resistant phenotype in human non-small-cell lung cancer and with tamoxifen or fulvestrant resistance in breast cancer.16–19 Which miR-221/-222 targets are involved in the development of therapy resistance in different disease states remain unknown. Currently, p27/kip1, p57/kip2, c-kit, Bim, ERα, PTEN, TIMP3, PUMA and etc. have been reported to be miR-221/-222 targets that are involved in cancer progression or development.14–16,18–21 We have determined in cell model systems that these known miR-221/-222 targets do not appear to be involved in the CRPC phenotype (Supplementary Figure S1).
In this study, we applied biochemical and bioinformatical approaches to systematically dissect miR target networks in LNCaP and LNCaP-Abl. We identified HECTD2 and RAB1A as miR-221/-222 targets involved in modifying AR signaling and stimulating AI growth during CRPC development.
RESULTS
MiR-221 overexpression enhances AI cell growth
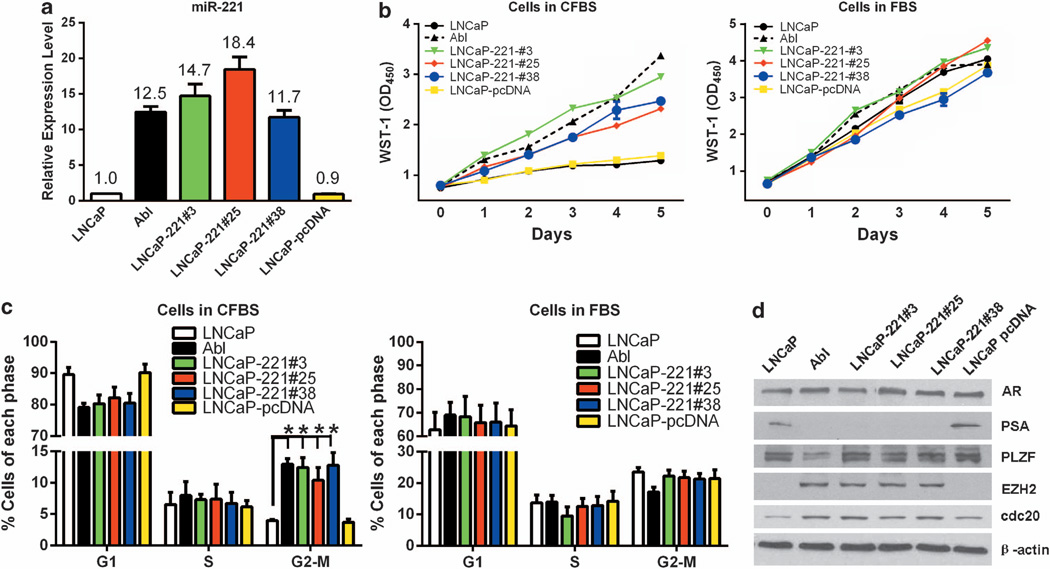
Using transient transfection assays, we previously demonstrated that overexpression of miR-221/-222 in LNCaP promoted the development of CRPC phenotype.7 We then generated LNCaP cells stably overexpressing miR-221 using a pcDNA3.1-miR-221 overexpression plasmid. Transfected cells were maintained and selected with Geneticin in a regular medium containing the serum level of androgen. Twenty-five out of 47 isolated clones had ≥10 fold higher miR-221 expression than that in the parental LNCaP. We chose three clones, designated as LNCaP-221#3, -#25 and -#38, which express miR-221 at a level comparable to that in LNCaP-Abl, for further study (Figure 1a).
Figure 1. MiR-221 expression promotes androgen independent growth.
(a) Quantitative real-time–PCR confirmation of miR-221 expression in LNCaP, LNCaP-Abl, LNCaP-miR-221 overexpressing cell lines (#3, #25 and #38), and LNCaP-pcDNA (LNCaP transfected with the vector control). The expression levels were relative to that in LNCaP, which was arbitrarily set as 1.0. (b) Growth curve of LNCaP (black solid lines), LNCaP-Abl (broken black solid lines) and LNCaP-miR-221 stable-expressing cell lines (#3, #25 and #38; green, red and blue lines, respectively), and LNCaP-pcDNA (yellow lines) in medium with charcoal-treated serum (CFBS, left) and regular medium (fetal bovine serum, right) over 5-day time course. Each point represents the mean of three independent values with s.d. (c) Cell cycle analysis by flow cytometry. LNCaP, LNCaP-Abl, LNCaP-miR-221 overexpressers and LNCaP-pcDNA cell lines were kept in charcoal-treated medium (CFBS, left) for 4 days before analysis or in regular medium (fetal bovine serum, right). Error bars indicate ± s.d., *P < 0.05 by student’s t-test. (d) Western blot analysis of AR, PSA, PLZF, EZH2 and cdc20 in LNCaP, LNCaP-Abl, LNCaP-miR-221 overexpressing cell lines (#3, #25 and #38). β-actin level was used as a loading control.
We first determined whether these miR-221 overexpressing cell lines grew well without androgen, and thus manifest growth characteristics of CRPC. Similar to LNCaP-Abl, LNCaP-221#3, #25 and #38 were able to grow in an androgen-depleted medium (Figure 1b, the left panel), while the parental LNCaP and LNCaP transfected with the vector control (LNCaP-pcDNA) grew poorly in the absence of androgen. All cell lines exhibited similar growth efficiencies in media containing the regular serum (Figure 1b, the right panel). The LNCaP-miR-221 overexpressing cell lines could be maintained and passaged in an androgen-depleted medium for 25–30 days. However, prolonged passage in the absence of androgen eventually led to death.
One model that could explain CRPC growth is that CRPC cells activate pathways bypassing the cell cycle arrest caused by the androgen deprivation.22 Therefore, we compared the ratios of cell populations at different cell cycle phases by flow cytometry, in the presence or absence of androgen in different cell lines. In the regular medium, the ratio of G1:S:G2-M in LNCaP was 6:1:2 (Figure 1c, the right panel); while in the androgen-depleted medium, most of LNCaP cells (>90%) were arrested at G1 phase. LNCaP-Abl, which is regularly maintained in the absence of androgen, exhibited 8:1:2 for G1:S:G2-M (Figure 1c, the left panel). LNCaP-miR-221 overexpressing cell lines are regularly maintained in the presence of androgen (regular medium). However, depleting androgen did not significantly change the distribution of LNCaP-miR-221 overexpressing cells at different cell cycle stages (G1:S:G2-M = 7–8:1:2 before and after removing androgen, Figure 1c). This result indicated that the elevated miR-221 expression rescued LNCaP cells from growth arrest at G1 phase due to the lack of androgen, as a result, promoting AI growth.
We had previously shown that transient upregulation of miR-221/-222 expression in LNCaP reduced AR-mediated PSA expression.7 To further explore the impact of miR-221 overexpression on AR-mediated transcription, we examined the expression levels of AR and several well-characterized androgen-responsive genes, PSA, PLZF,23 cdc2022 and EZH224 (Figure 1d). AR protein expression level was not significantly influenced by stable miR-221 overexpression. However, PSA expression was nearly abolished in the three LNCaP-miR-221 overexpressing cell lines even in the presence of androgen, similar to that seen in LNCaP-Abl. We also observed a elevation of EZH2 and cdc20 in miR-221 overexpressing cell lines comparable to the level in LNCaP-Abl. Surprisingly, expression of the AR-controlled PLZF in LNCaP-miR-221 over-expressing cell lines was not affected by the overexpression of miR-221, while the expression of PLZF was dramatically downregulated in LNCaP-Abl compared with LNCaP. The expression pattern of androgen-responsive genes indicated that LNCaP-miR-221 overexpressing cell lines are similar to but not exactly the same as LNCaP-Abl.
Upregulation of miR-221 altered the expression pattern of many cell cycle control genes
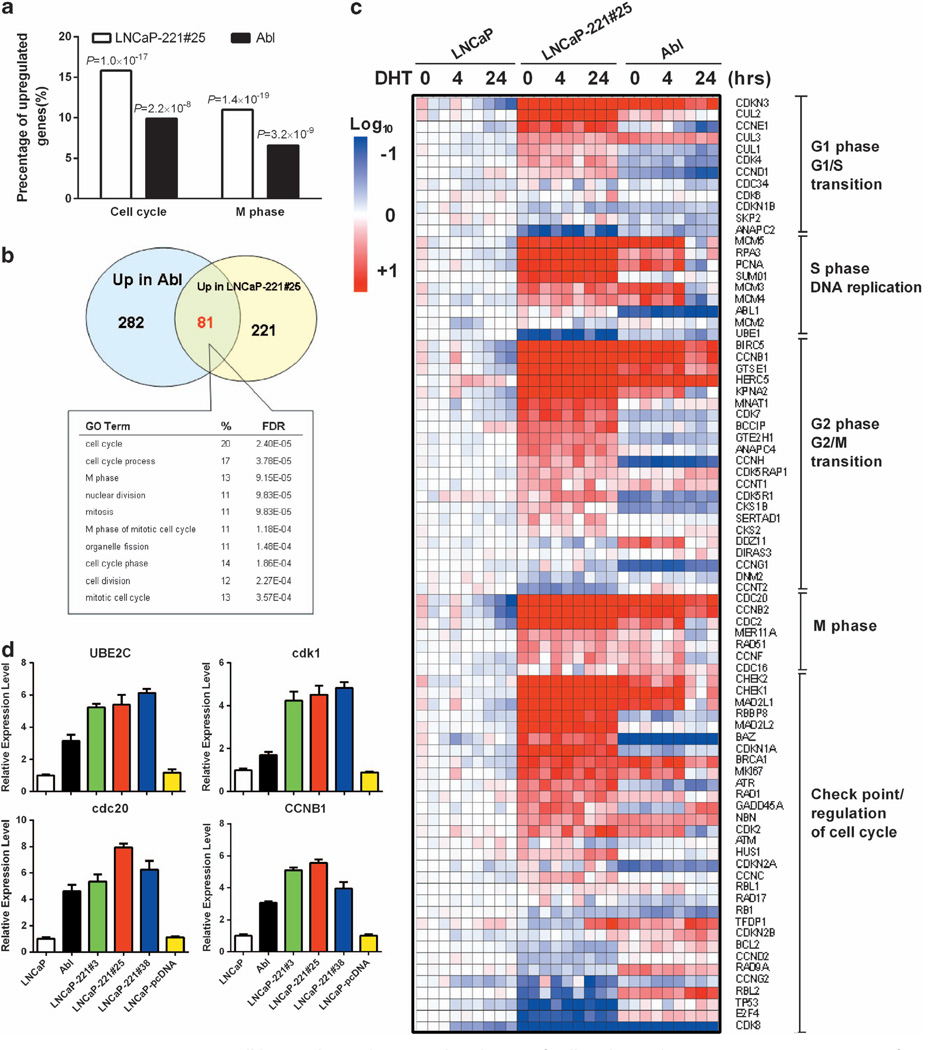
To thoroughly understand the impact of miR-221 overexpression on global gene expression, we compared the whole-genome gene expression profiles of LNCaP, and LNCaP-miR-221 overexpressing cell lines (LNCaP-Abl and LNCaP-221 #25), followed by Gene Ontology (GO) analyses. In all 302 upregulated genes (increased fold >2.0, P < 0.001) in LNCaP-221#25 compared with LNCaP, ‘cell cycle’ and ‘M phase’ were the top GO biological processes (P = 1.0 × 10−17 or 1.4 × 10−19, respectively, Fisher exact P-values, hereinafter, Figure 2a). Similarly, LNCaP-Abl also exhibited a significantly enriched level of ‘cell cycle’ (P = 2.2 × 10−8) or ‘M-phase’ genes (P = 3.2 × 10−9) in all 363 upregulated genes (Figure 2b). Additionally, majority of 81 genes, which are upregulated in both LNCaP-Abl and LNCaP-221#25, appeared to be involved in ‘cell cycle’ (false discovery rate (FDR) = 2.4 × 10 −5) and ‘M phase’ (FDR = 9.2 × 10−5; Figure 2b).
Figure 2. LNCaP-miR-221 overexpressing cell lines selectively upregulated a set of cell cycle/M-phase genes.
(a) Comparsion of percentages of cell cycle and M-phase genes in all upregulated genes in LNCaP-miR-221 overexpressing cell lines (#25) and LNCaP-Abl. P-values were from student’s t-test. (b) A Venn-diagram showing GO analysis of the upregulated genes both in LNCaP-Abl and LNCaP-221#25. (c) Heat map of normalized mRNA expression levels of selected cell cycle genes. The gene sets were taken out from LNCaP, LNCaP-Abl and LNCaP-221#25 without DHT (0) or with DHT treatment for 4 h4 or 24 h.24 Data from three independent experiments is presented. Red and blue color represents upregulation and downregulation, respectively. (d) Confirmation of expression by quantitative real-time–PCR analysis for UBE2C, cdk1, cdc20 and Cyclin B1. Error bars indicate ± s.d. N = 3.
The expression alternations of a signature of all key cell cycle or checkpoint regulator genes were further compared before and after androgen treatment and among different cell lines25,26 (Figure 2c). Notably, a large portion of cell cycle genes, involved in G2 phase and G2/M-phase transition or M-phase cell cycle genes were significantly elevated in LNCaP-Abl and LNCaP-221#25 (Figure 2c). Wang et al.22 has previously found that UBE2C, cdk1, cdc20 and Cyclin B1 were highly expressed in LNCaP-Abl, and their high expression was thought to contribute to AI growth. These genes were referred to as AR selectively upregulated M-phase cell cycle genes.27 We found that UBE2C, cdk1, cdc20 and Cyclin B1 were also dramatically upregulated in LNCaP-221#25, as validated by TaqMan quantitative PCR (Figure 2d). We hypothesized that upregulation of miR-221 downregulated some mir-221 targets, which may have involved in the elevated expression of a set of cyclins, leading to the enhanced AI cell growth.
Overexpression of miR-221 downregulated the androgen-induced and AR-mediated transcription without affecting the AR or AR-androgen integrity
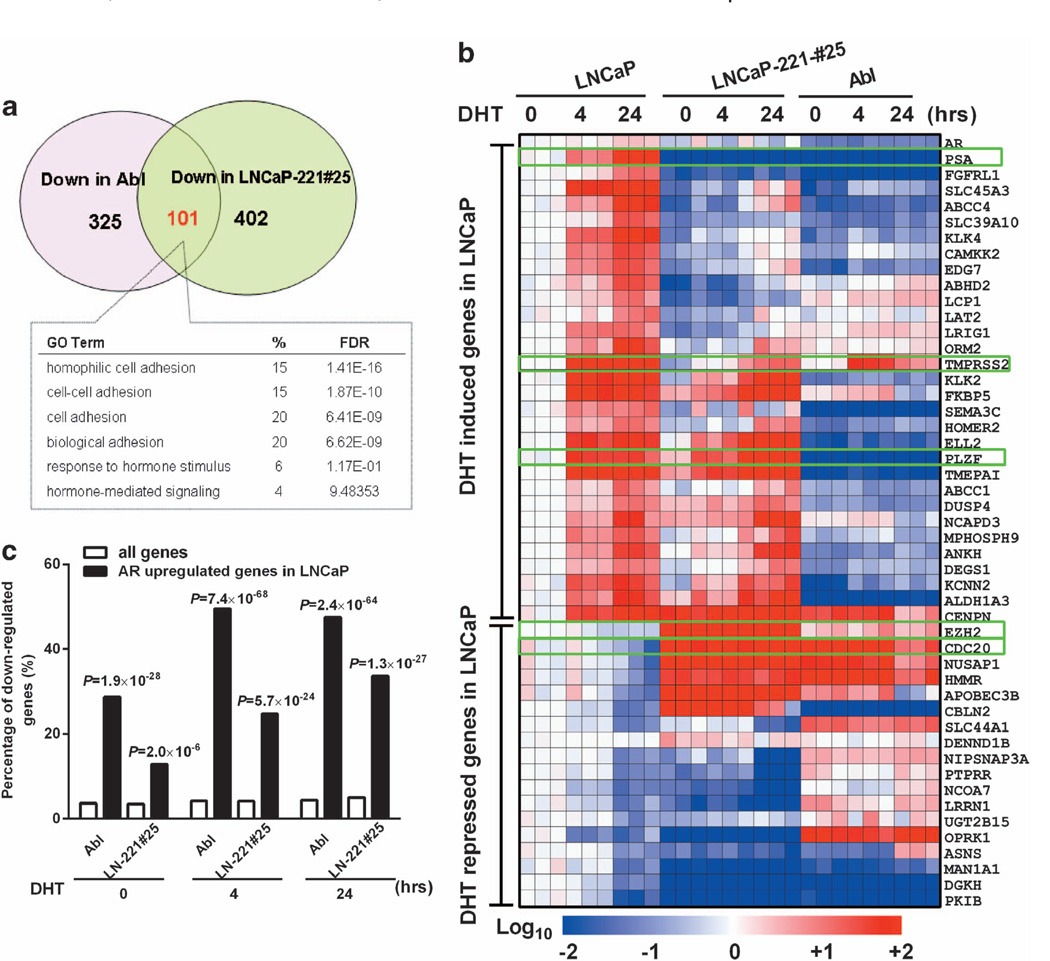
GO analysis of 101 genes that are both downregulated in LNCaP-Abl and LNCaP-221#25, versus those in LNCaP (decreased fold of expression >2.0, P < 0.001), revealed that genes involved in ‘cell-cell adhesion’ (for example, P-cadherin and cadherin-like, FDR <10−9) and ‘hormone-stimulated responses’ (for example, PSA and TMPRSS2, FDR = 0.1) were the top two broad GO categories (Figure 3a). Further comparing the expression pattern of all androgen-responsive genes, demonstrated that ~50% of the genes, which are DHT inducible in LNCaP and significantly repressed in LNCaP-Abl, were downregulated in LNCaP-221#25, including PSA and TMPRSS2 (Figure 3b).28,29
Figure 3. MiR-221 expression affects the expression of a subgroup of androgen-responsive genes.
(a) A venn-diagram showing GO analysis the genes which were both down-regulated in LNCaP-Abl and LNCaP-221#25. (b) A normalized heat map comparing the expression level of androgen-responsive genes in LNCaP, LNCaP-Abl and LNCaP-221#25 treated without DHT (0) or with 10 nm DHT for 4h4 or 24h.24 The 94 genes signature contains both androgen-induced and androgen-repressed genes. Data of three independent experiments is presented. Red and blue color represents upregulation and downregulation, respectively. (c) Comparison of the percentages of downregulation of AR upregulated signature genes in LNCaP-Abl and LNCaP-221#25. Significance was determined by the Student’s t-test.
Using an AR-regulated gene signature defined by the presence of AR-binding sites near the gene locus,30 we further analyzed downregulated genes in LNCaP-miR-221 overexpressors (Figure 3c). AR upregulated genes in LNCaP were statistically significantly repressed in both LNCaP-Abl and LNCaP-221#25 (all P-values<10−5). Although AR upregulated genes were more likely to be repressed in LNCaP-Abl than in LNCaP-221#25 (Figure 3c), indicating that AR activity was only partially impaired in LNCaP-miR-221 overexpressing cell lines. As it is known that LNCaP-Abl has undergone an extensive degree of epigenetic change, we anticipated that much of the altered AR program in LNCaP-Abl might not be driven by miR-221/-222.
We further investigated several potential possibilities that may affect processes in the AR function. We first compared the transcriptional efficiency driven by a PSA promoter or PSA-derived androgen-responsive elements using luciferase reporter constructs31 (Supplementary Figure S2A). In LNCaP-Abl and LNCaP-221#25, DHT treatment was unable to significantly stimulate transcriptional activities from these reporter constructs (Supplementary Figure S2B). We also compared the AR occupancy at the PSA (Supplementary Figure S2C, upper panel, Supplementary Table S2) and TMPRSS2 (Supplementary Figure S2C, lower panel, Supplementary Table S2) promoter regulatory regions in LNCaP, LNCaP-Abl and LNCaP-miR-221 overexpressing cell lines by AR chromatin immunoprecipitation (ChIP) assays. It showed that in LNCaP-miR-221 overexpressing cell lines, the ability of AR binding to androgen-responsive element at the PSA or TMPRSS2 locus was not affected, and DHT-enhanced AR binding to the PSA promoter/enhancer regions did not facilitate transcription efficiency at the PSA or TMPRSS2 genes. To exclude the possibility that the integrity of AR itself may be affected by overexpression of miR-221, we confirmed AR translocation into the nucleus with DHT treatment and phosphorylation of AR were not altered in LNCaP-miR-221 overexpressing cell lines (Supplementary Figures S3 and S4). All these results suggested that overexpression of miR-221 may not affect AR integrity. Thus, the impaired expression of a subset of AR-regulated genes caused by the upregulation of miR-221 most likely is not due to the defect of the fundamental function of AR, but may have resulted from the dysfunction of factors associated with the AR machinery.
Multiple miR-221 target genes are simultaneously involved in the development of androgen independency in LNCaP-miR-221 overexpressing cell lines
MiRs bind to the 3′-untranslated region (UTR) and downregulate their target genes’ expression by reducing target mRNAs’ stability, leading to a reduced level of steady-state mRNAs and/or by inhibiting translation efficiency of target mRNAs.32 We implemented two approaches to identify miR-221/-222 targets involved in the AI phenotype. The first approach was to filter and select genes whose mRNA abundance was decreased due to a high-level expression of miR-221 in LNCaP-miR-221 overexpressing cell lines and in LNCaP-Abl (repressed by twofold; P-value < 0.001) from the potential miR-221/222 targets identified by at least two prediction algorithms ‘miRanda’, ‘TargetScan’ and ‘PicTar’ programs.33–35 This approach identified one target, AMMECR1, which is an Alport syndrome related gene and is located on the X chromosome36 (Supplementary Table S1). Further functional studies indicated that the expression level of AMMECR1 did not significantly affect AR-mediated PSA transcription and cell growth of LNCaP (Supplementary Figure S5).
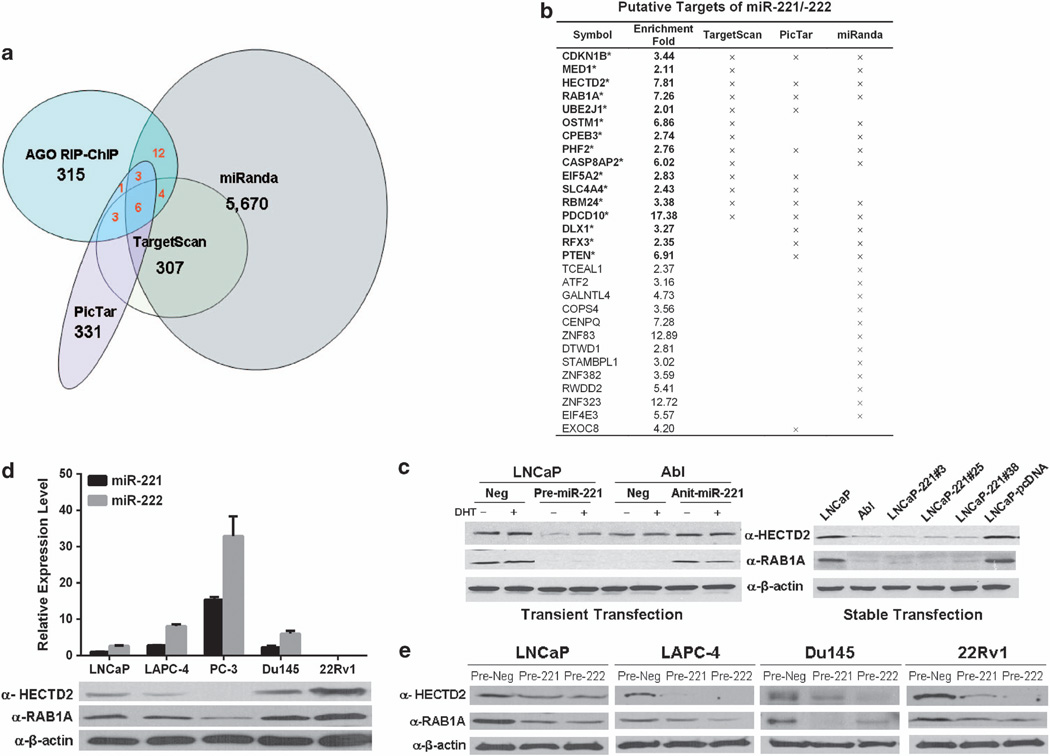
The second approach was the direct affinity purification of target mRNAs that are associated with the miR–RISC complex.32 We used the anti-AGO-RIP-ChIP approach described by Wang et al,37 which uses anti-AGO to IP AGO-containing miRNPs and associated mRNAs. MiRNPs in total cell lysates from LNCaP-miR-221 or −222 transient overexpressing cell lines were co-immunoprecipitated with anti-AGO and profiled. As controls, LNCaP transfected with a scrambled miR precursor control and IP-ed with a non-immune serum were performed in parallel. The miR-221/-222 known target, p27/kip1, was used to optimized the efficiency of the AGO-RIP-ChIP of miR-221RNPs (see Supplementary Figure S6). In total, 315 unique gene transcripts were enriched by >2.0 fold in anti-AGO-RIP-ChIP of LNCaP-miR-221/-222 overexpressing cell lines (biologically triplicate experiments, P < 0.001; Figure 4a). These 315 anti-AGO-pull-down genes were further filtered for miR-221/-222 targets predicted by ‘miRanda’, ‘TargetScan’ and ‘PicTar’ programs33–35 (Figure 4a). We identified 16 potential targets (Figure 4b) and further evaluated whether they indeed served as miR-221/-222 targets by determining whether their protein level could be downregulated by miR-221/-222.
Figure 4. Identification of miR-221/-222 targets involved in CRPC.
(a) Venn-diagram depicting the numbers of overlapped miR-221/-222 targets between those identified by anti-AGO-RIP-ChIP and those predicated by three miR target prediction programs (TargetScan, PicTar and miRanda). (b) Putative miR-221/-222 targets identified in LNCaP. Enrichment fold indicates the extent enrichment of the transcript in anti-AGO-pull-down materials. Check marks (x) indicate ones predicted by each of the target prediction programs. (c) Western blots analysis of the expression level of HECTD2 and RAB1A. Total protein extracts (50 µg each) were isolated from the LNCaP, LNCaP-Abl and LNCaP-miR-221 stable cell lines. Left panel: LNCaP and LNCaP-Abl cells were transiently transfected with precursors of miR-221 or anti-sense miR-221 and treated with or without DHT. Right panel: LNCaP, LNCaP-Abl, LNCaP-miR-221 overexpressing cell lines of #3, #25 and #38, and the control cell line, LNCaP-pcDNA. (d) Relative expression levels of miR-221/-222 (upper panel) and putative target of HECTD2 and RAB1A (lower panel) in five CaP cell lines, LNCaP, LAPC-4, PC-3 Du145 and 22Rv1. Expression levels of miRs were relative to that of miR-221 in LNCaP, which was arbitrarily set as 1.0. MiR expression levels were normalized by U6. The β-actin level was used as a loading control. (e) Western blots analyses of the expression level of HECTD2 (right panel) and RAB1A (middle panel). Total protein extracts (50 µg each) were isolated from LNCaP, LAPC-4, Du145 and 22Rv1 cell lines, which were transiently transfected with precursors of miR-221 or miR-222.
It appeared that protein levels of two miR-221 potential targets, HECTD2 and RAB1A, were significantly downregulated in LNCaP that transiently or stably overexpressed miR-221 and LNCaP-Abl (Figure 4c, right panel). Furthermore, knocking down miR-221 in LNCaP-Abl increased the expression levels of HECTD2 and RAB1A (Figure 4c left panel). The inverse correlation of the expression level of endogenous HECTD2, RAB1A with the miR-221/-222 expression levels was also observed in several other CaP cell lines including LAPC-4, PC-3, Du145 and 22Rv1 (Figure 4d). PC-3, which had the highest level of miR-221/-222 expression, exhibited the lowest expression level of HECTD2 and RAB1A among all the cell lines tested. In contrast, 22Rv1, which had the lowest level of miR-221/-222 expression, possessed the highest endogenous HECTD2 and RAB1A expression levels (Figure 4d). In addition, similar to the observation in LNCaP, transient overexpression of miR-221/-222 in LAPC-4, 22Rv1 or Du145 cell lines downregulated the expression levels of HECTD2 or RAB1A (Figure 4e). These results suggested that miR-221/-222 negatively regulates HECTD2 and RAB1A protein expression in these CaP cell lines.
Protein levels of CDKN1B (p27/kip1) and PHF2 can be downregulated by transiently overexpressing miR-221 in LNCaP, although their expression level remained unchanged in stable LNCaP-miR-221 overexpressing cell lines and LNCaP-Abl (Supplementary Figure S7). Protein levels of OSTM1, CEPB3, CASP8AP2, EIF5A2, RBM24, PDCD10 and RFX3, were not affected by transient or stable overexpression of miR-221 (Supplementary Figure S7). PTEN protein is undetectable in LNCaP (a PTEN-negative cell line). Expression levels of MED1 and UBE2J1 were slightly decreased in miR-221 stable overexpressing cell lines. However, changing of MED1 or UBE2J1 expression has no impact on LNCaP AI growth. Furthermore, a recent study showed that activation, instead of downregulation, of MED1 promotes CRPC development.38 Thus, it is unlikely that these mir-221/222 targets are involved in CRPC development.
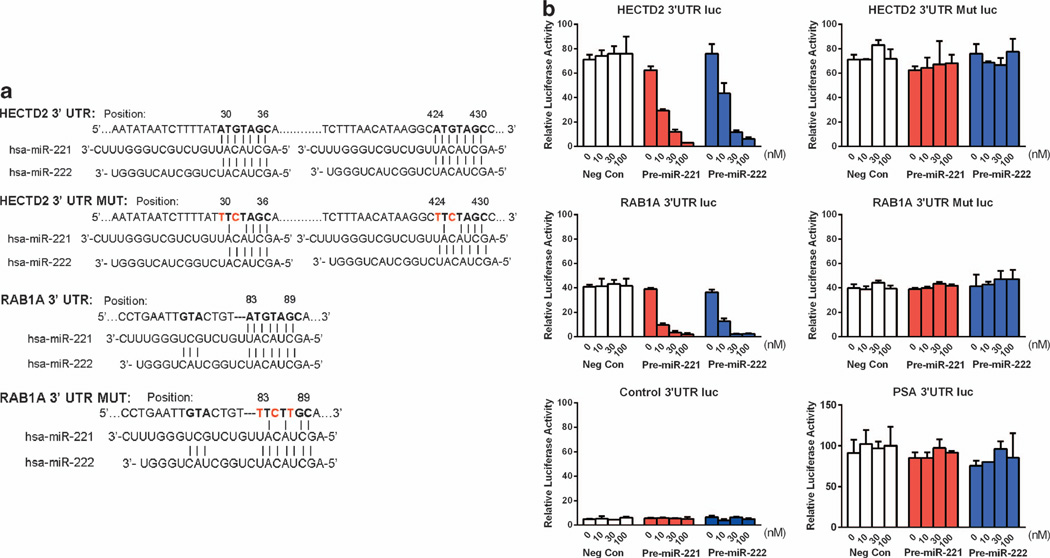
To confirm that HECTD2 or RAB1A are true targets of miR-221/-222, we tested whether expression of luciferase reporter constructs containing the 3′-UTR of HECTD2 or RAB1A, (containing miR-221/-222-seeding sequences), could be downregulated by overexpression of miR-221/-222 (Figure 5a). To demonstrate that the interaction of miR-221/-222 with the target sites predicted in the 3′-UTR of HECTD2 and RAB1A is essential for the function of miR-221/-222, we also tested luciferase reporter constructs containing 3′-UTR of HECTD2 or RAB1A with mutated miR-221/-222-seeding sequences (Figure 5a). Overexpression of miR-221 or −222 could notably repress the activity of luciferase reporter genes containing the intact 3′-UTRs of HECTD2 or RAB1A in a dose-dependent manner (Figure 5b), site mutations on miR-221/-222-seeding sequences in the 3′-UTR of HECTD2 or RAB1A completely abolished the response to miR-221/-222 overexpression (Figure 5b). The activities of the control luciferase reporter gene of a PSA 3′-UTR and the luciferase vector without insertions were not affected by the overexpression of miR-221/-222 (Figure 5b). Overexpression of the control miR precursors did not affect the activity of all constructs (Figure 5b). These results demonstrated that HECTD2 or RAB1A are true miR-221/-222 targets.
Figure 5. Validation of HECTD2 and RAB1A as miR-221/-222 targets.
(a) Wild-type and mutated sequences of predicated miR-221/-222-binding site in the 3′-UTR of HECTD2 (NM_182765) and RAB1A (NM_016021). Red nucleotides indicate site-directed mutations. (b) Luciferase activities derived from constructs containing different 3′-UTR as indicated on the top of each panels. Each construct was co-transfected with different amounts (0, 10, 30 and 100 nM) of Pre-miR-221 or Pre-miR-222 or miR-scramble RNA (negative control) into LNCaP. Data are mean ±s.d. of three independent experiments.
Reduction of RAB1A and HECTD2 promoted the CRPC phenotype
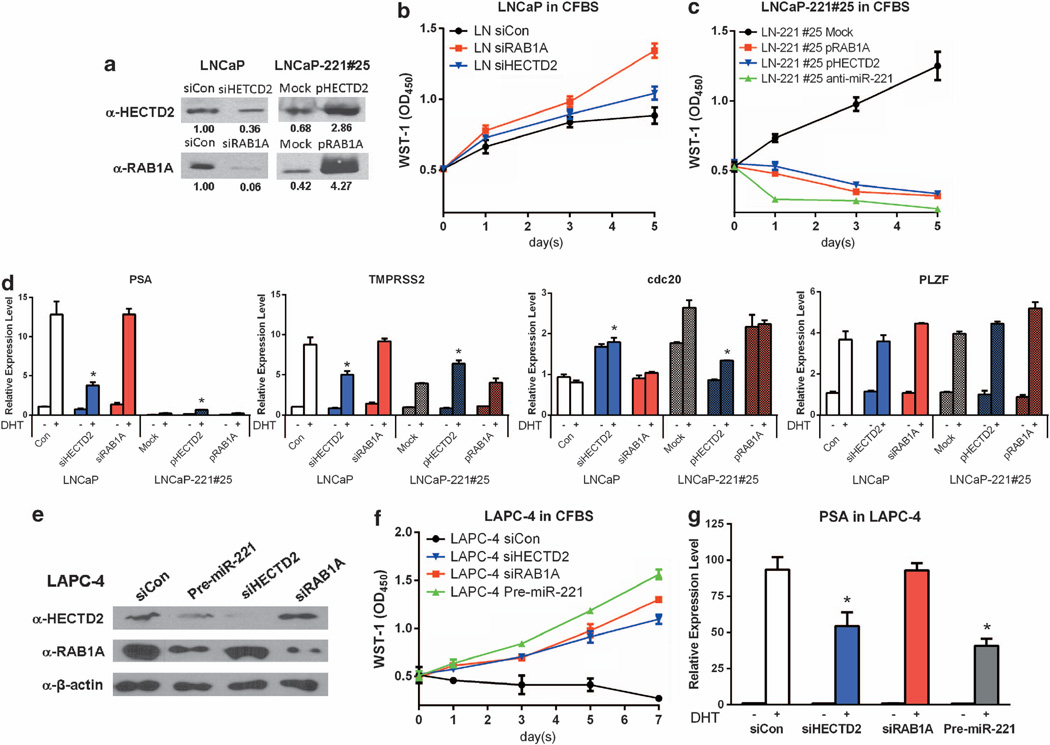
To determine the role of miR-221 targets in CRPC development, we investigated the impact of their expression levels on cell growth and PSA expression in response to DHT treatment. We first determined whether knocking down HECTD2 or RAB1A in LNCaP could affect cell growth. The efficiency of each small interfering RNA (siRNA) is shown in Figure 6a. Knocking down HECTD2 did not have a significant impact on cell growth in the presence of androgen, although it significantly enhanced cell growth in the absence of androgen (Figure 6b). Interestingly, knocking down RAB1A increased the LNCaP growth in either the presence or absence of androgen (Figure 6c). Furthermore, overexpressing RAB1A or HECTD2, which were repressed by miR-221 expression in LNCaP-221#25 cells, or knocking down miR-221 expression, could abolish previous established AI growth in LNCaP-221#25 cells (Figure 6c). As controls, co-transfections of the expression construct and siRNA of each target gene were performed to demonstrate the specificity of the siRNA effects (Supplementary Figure S8). The growth study suggested that downregulation of HECTD2 and RAB1A by miR-221/-222 may have sustained the AI cell growth.
Figure 6. The impact of HECTD2 and RAB1A on androgen independent growth and DHT-induced transcription.
(a) Expression levels of HECTD2 and RAB1A measured by western blots after knocking down by siRNAs in LNCaP or overexpressing in LNCaP-221#25. Relative expression levels were indicated under individual gel images. All protein expression levels were normalized with that of β-actin, and compared with LNCaP transfected with the control siRNA. (b, c) Growth Curve of LNCaP (b) or LNCaP-221#25 (c) in androgen-free medium (CFBS), after knocking down by siRNA or overexpressing RAB1A (red lines), HECTD2 (blue lines), or transfected with the transfection control (black lines) over a 5-day time course. MiR-221 was also knocked down in LNCaP-221#25 (right panel, green line). Each dot with a bar represents the mean of triplicates±s.e.m.. (d) The impact of expression levels of HECTD2 and RAB1A on the expression of androgen-responsive signature genes, PSA, TMPRSS2, cdc20 and PLZF in LNCaP or LNCaP-221#25. Twenty-four hours after transfection with siRNA or with expressing-constructs, LNCaP or LNCaP-221#25 were treated with or without DHT for additional 24 h. (e) Expression levels of HECTD2 and RAB1A were measured by western blots after knocking down by siRNAs or overexpressing miR-221 in LAPC-4. β-actin level was used as a loading control. (f) Growth Curve of LAPC-4 in CFBS medium after knocking down RAB1A (red line), HECTD2 (blue line), or overexpressing miR-221 (green line) or transfected with siRNA control (siCon, black lines). (g) The impact of expression levels of HECTD2 and RAB1A on the expression of PSA in LAPC-4. Twenty-four hours after transfection with siRNA or with miR-221 precursors, LAPC-4 cells were treated with or without DHT for additional 24 h. The mRNA levels of different genes in each cell lines were analyzed by TaqMan quantitative real-time–PCR. The relative expression level of each gene was normalized with that of glyceraldehyde 3-phosphate dehydrogenase, and compared with cells transfected the negative control siRNA, which was arbitrarily set as 1.0 (mean (N = 3) ±s.d.). *P-value of one-way ANOVA < 0.05.
We further evaluated the role of the putative miR-221/-222 targets on the AR-mediated transcription. Each of the target genes was transiently overexpressed in LNCaP-221#25 (Figure 6a) or transiently knocked down by siRNA in LNCaP (Figure 6a). Following treatment with DHT, we examined the impact of mRNA expression level of selected AR-mediated genes, including AR-mediated ligand-dependent PSA, TMPRSS2 and PLZF, and the AR-mediated ligand-independent cdc20. As described above, PSA, TMPRSS2 and PLZF are DHT inducible in LNCaP and downregulated in LNCaP-Abl. In LNCaP-221#25, PSA, TMPRSS2 were dramatically downregulated, while PLZF expression was not altered. Changing the expression level of RAB1A did not affect mRNA expression of all genes analyzed (Figure 6d). Expression of PLZF was not affected by altered expression of these two miR-221/-222 targets (Figure 6d). Knocking down HECTD2, which is a ubiquitin E3 ligase, significantly reduced DHT-induced PSA and TMPRSS2 and increased the cdc20 expression in LNCaP (Figure 6d). Overexpressing HECTD2 slightly rescued PSA and TMPRSS2 expression and significantly downregulated the expression of cdc20 in LNCaP-221#25 (Figure 6d). These results indicated that changing expression levels of HECTD2 modified the expression of some AR-mediated genes in response to DHT treatment. The effect of expression level alterations of RAB1A, HECTD2 on PSA transcription or AI cell growth were also demonstrated in LAPC-4 cell line (Figures 6e–g). In summary, we demonstrated that the expression level of two miR-221/-222 target genes, HECTD2 and RAB1A, is of importance in the development of the CRPC phenotype.
Upregulation of miR-221/-222 activates pathways involved in metastasis or epithelial to mesenchymal transition
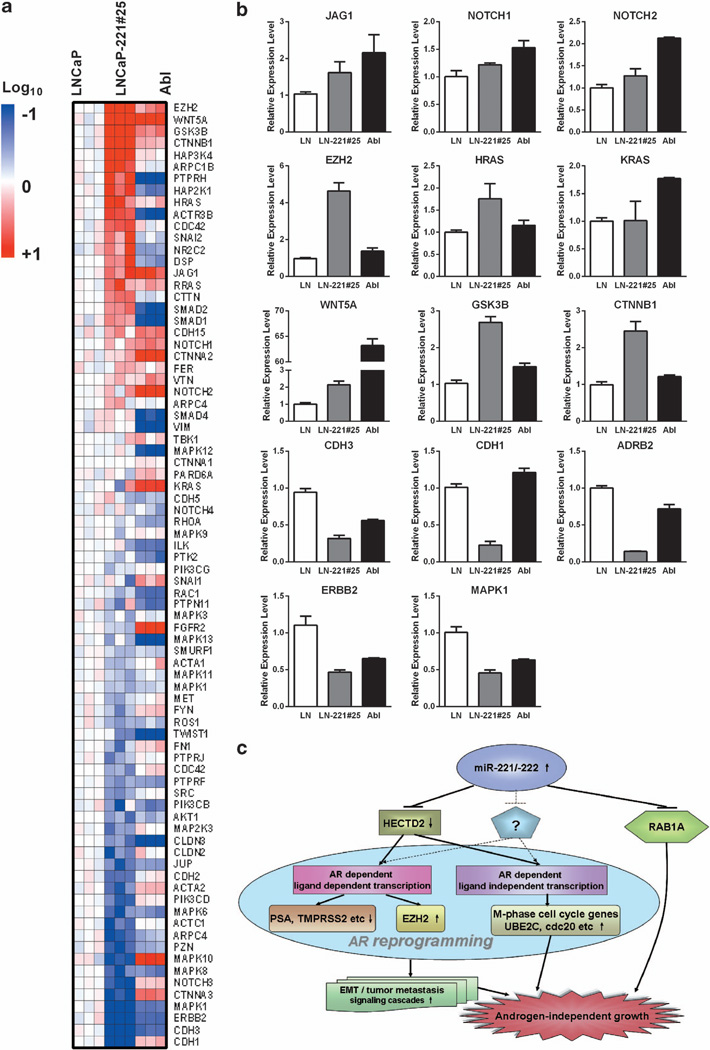
GO analyses of genes downregulated by elevated miR-221 expression also revealed that the expression of genes in ‘cell-cell adhesion’ process was reduced (FDR<10−8; Figure 3a), indicating that pathways involved in metastasis or epithelial to mesenchymal transition (EMT) may be activated in LNCaP-miR-221 overexpressing cells. The analysis of EMT pathway genes revealed that expression of several important EMT or tumor metastasis regulators, including GSK3β, EZH2, Wnt5A and RAS, JAGG1/Notch1/Notch2 were upregulated both in LNCaP-Abl and LNCaP-221#25 (Figure 7a). Expression of cadherin (CDH3 or CDH1), ERBB2, MAPK1 and beta-2-adrenergic receptor (ADRB2) were downregulated in LNCaP-Abl and/or LNCaP-221#25. The relative expression level of selected genes in EMT pathways was further confirmed by quantitative real-time-PCR, shown in Figure 7b. We hypothesized that downregulation of miR-221/-222 target molecules in LNCaP-Abl and/or LNCaP-221#25 may have activated the EZH2, WNT, GSK3β, RAS and Notch pathways, and downregulated the β-adrenergic pathway, as a result promoting cell proliferation, cell transformation and cell invasion. The detailed relationship among these pathways in CaP or CRPC remains to be further characterized.
Figure 7. The impact of miR-221/-222 expression on genes involved in EMT pathways.
(a) Heat map of the expression of EMT signature genes in LNCaP, LNCaP-Abl and LNCaP-221#25. Data from three independent experiments is presented. Red and blue color represents upregulation and downregulation, respectively. (b) Quantitative real-time–PCR confirmation of JAG1, NOTCH1, NOTCH2, EZH2, HRAS, KRAS, WNT5A, GSK3B, CTNNB1, CDH3, CDH1, ERBB1, MAPK1 and ADRB2 mRNA expression in LNCaP (LN, white bars), LNCaP-221#25 (LN-miR-221, gray bars) and LNCaP-Abl (Abl, black bars). Relative expression levels were normalized with that of glyceraldehyde 3-phosphate dehydrogenase, and compared with LNCaP, which was arbitrarily set as 1.0 (mean (N = 3) ±s.d.). (c) A model for miR-221/-222-mediated pathways in CaP cells. Expression of miR-221/-222 downregulates multiple target genes expression, including HECTD2, RAB1A and perhaps unidentified factors (?). HECTD2 are essential factors required for AR-mediated and androgen-dependent transcription, that is, PSA and TMPRSS2 expression. HECTD2 is also involved in ligand independent AR-mediated transcription that is, cdc20 expression. As results of AR reprogramming, pathways involved in EMT/tumor metastasis, other signal cascades, and androgen independent growth are promoted. Downregulation of RAB1A–enhanced androgen independent growth, although molecular mechanisms involved remained to be determined.
DISCUSSION
Taken together with our prior work, we demonstrated that increased miR-221/-222 expression is associated with CRPC (Figure 7c). Using unbiased approaches, we identified miR-221/-222 targets, HECTD2 and RAB1A that are potentially involved in the development of the CRPC phenotype (Figure 7c). The reduced HECTD2 expression level significantly decreased androgen-induced and AR-mediated transcription, potentially serving roles in modifying the AR signaling pathways. Downregulation of HECTD2 and RAB1A contributed to AI cell growth. Our expression profiling data revealed that accompanying with the development of CRPC, expression of a set of cyclins was activated to sustain the AI cell growth. We hypothesized that upregulation of miR-221/-222 downregulated HECTD2 and RAB1A, which subsequently resulted in reprogramming of AR pathways and activation of new cyclins, leading to the development of the CRPC phenotype. In particular, elevation of cdc20, an AR-controlled G2-M-phase cell cycle regulator, was induced by downregulation of HECTD2, which may contribute to AI cell growth. The possibility that we might have missed some miR-221/-222 target molecules involved in CRPC, due to the stringency of pull-down methods or due to the capacity of target predicting algorithms cannot be excluded (Figure 7c).
Currently, it is not yet completely clear about the exact mechanisms by which HECTD2 and RAB1A mediate the miR-221/-222-induced phenotype-CRPC. Very little information is available for the role of HECTD2 and RAB1A in the AR-mediated pathways and cell cycle controls. HECTD2, is a HECT domain containing ubiquitin E3 ligase, had a significant effect on the androgen-induced PSA transcription and on AI cell growth. Recently, investigators found that ubiquitin-mediated degradation of crucial tumor suppressor molecules was catalyzed by HECT-type E3, indicating that various HECT family members could be essential contributors to cancer development through crucial signaling pathways.39 We hypothesized that HECTD2 may post-translationally control the stability or abundance of specific AR machinery-associated factors and/or cell cycle proteins essential for the development of CRPC. RAB1A is a member of the Ras superfamily of GTPases, cycling between inactive GDP-bound and active GTP-bound forms.40 It has been shown that this specific GTPase controls vesicle traffic from the endoplasmic reticulum to the Golgi apparatus.40 Multiple alternatively spliced transcript variants encoding different protein isoforms have been identified for RAB1A.41 It is not known whether RAB1A potentially might also be involved in the RAS/RAF pathway, which could contribute to the activation of EMT and the development of aggressive cancer types.
The connection between miR-221/-222 expression and EMT was also found in the context of breast cancer. Stinson et al.42 found that TRPS1 targeting by miR-221/222 promoted EMT. However, TRPS1 is not a miR-221/222 target in LNCaP and LNCaP-derived AI cell lines. Recent evidence indicated that the Notch pathway is often recruited to stimulate growth or reprogramming tumor cells via EMT.43 It has been shown that EZH2 elevation repressed the expression of multiple tumor-suppressors and increased tumor aggressiveness.44 Based on our data, we think that the upregulation of the JAGGED1/NOTCH pathway and EZH2-mediated pathways due to the high expression of miR-221/222 may have a significant impact on promoting EMT/metastasis in CaP.44,45 Whether EMT promotion is a direct result of the AR pathway reprogramming or a consequence of the downregulation of identified miR-221/222 targets remains to be determined.
In summary, we demonstrated the involvement of miR-221/-222 in the promotion of the development of the CRPC phenotype. We are currently applying bioinformatics and biochemical approaches to delineate the molecular components and mechanisms by which HECTD2 and RAB1A operate during the transition to CRPC. New insights into the role of miR-221/222-mediated pathways during the CaP progression from hormone-sensitive CaP to CRPC may provide new therapeutic target designs.
MATERIALS AND METHODS
Cell culture
LNCaP and LNCaP-Abl cells (provided by Zoran Culig, Innsbruck Medical University, Austria)46 were maintained in RPMI-1640 plus 10% fetal bovine serum or 10% charcoal-stripped fetal bovine serum, respectively. LAPC-4 cell line was provided by Dr Robert Reiter.47 PC-3, Du145 and 22Rv1 cell lines were obtained from American Type Culture Collection. For flow cytometry analyses, all cells were cultured in either regular medium or charcoal-treated medium for 4 days, collected and stained with propidium iodine for sorting analyses at Dana-Farber Cancer Institute Cytometry Core Facility.
Materials and reagents
Synthetic, chemically modified short single-stranded RNA oligonucleotides: Pre-miR-221, Pre-miR-222 and Pre-miR-negative control, were purchased from Ambion (Austin, TX, USA). Antibodies against AR (441), PSA(C-19), cdk1,17 MED1(M-255), UBE2J1(18-Y) and RAB1A (C-19) were from Santa Cruz Biotechnology (Dallas, TX, USA), PLZF (2A9) was from Calbiochem (Billerica, MA, USA), EZH2 (AC22) was from Cell Signaling (Danvers, MA, USA), p27/kip (57) from BD Bioscience (San Jose, CA, USA), cdc20 and HECTD2 were from (Abcam, Cambridge, MA, USA), and β-actin was from Sigma-Aldrich (St Louis, MO, USA). All TaqMan primers and probes were purchased from Applied Biosystem (Carlsbad, CA, USA). A control siRNA (siControl) and siRNAs targeting HECTD2, RAB1A and other genes (ON TARGETplus SMARTpool siRNA) were purchased from Dharmacon (Dharmacon, Lafayette, CO, USA). Overexpression of each HECTD2 or RAB1A was used the Myc-DDK-tagged open reading frame (ORF)-derived clones, purchased from Origene (Rockville, MD, USA). WST-1 kit (Roche, Indianapolis, IN, USA) was used to measure cell proliferation.
Gene expression array experiments
LNCaP, LNCaP-Abl or LNCaP-221#25 cells were grown in the absence or presence of 10 nm DHT for 4 or 24 h. Total RNAs were isolated using an RNeasy kit (QIAGEN, Valencia, CA, USA). Biological triplicate total RNAs were analyzed by Affymetrix human U133 plus 2.0 expression array (Affymetrix, Santa Clara, CA, USA) at the Dana-Farber Cancer Institute Microarray Core Facility.
Luciferase reporter assays
pGL3-PSA promoter, pGL3-PSA enhancer and four copies of androgen-responsive elements promoter luciferase reporter vectors were provided by Bristol-Myers Squibb (Princeton, NJ, USA). pGL3-PSA 3′-UTR luciferase reporter and empty pGL3 Basic vectors were provided by Qianben Wang (Columbus, OH, USA). The 3′-UTR reporter clones of HECTD2 in pMirTarget plasmids were purchased from Origene. The 3′-UTR fragment of RAB1A were amplified from human genomic DNA and inserted into pMIR-REPORT miRNA reporter vector (Ambion Inc.). Site-directed mutations at miR-221/-222-seeding sequences of 3′-UTRs of HECTD2 or RAB1A were introduced by using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Inc., Santa Clara, CA, USA). LNCaP, LNCaP-Abl or LNCaP-221#25 was transfected with above luciferase reporter constructs and the control renilla luciferase expression construct, pRL-SV40 (Promega, Madison, WI, USA). Cells were harvested 48 h post transfection and assayed with Dual Luciferase Assay Kit (Promega).
AGO-RIP-ChIP and microarray analysis
AGO-RIP-ChIP was performed as described by Wang et al.37 The monoclonal antibody against AGO (2A8) was a gift from Peter T Nelson (University of Kentucky, Lexington, Kentucky50). Briefly, 107 LNCaP were transiently transfected with 30 nm Pre-miR-221 or Pre-miR-222 or Pre-miR-negative control (Ambion). Forty-eight hours after transfections, cells were rinsed and lysed on ice with fresh lysis buffer with protease inhibitors. Cell lysates were collected and cleared with pre-blocked Protein G beads (Invitrogen, Grand Island, NY, USA), and proceeded for co-immunoprecipitation with either anti-AGO G beads, NMS (Pierce biotechnology, Rockford, IL, USA) G beads at 4°C for 90 min. RNAs that co-immunoprecipitated with anti-AGO antibodies were extracted using TRIzol (Invitrogen). Microarray analyses of RNAs isolated from RIP-ChIP or from total cell lysates were performed using Affymetrix human U133 plus 2.0 expression array. Biological triplicates were carried out.
Statistical and bioinformatics analysis
All gene expression microarray experiments were carried out in biological triplicates. Data were normalized and summarized using multi-array average (RMA).48 The differentially expressed genes were identified using SAM algorithm that is implemented in the MeV program.49 GO analyses (DAVID Bioinformatics Resources) were employed to study the impact of miR-222 overexpression. Only top GO biological processes with significant fisher exact P-values and FDRs were considered.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by a SPORE in Prostate Cancer 2 P50 CA090381-06 and TS was supported by a Department of Defense (DoD) Prostate Cancer Training Award W81XWH-09-1-0372. We thank Dr Peter T Nelson at University of Kentucky and Dr Zissimos Mourelatos at University of Pennsylvania for kindly providing us the anti-AGO antibody. We also thank Dr Wang-xia Wang for technical suggestions on AGO-RIP-Chip experiments.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
REFERENCES
- 1.American Cancer Society. Cancer Facts and Figs 2012. Atlanta, GA, USA: American Cancer Society; 2012. [Google Scholar]
- 2.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 3.Rini BI, Small EJ. Hormone-refractory Prostate Cancer. Curr Treat Options Oncol. 2002;3:437–446. doi: 10.1007/s11864-002-0008-1. [DOI] [PubMed] [Google Scholar]
- 4.Yuan X, Balk SP. Mechanisms mediating androgen receptor reactivation after castration. Urol Oncol. 2009;27:36–41. doi: 10.1016/j.urolonc.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 6.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A novel androgen receptor splice variant is up regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun T, Wang Q, Balk S, Brown M, Lee GS, Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69:3356–3363. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun T, Yang M, Chen S, Balk S, Pomerantz M, Hsieh CL, et al. The altered expression of MiR-221/-222 and MiR-23b/-27b is associated with the development of human castration resistant prostate cancer. Prostate. 2012;72:1093–1103. doi: 10.1002/pros.22456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 10.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 11.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 12.Coppola V, De Maria R, Bonci D. MicroRNAs and prostate cancer. Endocr Relat Cancer. 2010;17:F1–F17. doi: 10.1677/ERC-09-0172. [DOI] [PubMed] [Google Scholar]
- 13.Watahiki A, Wang Y, Morris J, Dennis K, O’Dwyer HM, Gleave M, et al. MicroRNAs associated with metastatic prostate cancer. PLoS One. 2011;6:e24950. doi: 10.1371/journal.pone.0024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepato-cellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 15.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenecity through PTEN and TIMP3 down regulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30:1082–1097. doi: 10.1038/onc.2010.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terasawa K, Ichimura A, Sato F, Shimizu K, Tsujimoto G. Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells. FEBS J. 2009;276:3269–3276. doi: 10.1111/j.1742-4658.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, et al. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer. 2010;9:229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang F, Wang Z. Identification and characterization of PLZF as a prostatic androgen-responsive gene. Prostate. 2004;59:426–435. doi: 10.1002/pros.20000. [DOI] [PubMed] [Google Scholar]
- 24.Bohrer LR, Chen S, Hallstrom TC, Huang H. Androgens suppress EZH2 expression via retinoblastoma (RB) and p130-dependent pathways: a potential mechanism of androgen-refractory progression of prostate cancer. Endocrinology. 2010;151:5136–5145. doi: 10.1210/en.2010-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 26.Mizuno H, Nakanishi Y, Ishii N, Sarai A, Kitada K. A signature-based method for indexing cell cycle phase distribution from microarray profiles. BMC Genomics. 2009;10:137. doi: 10.1186/1471-2164-10-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taneja SS, Ha S, Garabedian MJ. Androgen stimulated cellular proliferation in the human prostate cancer cell line LNCaP is associated with reduced retinoblastoma protein expression. J Cell Biochem. 2001;84:188–199. doi: 10.1002/jcb.1278. [DOI] [PubMed] [Google Scholar]
- 28.Love HD, Booton SE, Boone BE, Breyer JP, Koyama T, Revelo MP, et al. Androgen regulated genes in human prostate xenografts in mice: relation to BPH and prostate cancer. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci USA. 2002;99:11890–11895. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 31.Jia L, Kim J, Shen H, Clark PE, Tilley WD, Coetzee GA. Androgen receptor activity at the prostate specific antigen locus: steroidal and non-steroidal mechanisms. Mol Cancer Res. 2003;1:385–392. [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 35.Krek A, Gru¨n D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 36.Vitelli F, Piccini M, Caroli F, Franco B, Malandrini A, Pober B, et al. Identification and characterization of a highly conserved protein absent in the Alport syndrome (A), mental retardation (M), midface hypoplasia (M), and elliptocytosis (E) contiguous gene deletion syndrome (AMME) Genomics. 1999;55:335–340. doi: 10.1006/geno.1998.5666. [DOI] [PubMed] [Google Scholar]
- 37.Wang WX, Wilfred BR, Hu Y, Stromberg AJ, Nelson PT. Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA. 2010;16:394–404. doi: 10.1261/rna.1905910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z, Zhang C, Wu D, Chen H, Rorick A, Zhang X, et al. Phospho-MED1-enhanced UBE2C locus looping drives castration-resistant prostate cancer growth. EMBO J. 2011;30:2405–2419. doi: 10.1038/emboj.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Haas AK, Yoshimura S, Stephens DJ, Preisinger C, Fuchs E, Barr FA. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- 41.Weide T, Teuber J, Bayer M, Barnekow A. MICAL-1 isoforms, novel rab1 interacting proteins. Biochem Biophys Res Commun. 2003;306:79–86. doi: 10.1016/s0006-291x(03)00918-5. [DOI] [PubMed] [Google Scholar]
- 42.Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O’Brien C, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4 doi: 10.1126/scisignal.2001538. ra41. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS, et al. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009;69:2400–2407. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu J, Cao Q, Mehra R, Laxman B, Yu J, Tomlins SA, et al. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12:419–431. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 45.Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64:6854–6857. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- 46.Pfeil K, Eder IE, Putz T, Ramoner R, Culig Z, Ueberall F, et al. Long-term androgen-ablation causes increased resistance to PI3K/Akt pathway inhibition in prostate cancer cells. Prostate. 2004;58:259–268. doi: 10.1002/pros.10332. [DOI] [PubMed] [Google Scholar]
- 47.Klein KA, Reiter RE, Redula J, Moradi H, Zhu XL, Brothman AR, et al. Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nat Med. 1997;3:402–408. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- 48.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 49.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 50.Nelson PT, De Planell-Saguer M, Lamprinaki S, Kiriakidou M, Zhang P, O’Doherty U, et al. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.