Abstract
Purpose
There is a need to identify better glycan biomarkers for diagnosis, early detection and treatment monitoring in lung cancer using biofluids such as blood. Biofluids are complex mixtures of proteins dominated by a few high abundance proteins that may not have specificity for lung cancer. Therefore two methods for protein enrichment were evaluated; affinity capturing of IgG and enrichment of medium abundance proteins, thus allowing us to determine which method yields the best candidate glycan biomarkers for lung cancer.
Experimental design
N-glycans isolated from plasma samples from 20 cases of lung adenocarcinoma and 20 matched controls were analyzed using nLC-PGC-chip-TOF-MS. N-glycan profiles were obtained for five different fractions: total plasma, isolated IgG, IgG depleted plasma, and the bound and flow-through fractions of protein enrichment.
Results
Four glycans differed significantly (FDR<0.05) between cases and controls in whole unfractionated plasma, while four other glycans differed significantly by cancer status in the IgG fraction. No significant glycan differences were observed in the other fractions.
Conclusions and clinical relevance
These results confirm that the N-glycan profile in plasma of lung cancer patients is different from healthy controls and appears to be dominated by alterations in relatively abundant proteins.
Keywords: N-glycan, mass spectrometry, non-small cell lung cancer, biomarker, protein enrichment
INTRODUCTION
Lung cancer is the leading cause of cancer deaths in both men and women in the United States [1]. Currently no FDA approved test is available to assist with screening, diagnosis or early detection of lung cancer. Therefore, there is a need to identify biomarkers for risk assessment, early detection and disease monitoring [1]. Reports of proteomic, metabolomic, gene expression, miRNA and autoantibody biomarkers have all shown promise for lung cancer [2], however, they have not made it to the clinic yet. Biomarkers may also be provided by protein glycosylation patterns. Glycosylation is one of the most prevalent post-translational modifications, and affects more than 50% of the human proteome [3]. Cell membrane proteins, as well as proteins secreted and shed from cells are all highly glycosylated, and it is widely known that the glycosylation of these proteins play important roles in cell-cell communication and cell-matrix microenvironment interactions [4, 5]. It has been observed that N-glycan patterns of cancer tissue are different compared to healthy tissue [6, 7]. This is particularly the case with respect to increased branching of N-glycans in tumor cells, which has the potential to yield a glycan signature for malignancy and metastasis [6, 8, 9].
While clear alterations in the N-glycosylation of cancer tissue may be observed, there is a need to determine a glycan signature for cancer in biological fluids that may have utility for screening towards early detection of cancer, monitoring response to treatment and other applications where informative tissue material may not be readily available. Biofluids, especially blood, represent a readily available source of specimens, which would be ideal to develop cancer related glycan biomarker applications (e.g. [10–14]). While most proteins in serum are produced in the liver and B-cells, and are thus not directly linked to the cancer-site, cancer-specific proteins may be shed or secreted into the bloodstream and contribute to altered glycan composition of plasma or serum.
There have so far been only few studies aimed at identifying altered glycosylation in lung cancer. Hoagland et al. reported increased levels of sialylated forms of haptoglobin in lung cancer using 2D-DIGE methodology [15]. In a separate study, Arnold et al. compared the serum N-glycome of lung cancer patients to controls by hydrophilic interaction chromatography (HILIC-HPLC) with fluorescence detection [16]. In this study altered glycan levels were observed for five glycan peaks, with an increase in tri- and tetra-antennary structures and a decrease in bi-antennary structures, but due to co-eluting N-glycans it was not possible to identify individual glycan structures.
Although altered N-glycan patterns have been observed in different types of cancer, their specificity remains unclear. Some are likely associated with IgG, and thus may represent a host response to cancer. Ig based response to cancer may be in the form of autoantibodies directed against glycopeptides as recently reported for mucin-type O-glycopeptides directed autoantibodies in early stage breast cancer [17]. The serum N-glycan patterns are dominated by glycans derived from the top 5–10% abundant glycoproteins, including, but not limited to IgG, transferrin, haptoglobin, IgA, alpha-1-antitrypsin and alpha-2-macroglobulin [18]. More in-depth analysis of the N-glycome would require targeted enrichment techniques as applied to proteomics [19].
Traditionally, enrichment of glycoproteins has been performed using lectins with a broad variety of specificities [20–22]. Especially ConA and WGA lectins, which bind α-linked mannose [23, 24] and GlcNAcβ(1–4)GlcNAc with secondary affinity for sialic acids [25, 26], may be used for the general enrichment of glycoproteins. However, lectins have low specificities [20], especially for large intact proteins or protein complexes, such that their use is associated with limited reproducibility. Moreover, this technology does not overcome the wide dynamic range of variability in protein concentrations. Alternatively, immune-capture of specific glycoproteins may be performed. This strategy has mostly been applied to IgG using Protein A or G [27–32], but also to other proteins, such as haptoglobin [31] and IgA [33]. Immunoprecipitation is highly dependent on the antibody avidity and affinity for the targeted epitope and non-specific binding may occur. Therefore, it may be difficult to target lower abundance proteins using this technology. The use of depletion columns such as MARS-6 or MARS-14 has also been applied for N-glycan analysis [31, 34]; however, such depletion columns only remove the top 6 or 14 most abundant proteins, respectively, and do not substantially alter protein concentrations of the lower abundance proteins. Recently, the use of hexapeptide coated beads for the enrichment of medium to lower abundance proteins has been introduced [35]. This method was reported to enrich for medium and low abundance proteins and was recently applied to N-glycan analysis [36]. However, it has not been evaluated for its potential to identify candidate glycan biomarkers.
Mass spectrometric technology combined with separation using carbon-based stationary phases now allows identification and relative quantitation of N-glycans in a single run [37–41]. Our group introduced chip-based nano-liquid chromatography coupled with time-of-flight mass spectrometry (nLC-TOF-MS) for the separation, identification and quantitation of N-glycans to identify glycan-based biomarkers (e.g. [13, 41]). nLC-PGC-chip-TOF-MS has been proven to allow effective separation of glycan isomers in a highly repeatable way [13, 39] while facilitating relative quantitation of glycans and glycan compositions with good repeatability. An average inter-day CV of 0.04, determined on log10 transformed integrals was observed (Manuscript in preparation).
In this study, we applied two different methods for protein enrichment, 1) IgG immunocapturing using Prot G and 2) enrichment of lower abundance proteins using hexapeptide coated beads, and evaluated their efficacy for glycan-based biomarker discovery. To this end, we conducted a pilot study using a small sample set of 20 adenocarcinoma lung cancer cases and 20 matched controls and compared N-glycan patterns that were obtained from immunocaptured IgG, the IgG flow-through, the hexapeptide enriched fraction and the hexapeptide flow-through using carbon separation followed by mass spectrometry to those obtained from whole plasma.
EXPERIMENTAL SECTION
Clinical samples
Patient samples (plasma) from early stage resectable lung adenocarcinoma (cases) and healthy controls were obtained from the Fred Hutchison Cancer Research Center (FHCRC). Consent was obtained from each subject following an approved IRB protocol. Plasma was prepared using a standard procedure that involved centrifugation of blood collected with anti-coagulant followed by storage of plasma samples at −80°C until analysis. Controls were current or former smokers that were age (60.35 ± 10.29) and gender (60% of females) matched to cases for this study. Plasma samples from controls were collected in the same manner as the patient cases to prevent any potential bias in collection of these samples.
N-glycan release from plasma samples
N-glycan release of blood derived plasma samples was performed as described previously[42], with slight modifications. Briefly, 50 μL of a 200mM ammonium bicarbonate (Sigma-Aldrich, St. Louis, MO) solution with 10 mM dithiothreitol (DTT, Promega, Madison, WI) was added to 50μL of plasma. Proteins in the samples were denatured using six cycles alternating between 100°C and room temperature (RT) for 10 seconds each. Two μL of PNGaseF (New England Biolabs, Ipswich, MA, corresponding to 1000 NEB units or 15 IUB mU) was added to the samples, and enzymatic glycan release was performed in a CEM (Matthews, NC) microwave at 20W for 10 min. Deglycosylated proteins were precipitated using 400 μL of ice-cold ethanol, and the samples were chilled at −80°C for 1 hour. Upon centrifugation, the supernatant was transferred to new Eppendorf tubes, and dried in vacuo.
Isolation of IgG from Plasma
Immunoglobulin G was captured from 10 μL of plasma using a Protein G affinity purification step as previously published [27]. In short, 10 μL of plasma was added to 50 μL Protein G coated beads in 190 μL DPBS in a 96-well filter plate (Orochem, Lombard, IL). The IgG’s were allowed to bind at room temperature for 1h while shaking continuously, after which the flow-through (IgG-depleted fraction) was collected using a vacuum manifold. The IgG’s bound to the Prot G were washed four times using DPBS, followed by two times using water to remove excess salt. IgG’s were eluted using 200 μL of 100 mM formic acid in water and collected using a vacuum manifold. Both the IgG depleted and the IgG fraction were brought to dryness in vacuo and subsequently resuspended in 100 μL of a 100mM ammonium bicarbonate solution with 5 mM DTT for N-glycan release using the procedure described for plasma.
Enrichment of proteins with low abundance from plasma samples
Protein enrichment using peptide-coated beads was performed as described [36], with slight modifications. The dry beads (Bio-Rad, Hercules, CA) were rehydrated using 20% aqueous ethanol according to the manufacturers protocol. 15 μL of bead slurry was transferred to each of 40 wells of a 96-well filter plate (Orochem), and washed 3 times using 200 μL of PBS. 25 μL of plasma were loaded onto the wells and proteins were allowed to bind to the beads for 2h at room temperature under continuous rotation. Using a vacuum manifold, the non-bound fraction was removed and collected into a 96-well collection plate (Corning, Corning, NY). Peptide beads were washed using 2 × 400 μL of PBS, followed by 2 × 400 μL 200mM ammonium bicarbonate. On-bead N-glycan release was subsequently performed by addition of 1 μL PNGaseF in 100 μL 200mM ammonium bicarbonate, followed by mixing and overnight incubation at 37°C. Released N-glycans were collected in a 96-well collection plate using a vacuum manifold. The non-bound fraction was brought to dryness in vacuo and subsequently resuspended in 100 μL of a 100mM ammonium bicarbonate solution with 5 mM DTT for N-glycan release using the procedure described for plasma.
N-glycan purification using graphitized carbon SPE
Oligosaccharides released by PNGaseF were purified using graphitized carbon SPE cartridges (total plasma samples, Grace, Deerfield, IL) or 96-well PGC filter plates (All depletion samples, 40 μL PGC, Glygen, Columbia, MD). [13, 39, 43] Briefly, cartridges were conditioned using 4 mL of 80% ACN containing 0.05% TFA (EMD chemicals, Gibbstown, NJ), followed by 4 mL of water containing 0.05% TFA. Oligosaccharide samples were reconstituted in 500 μL of water and subsequently loaded onto the cartridges. Cartridges were washed using 3 × 4 mL of water and N-glycans were eluted using 4 mL of 40% ACN containing 0.05% TFA.
Wells of the 96-well PGC SPE plate were conditioned using 2 × 200 μL of 80% ACN containing 0.05% TFA, followed by 2 × 200 μL of water containing 0.05% TFA. Oligosaccharide samples were reconstituted in 200 μL of water and subsequently loaded onto the wells. Wells were washed using 4 × 200 μL of water and N-glycans were eluted using 2 × 200 μL of 40% ACN containing 0.05% TFA. All eluates were dried in vacuo prior to analysis.
nHPLC-chip-TOF-MS analysis
N-glycans were analyzed using an Agilent (Santa Clara, CA) 6200 series nanoHPLC-chip-TOF-MS, consisting of an autosampler, which was maintained at 8°C, a capillary loading pump, a nanopump, HPLC-chip-MS interface and an Agilent 6210 time of flight (TOF) mass spectrometer. The microfluidic chip (glycan chip II, Agilent) contained a 9 × 0.075 mm i.d. enrichment column coupled to a 43 × 0.075 mm i.d. analytical column; both packed with 5 μm porous graphitized carbon (PGC). N-glycans from plasma, IgG depleted plasma and flow-through of the protein enrichment were reconstituted in 45 μL of water and diluted 1:5 with water prior to analysis; 1 μL of sample was used for injection. N-glycans from IgG and enriched low abundance proteins were reconstituted in 50 μL of water and, without further dilution, 1 μL of sample was used for injection. Upon injection, the sample was loaded onto the enrichment column using 3% ACN containing 0.1% Formic acid (FA, Fluka, St. Louis, MO). After the analytical column was switched in-line, the nano-pump delivered a gradient of 3%ACN with 0.1% FA (solvent A) and 90% ACN with 0.1% FA (solvent B).
Data processing
Data processing was performed using Masshunter® qualitative analysis (version B.03.01, Agilent) and Microsoft® Excel® for Mac 2011 (version 14.1.3, Microsoft), according to Hua et al. [13] with modifications. Data was loaded into Masshunter qualitative analysis, and glycan features were identified and integrated using the Molecular Feature Extractor algorithm. First, signals above a signal to noise threshold of 5.0 were considered. Then, signals were deconvoluted using a tolerance of 0.0025 m/z ± 10 ppm. The resulting deconvoluted masses were subsequently annotated using a retrosynthetic theoretical glycan library [44], where a 15 ppm mass error was allowed. Glycan compositions and peak areas were exported to csv-format for further statistical evaluation.
Statistical analysis
Prior to statistical analysis, raw peak areas were total quantity (also so called “total-ion-current”) normalized based on the underlying assumption that the total amount of ionized glycans that reach the detector is similar for different samples and glycan profiles for each dataset. Glycans detected in fewer than 70% of samples were discarded from downstream analysis to reduce the bias that could be induced by imputation for missing not at random. Unobserved values for each glycan below the predefined detection limit were imputed as one-half of the glycan-specific minimum of the observed values. For the total plasma data set, each sample had two reads, which were averaged to yield a single value for each glycan. Finally, the data were log2 transformed to reduce the influence of extreme values to meet homogeneity of variance assumptions. All statistical analyses were conducted in R 2.12.0 language and environment (http://www.r-project.org/).
Partial least squares regressions (PLS) were used to determine if global glycomic profiles could separate cancer from control patients. Because of potential confounding effects of gender and age, we adjusted for differences in gender and age between subjects in these analyses. In addition, batch was included as a covariate in analyses of the total plasma samples because these samples were processed in two randomized batches, while other samples were done in a single 96-wall batch.
For the differential analyses, we identified glycans that significantly differ between cancer and control patients by a permutation t-test, while adjusting for inter-patient differences in gender and age, and for the total plasma data set, batch. We used 10,000 permutations to compute P-values for significance. False discovery rates (FDRs) were calculated to account multiple testing. Glycan expression change by cancer status was considered significant at FDR < 0.05.
RESULTS
A feasibility study was performed to determine whether protein enrichment can improve sensitivity and specificity for the development of glycan-based biomarkers for non-small cell lung cancer. We evaluated two different protein enrichment strategies: 1) IgG affinity capturing with Protein G and 2) the use of a non-specific hexapeptide library (Proteominer, PM) and compared them to whole plasma N-glycan analysis. First, N-glycan profiles were obtained from whole plasma samples from 20 cases and 20 controls. In parallel, immunoglobulin G (IgG) was immuno-purified from aliquots from the same sample set, and N-glycan profiles were obtained from both the IgG protein and IgG depleted serum fractions. Finally, enrichment of medium-abundance proteins was performed using hexapeptide coated beads (Proteominer, PM, Bio-rad). Here, N-glycan profiles were obtained for both the flow-through (PM-flow-through), containing mostly highly abundant proteins, and the bound fraction (PM-bound), comprising both high-abundance and medium abundance proteins.
Glycan features observed in the enrichment methods
An average of 327 glycan features per run was identified in each of the whole plasma samples. It has to be noted that the anomers originating from the different confirmations of the OH group at the reducing end are often separated on PGC stationary phases, and may thus be identified as individual glycan features in the analysis. For the IgG and IgG depleted and the PM-bound and PM-flow-through fractions an average of 133, 264, 47 and 241 glycan features were identified, respectively. These numbers are summarized in Table 1.
Table 1.
Characteristics of the glycan profiles observed and analyzed for the different enrichment strategies.
| Average no. of features per run | Average no. of compositions per run | No. of compositions used in statistical analysis | |
|---|---|---|---|
| Whole plasma | 327 | 94 | 79 |
| IgG | 133 | 62 | 40 |
| IgG depleted | 294 | 91 | 62 |
| PM-bound | 47 | 27 | 20 |
| PM-flow-through | 241 | 82 | 65 |
The glycan features originate from a more limited number of glycan compositions. The average number of compositions in each of the runs of the plasma samples was determined to be 94, while the IgG, IgG depleted and the PM-bound and PM-flow-through fractions contained an average number of 62, 91, 27 and 82 glycan compositions, respectively. Not all compositions are observed in every sample and therefore only compositions that were observed in at least 70% of the samples were used for statistical evaluation. Thus, 79, 40, 62, 20 and 65 compositions were included in the statistical analysis for whole plasma, IgG, IgG depleted and the PM-bound and PM-flow-through fractions, respectively. It has to be noted that the number of glycans and glycan compositions observed in the PM-bound fraction is relatively low. The generally low number of glycan compositions in the PM-bound fractions is consistent with two possibilities: 1) the capture is inefficient and only a small amount of protein is retained and 2) the capture is specific and retains a small subset of proteins. The former is the likely explanation as enrichment of even a single protein such as IgG yields a large number of glycans.
Glycosylation patterns are altered with different enrichment techniques
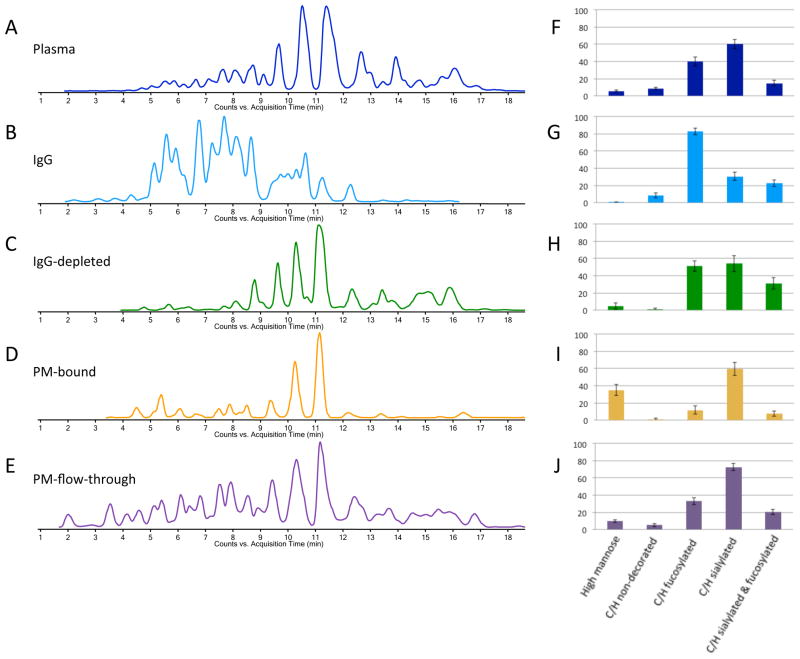
Typical extracted glycan chromatograms (EGC) for the different fractions (plasma, IgG, IgG depleted, PM-bound and PM-flow-through) are depicted in Figure 1A–E. As clearly illustrated in Figure 1, the glycosylation pattern of IgG is quite different than the overall N-glycan profile. There are much higher abundances of early-eluting glycans in the IgG-bound fraction, which is complemented by the depletion of these species in the IgG-depleted plasma.
Figure 1. Comparison between total plasma (A,F), IgG (B,G), IgG depleted plasma (C,H), medium- abundant protein enriched fraction (D,I) and protein enrichment flow-through (E,J).
Extracted glycan chromatograms, obtained using Masshunter software are depicted (A–E), as well as bar graphs showing the relative abundances (%) of different compositional features of the N-glycans observed in each fraction (F–J).
To further illustrate the altered glycosylation patterns, bar graphs of the relative abundances of the major types of glycosylation (high mannose, complex/hybrid non-decorated, complex/hybrid fucosylated, complex/hybrid sialylated and complex/hybrid fucosylated and sialylated) are depicted in Figure 1F–J. Clearly, there are fewer high-mannose type glycans and sialylated glycans in the IgG fraction; corresponding changes are observed in the IgG-enriched fraction (PM-bound) also shows an altered glycosylation pattern, in which the high-mannose type glycans are enriched, while the levels of fucosylated glycans are decreased (Figure 1I).
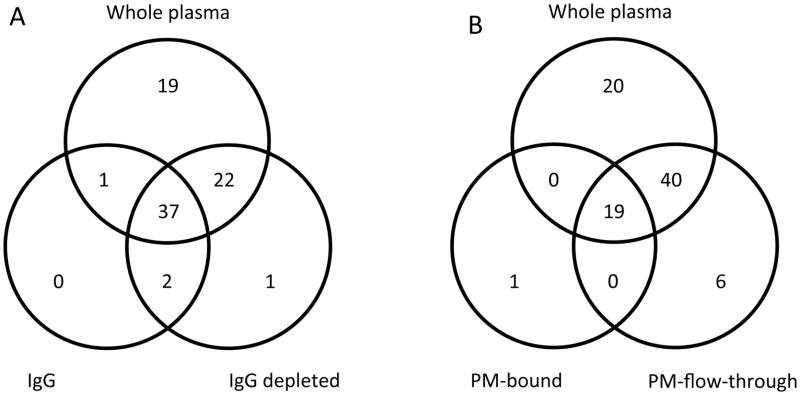
Changes at the level of individual glycans were also observed. Venn-diagrams show a large overlap in the glycan compositions across the different methods, while also showing the presence of some glycan compositions specific to some of the fractions (Figure 2). Interestingly 37 glycan compositions were common to whole plasma, IgG and IgG depleted fractions and 22 compositions were present in both whole plasma and IgG depleted fraction, but not in the IgG-bound fraction. In contrast, 19 glycan compositions were common to whole plasma and as well as the PM-bound and PM-flow-trough fractions, while 40 compositions were found to be present in both the whole plasma and PM-flow-through fraction. Only one glycan composition was found to be unique to the PM-bound fraction. As stated earlier, it is highly likely that the inefficiency of the capturing procedure results in low levels of protein attached, thus allowing only smaller numbers of glycans to be detected.
Figure 2. Venn diagrams of the glycan compositions of the five fractions.
Compared are whole plasma, IgG and IgG depleted plasma (A) and whole plasma, PM- bound and PM-flow-through (B).
Differential analysis separates cases from controls using enrichments strategies
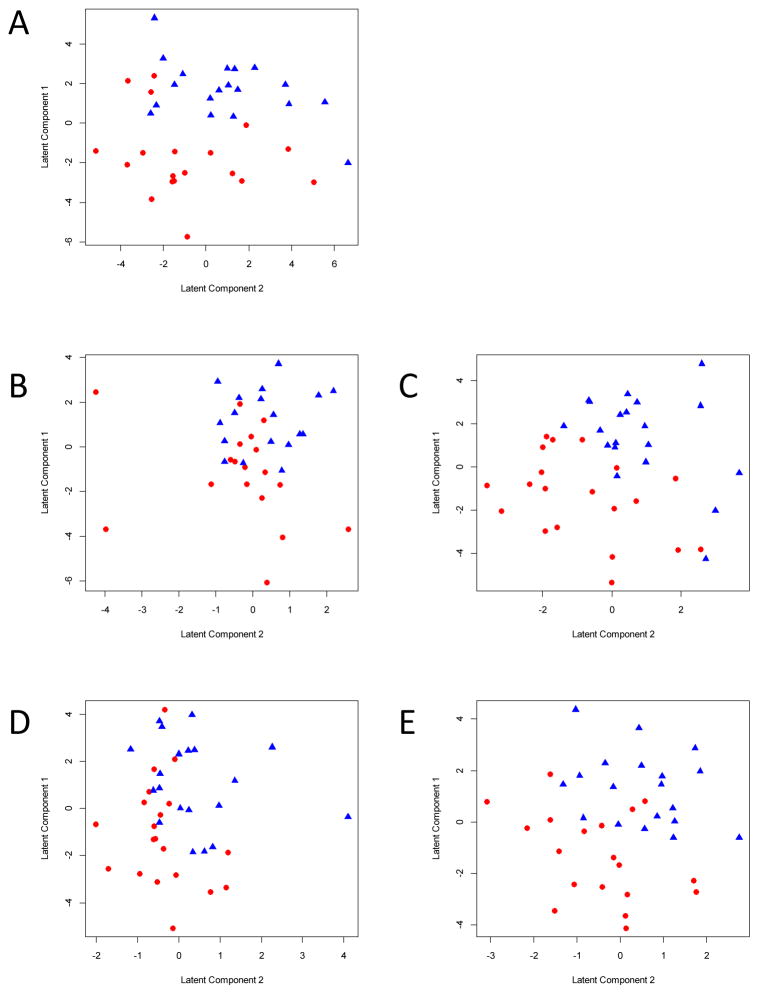
To evaluate the potential of each of the five fractions to separate cancer patients from controls based on global glycomic profiles, partial least squares regression analyses were performed. For all five data sets generated by different protein enrichment methods, cancer samples could be separated from control samples based on glycomic profiles (Figure 3). However, the separation was somewhat less pronounced in the PM-bound fraction (Figure 3D), where a greater overlap is observed between the cases and controls. These results may suggest that the glycosylation of the medium-abundance proteins targeted in this analysis could be less influenced by the disease process. But there are several alternative reasons for the poor differentiation in this fraction. It could mean that the small number of glycan compositions observed in this fraction is insufficient to yield significant differentiation, or that the capture is not suitably reproducible to yield disease-specific glycans.
Figure 3. Cancer cases can be separated from controls using N-glycan profiles.
PLS-DA plots show the separation of cancer cases and controls using N-glycan profiles obtained from whole plasma (A), plasma IgG (B), IgG depleted plasma (C), PM-bound (D) and the PM-flow-through fraction (E).
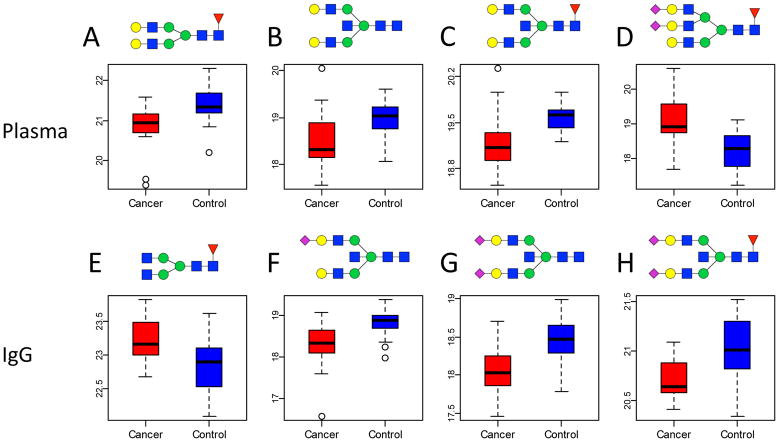
A differential analysis was used to identify individual glycan compositions that differ significantly between cancer and control samples for each of the protein enrichment methods (total, IgG, IgG depleted, PM-bound and PM-flow-through). Four glycans in whole plasma, including Hex5HexNAc4Fuc1, Hex5HexNAc5, Hex5HexNAc5Fuc1 and Hex6HexNAc5Fuc1Sia2, showed a significant (FDR <0.05) difference in peak area between cancer cases and controls (Table 2). The three biantennary neutral glycans, which are structurally closely related, were down regulated with lung cancer, while the sialylated triantennary glycan showed increased levels in cancer patients compared to controls. Four other glycans were observed to be statistically significant different in the IgG fraction: Hex3HexNAc4Fuc1, Hex5HexNAc5Sia1, Hex5HexNAc5Sia2 and Hex5HexNAc5Fuc1Sia2. Here, a truncated biantennary glycan is upregulated with lung cancer, while decreased levels were observed for the structurally closely related sialylated glycans containing a bisecting GlcNAc. The differences in the areas are depicted by box-whisker plots in Figure 4. Interestingly, the levels of the glycans that were altered significantly on IgG were not altered significantly in the total plasma analysis. This illustrates the complexity of biofluids such as plasma, where glycan levels on one protein might be increasing, while the levels on other proteins might be decreasing, thus washing-out the significance of certain glycan levels on individual proteins
Table 2.
Glycan compositions in the five different protein enrichment methods with a raw (unadjusted for FDR) p-value <0.05 in at least one of the enrichment strategies. Glycans with significance in FDR adjusted p-values are highlighted in bold.
| Glycan Composition s | Whole plasma | IgG | IgG depleted | PM-bound | PM-flow-through | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw P-value | FDR | Fold Change | Raw P-value | FDR | Fold Change | Raw P-value | FDR | Fold Change | Raw P-value | FDR | Fold Change | Raw P-value | FDR | Fold Change | |
| H10N2 | 0.221 | 0.53 | 1.149 | - | - | - | 0.251 | 0.698 | 1.114 | 0.88 | 0.942 | 1.058 | 0.006 | 0.201 | 1.371 |
| H3N2 | 0.01 | 0.112 | 0.882 | - | - | - | 0.81 | 0.913 | 1.024 | 0.065 | 0.278 | 1.458 | 0.877 | 0.953 | 1.008 |
| H3N3F1 | 0.046 | 0.235 | 1.206 | 0.309 | 0.467 | 1.173 | 0.57 | 0.851 | 1.174 | - | - | - | 0.23 | 0.548 | 1.3 |
| H3N4F1 | 0.168 | 0.473 | 1.233 | 0.004 | 0.041 | 1.281 | 0.884 | 0.962 | 1.224 | - | - | - | 0.025 | 0.203 | 1.449 |
| H4N3 | 0.05 | 0.235 | 0.914 | 0.28 | 0.467 | 0.904 | 0.224 | 0.669 | 0.866 | - | - | - | 0.598 | 0.818 | 0.923 |
| H4N3F1 | 0.01 | 0.112 | 0.826 | 0.612 | 0.699 | 1.068 | 0.584 | 0.851 | 0.899 | - | - | - | 0.252 | 0.562 | 0.697 |
| H4N3F1S1 | 0.892 | 0.91 | 0.968 | 0.025 | 0.143 | 3.292 | 0.382 | 0.718 | 1.082 | - | - | - | 0.487 | 0.753 | 1.098 |
| H4N4 | 0.011 | 0.112 | 0.848 | 0.114 | 0.286 | 0.822 | 0.303 | 0.716 | 0.901 | - | - | - | 0.083 | 0.341 | 0.791 |
| H4N5 | 0.009 | 0.112 | 0.853 | 0.555 | 0.672 | 0.897 | 0.643 | 0.866 | 0.878 | - | - | - | 0.596 | 0.818 | 1.017 |
| H4N5F1 | 0.026 | 0.174 | 0.872 | 0.315 | 0.467 | 0.94 | 0.744 | 0.908 | 1.059 | 0.513 | 0.855 | 1.011 | 0.649 | 0.844 | 0.992 |
| H5N3 | 0.017 | 0.134 | 0.872 | - | - | - | 0.132 | 0.547 | 0.814 | - | - | - | 0.338 | 0.646 | 0.772 |
| H5N4 | 0.046 | 0.235 | 0.924 | 0.037 | 0.151 | 0.733 | 0.004 | 0.267 | 0.841 | - | - | - | 0.013 | 0.203 | 0.844 |
| H5N4S2 | 0.341 | 0.597 | 1.079 | 0.308 | 0.467 | 1.084 | 0.045 | 0.397 | 1.223 | 0.294 | 0.649 | 0.761 | 0.879 | 0.953 | 1 |
| H5N4F1 | 0.002 | 0.036 | 0.703 | 0.041 | 0.151 | 0.834 | 0.024 | 0.315 | 0.746 | - | - | - | 0.012 | 0.203 | 0.753 |
| H5N4F1S1 | 0.013 | 0.116 | 0.886 | 0.11 | 0.286 | 0.882 | 0.31 | 0.716 | 0.898 | 0.942 | 0.942 | 1.119 | 0.128 | 0.38 | 0.92 |
| H5N5 | 0.001 | 0.018 | 0.776 | 0.059 | 0.18 | 0.726 | 0.03 | 0.315 | 0.776 | - | - | - | 0.017 | 0.203 | 0.819 |
| H5N5S1 | 0.059 | 0.247 | 0.867 | <0.001 | 0.008 | 0.699 | 0.011 | 0.315 | 0.789 | 0.071 | 0.278 | 1.725 | 0.086 | 0.341 | 0.87 |
| H5N5S2 | 0.525 | 0.741 | 1.179 | <0.001 | 0.008 | 0.766 | 0.476 | 0.797 | 1.396 | - | - | - | 0.604 | 0.818 | 0.967 |
| H5N5F1 | <0.001 | 0.016 | 0.793 | 0.041 | 0.151 | 0.79 | 0.346 | 0.716 | 0.885 | 0.054 | 0.278 | 1.524 | 0.193 | 0.501 | 0.897 |
| H5N5F1S1 | 0.186 | 0.473 | 0.908 | 0.008 | 0.066 | 0.812 | 0.175 | 0.639 | 1.412 | 0.058 | 0.278 | 1.387 | 0.445 | 0.727 | 1.006 |
| H5N5F1S2 | 0.267 | 0.551 | 0.896 | 0.002 | 0.029 | 0.809 | 0.329 | 0.716 | 0.841 | 0.348 | 0.649 | 1.547 | 0.043 | 0.258 | 0.832 |
| H5N5F2S1 | 0.671 | 0.816 | 1.268 | - | - | - | 0.806 | 0.913 | 0.906 | - | - | - | 0.048 | 0.258 | 0.769 |
| H6N3 | 0.26 | 0.551 | 0.919 | - | - | - | 0.024 | 0.315 | 0.806 | - | - | - | 0.042 | 0.258 | 0.875 |
| H6N3F1 | 0.242 | 0.542 | 0.639 | - | - | - | 0.026 | 0.315 | 0.654 | - | - | - | - | - | - |
| H6N3F1S1 | 0.057 | 0.247 | 1.783 | 0.016 | 0.106 | 2.036 | 0.116 | 0.523 | 0.708 | - | - | - | - | - | - |
| H6N5F1S2 | <0.001 | 0.016 | 1.979 | 0.034 | 0.151 | 0.715 | 0.198 | 0.669 | 1.204 | - | - | - | 0.02 | 0.203 | 1.453 |
| H6N5F1S3 | - | - | - | 0.046 | 0.153 | 1.718 | 0.762 | 0.908 | 0.842 | - | - | - | 0.63 | 0.835 | 1.357 |
| H6N5F2S3 | - | - | - | - | - | - | - | - | - | - | - | - | 0.004 | 0.201 | 2.356 |
| H7N6F1S2 | 0.037 | 0.227 | 2.159 | - | - | - | - | - | - | - | - | - | 0.022 | 0.203 | 1.795 |
| H9N2 | 0.796 | 0.85 | 0.967 | 0.31 | 0.467 | 1.13 | 0.944 | 0.981 | 0.985 | 0.742 | 0.927 | 1.091 | 0.044 | 0.258 | 1.163 |
Figure 4. Four glycans in plasma and four glycans on IgG associate significantly with lung cancer.
Boxplots are depicted for Hex5HexNAc4Fuc1 (A), Hex5HexNAc5 (B), Hex5HexNAc5Fuc1 (C), Hex6HexNAc5Fuc1Sia2 (D), Hex3HexNAc4Fuc1 (E), Hex5HexNAc5Sia1 (F), Hex5HexNAc5Sia2 (G), Hex5HexNAc5Fuc1Sia2 (H). Differences in glycans signals in the upper row were observed in whole plasma, while the lower row reflects glycan changes observed on IgG. Key: square: N-acetylhexosamine, circle: hexose, red triangle: fucose, blue square: N-acetylglucosamine, green circle: mannose, yellow circle: galactose, purple diamond: sialic acid.
No significantly altered glycan levels were observed in the IgG depleted and PM-bound and flow-through samples, suggesting that PM enrichment strategies may have limited value in biomarker discovery. However, the absence of significant results may also be caused by the relatively low numbers of samples (20 cases and 20 controls) used in the study.
DISCUSSION
The presented work aims to compare the use of two different protein enrichment strategies: 1) IgG affinity capturing with Protein G and 2) the use of a non-specific hexapeptide library (Proteominer PM), with whole plasma N-glycan analysis to further investigate the potential of glycans in blood plasma as diagnostic biomarkers for lung cancer.
The glycosylation pattern of immunoglobulin G has been studied extensively (e.g. [27, 45–47]), however, not in relation to lung cancer. It is widely known that the N-glycan profile present on IgG differs dramatically compared to the total plasma or serum N-glycan profile, with the former having more smaller, neutral glycans and the latter containing more larger sialylated glycans. This same pattern was observed in this study (Figure 1). In contrast, the glycosylation profile of the enriched lower abundant proteins did not appear to be altered much compared to whole serum, which confirms previous reports using PM enrichment [36] as well as other protein depletion strategies [20, 31, 34] where N-glycan patterns on lower abundant proteins were reported to be similar to their total plasma or serum parent fluids. It is therefore proposed that the N-glycans attached to proteins in the bloodstream mostly reflect the glycosylation biosynthesis machinery of their site of production, e.g. plasma cells for IgG and liver cells for most non-immunoglobulins. The fact that the glycosylation changes observed in total plasma were mostly reflected by IgG glycosylation (Table 2) whereas the glycosylation pattern of the PM-bound fraction did not alter significantly with lung cancer support this hypothesis.
Four glycans in the whole plasma samples were shown to alter significantly with lung cancer (adenocarcinoma). The levels of three structurally closely related biantennary, non-sialylated glycans, of which two are highly likely to contain a bisecting GlcNAc are decreased with lung cancer, while the levels of a sialylated triantennary glycan is significantly increased. Similar results were obtained by Arnold et al. [16] who concluded that fractions containing tri-and tetra antennary structures with sialic acid were increased, while fractions with mostly biantennary structures were decreased in lung cancer [16]. Altered levels of individual glycan compositions in plasma may be caused by altered protein levels, but also altered glycosylation patterns on specific proteins, where the protein expression is not altered, but the extent and type of glycosylation does change. The importance and functions of the glycans are highly dependent on the protein to which they are attached. Current technology does not allow immediate identification of the proteins to which the altered glycans are attached. However, it has been reported that in healthy individuals the triantennary glycan is associated to the high abundant plasma proteins haptoglobin [48] and alpha-1-antitrypsin [49], which are produced in the liver while the biantennary structures have been found on IgG [50] and IgA [33], two proteins produced by plasma cells. Small amounts of Hex5HexNAc4Fuc1 have also been found on alpha-1-antitrypsin [49], which is a liver protein. These findings indicate that the glycosylation machinery in both liver cells as well as plasma cells is altered, but studies towards the functionality of the altered glycosylation remain difficult.
Therefore, it is very reassuring that we could for the first time report that the levels of four glycans on the specific protein IgG were significantly altered with IgG. It is long known that the level of galactosylation of the Fc portion of IgG decreases with age [51, 52] and rheumatoid arthritis [45, 53]. More recently, altered IgG Fc glycosylation was observed in other autoimmune diseases (e.g. [47, 54]) and cancer (e.g. [32, 50]). The effects of the glycosylation on the effectiveness of IgG molecules have been studied extensively, especially in the light of the development of antibody drugs [55]. It is widely known that aglycosylated antibodies are considerably less functional [56]; more recently, studies indicate that absence of the core fucose increases binding of the IgG to the Fc gamma III receptor and antibody-dependent cellular cytotoxicity [57]. While there is much less evidence for the impact of the level of galactosylation on IgG, and their results are not univocal, one study reported lower Fc receptor binding of non-galactosylated IgG relative to IgG with higher levels of galactosylation [58], indicating that the altered glycosylation profiles found in this study might have biological implications, such as impaired binding of the IgG molecules to the Fc receptor in cancer patients. Further studies into the functionality of IgG in cancer patients will be necessary to further investigate this hypothesis.
This feasibility study was performed on a relatively small sample set consisting of 20 lung cancer cases and 20 matched controls. The lung cancer cases were mostly late stage NSCLC so the biomarkers identified may only be suitable for late stage and not early stage disease. It will be necessary to conduct these types of studies in samples obtained from patients with early stage disease. Further, much larger studies will then be needed to validate our data and evaluate the capability of the aberrant glycans as candidate biomarkers for lung cancer and their suitability for clinical use. Moreover, the analysis was performed at the level of glycan compositions, but due to the different linkage isomers, higher specificity may be reached when compound specific analysis of larger sample sets is feasible in the near future. Such studies will also assist to address the sensitivity and specificity of N-glycans as biomarkers for lung cancer.
Overall, this study shows that the N-glycosylation pattern of blood-derived proteins is altered in patients with lung cancer. The changes are most prevalent on IgG, but the glycosylation pattern on liver-derived proteins is also affected likely demonstrating a systemic response to tumor growth. For future biomarker discovery studies, the additional analysis of protein specific N- and potentially O-glycosylation profiles will be highly valuable, while the use of non-specific protein enrichment methods was not observed to yield additional value and may instead further complicate the analysis.
CLINICAL RELEVANCE.
Lung cancer is currently the most deathly type of cancer in the Unites States and early detection increases chances of survival dramatically. There is currently no FDA approved test available that could aid in screening and early detection of lung cancer, and therefore there is a need to identify biomarkers to develop such a test. Protein N-glycosylation patterns are know to alter in cancer tissue, and altered glycosylation patterns have also been observed in serum of cancer patients. However, the specificity of these markers remains unclear, especially since serum and plasma are complex protein mixtures, which are dominated by a few high abundance proteins. It is therefore highly likely that more specific markers can be obtained using protein enrichment techniques.
In this study, two protein enrichment techniques were evaluated for their efficacy for future biomarker studies for glycan-based biomarker discovery in clinical samples. It could be shown that glycosylation patterns are altered in patients with lung cancer. Changes were also prevalent on isolated immunoglobulin IgG, while glycosylation patterns of liver-derived proteins were also affected. This indicates that the additional analysis of N-glycosylation of specific proteins will likely yield more valuable information than non-specific protein enrichment methods in future biomarker studies.
Acknowledgments
The authors are thankful for the funding provided by the National Institutes of Health (R21 CA135240, HD061923, HD059127, R01 GM049077, and UL1 TR 000002), the Department of Defense (CDMRP LCRP W81XWH1010635), the Tobacco Related Disease Research Program and the LUNGevity Foundation. The authors also would like to acknowledge the assistance of Jon Ladd (FHCRC) from the Hanash laboratory for his help with the acquisition of the blood samples.
Footnotes
The authors declare no conflict of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Hassanein M, Callison JC, Callaway-Lane C, Aldrich MC, et al. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev Res. 2012;5:992–1006. doi: 10.1158/1940-6207.CAPR-11-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 4.Gu J, Isaji T, Xu Q, Kariya Y, et al. Potential roles of N-glycosylation in cell adhesion. Glycoconj J. 2012;29:599–607. doi: 10.1007/s10719-012-9386-1. [DOI] [PubMed] [Google Scholar]
- 5.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Mehta A, Norton P, Liang H, Comunale MA, et al. Increased Levels of Tetra-antennary N-Linked Glycan but Not Core Fucosylation Are Associated with Hepatocellular Carcinoma Tissue. Cancer Epidemiol Biomarkers Prev. 2012;21:925–933. doi: 10.1158/1055-9965.EPI-11-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balog CI, Stavenhagen K, Fung WL, Koeleman CA, et al. N-glycosylation of colorectal cancer tissues: a liquid chromatography and mass spectrometry-based investigation. Mol Cell Proteomics. 2012;11:571–585. doi: 10.1074/mcp.M111.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asada M, Furukawa K, Segawa K, Endo T, Kobata A. Increased expression of highly branched N-glycans at cell surface is correlated with the malignant phenotypes of mouse tumor cells. Cancer Res. 1997;57:1073–1080. [PubMed] [Google Scholar]
- 9.de Leoz ML, Young LJ, An HJ, Kronewitter SR, et al. High-mannose glycans are elevated during breast cancer progression. Mol Cell Proteomics. 2011;10:M110 002717. doi: 10.1074/mcp.M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruhaak LR, Uh HW, Beekman M, Hokke CH, et al. Plasma protein N-glycan profiles are associated with calendar age, familial longevity and health. J Proteome Res. 2011;10:1667–1674. doi: 10.1021/pr1009959. [DOI] [PubMed] [Google Scholar]
- 11.Blomme B, Francque S, Trepo E, Libbrecht L, et al. N-glycan based biomarker distinguishing non-alcoholic steatohepatitis from steatosis independently of fibrosis. Dig Liver Dis. 2012;44:315–322. doi: 10.1016/j.dld.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Alley WR, Jr, Vasseur JA, Goetz JA, Svoboda M, et al. N-linked glycan structures and their expressions change in the blood sera of ovarian cancer patients. J Proteome Res. 2012;11:2282–2300. doi: 10.1021/pr201070k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hua S, An HJ, Ozcan S, Ro GS, et al. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst. 2011;136:3663–3671. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saldova R, Fan Y, Fitzpatrick JM, Watson RW, Rudd PM. Core fucosylation and alpha2-3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology. 2011;21:195–205. doi: 10.1093/glycob/cwq147. [DOI] [PubMed] [Google Scholar]
- 15.Hoagland LF, Campa MJ, Gottlin EB, Herndon JE, Patz EF., Jr Haptoglobin and posttranslational glycan-modified derivatives as serum biomarkers for the diagnosis of nonsmall cell lung cancer. Cancer. 2007;110:2260–2268. doi: 10.1002/cncr.23049. [DOI] [PubMed] [Google Scholar]
- 16.Arnold JN, Saldova R, Galligan MC, Murphy TB, et al. Novel glycan biomarkers for the detection of lung cancer. J Proteome Res. 2011;10:1755–1764. doi: 10.1021/pr101034t. [DOI] [PubMed] [Google Scholar]
- 17.Blixt O, Bueti D, Burford B, Allen D, et al. Autoantibodies to aberrantly glycosylated MUC1 in early stage breast cancer are associated with a better prognosis. Breast Cancer Res. 2011;13:R25. doi: 10.1186/bcr2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Faca V, Hanash S. Mining the plasma proteome for disease applications across seven logs of protein abundance. J Proteome Res. 2011;10:46–50. doi: 10.1021/pr101052y. [DOI] [PubMed] [Google Scholar]
- 20.Lee A, Nakano M, Hincapie M, Kolarich D, et al. The lectin riddle: glycoproteins fractionated from complex mixtures have similar glycomic profiles. OMICS. 2010;14:487–499. doi: 10.1089/omi.2010.0075. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z, Hancock WS. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. J Chromatogr A. 2004;1053:79–88. [PubMed] [Google Scholar]
- 22.Lee A, Kolarich D, Haynes PA, Jensen PH, et al. Rat liver membrane glycoproteome: enrichment by phase partitioning and glycoprotein capture. J Proteome Res. 2009;8:770–781. doi: 10.1021/pr800910w. [DOI] [PubMed] [Google Scholar]
- 23.Goldstein IJ, Hollerman CE, Merrick JM. Protein-Carbohydrate Interaction. I. The Interaction of Polysaccharides with Concanavalin A. Biochim Biophys Acta. 1965;97:68–76. doi: 10.1016/0304-4165(65)90270-9. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein IJ, Hollerman CE, Smith EE. Protein-Carbohydrate Interaction. Ii. Inhibition Studies on the Interaction of Concanavalin a with Polysaccharides. Biochemistry. 1965;4:876–883. doi: 10.1021/bi00881a013. [DOI] [PubMed] [Google Scholar]
- 25.Nagata Y, Burger MM. Wheat germ agglutinin. Molecular characteristics and specificity for sugar binding. J Biol Chem. 1974;249:3116–3122. [PubMed] [Google Scholar]
- 26.Iskratsch T, Braun A, Paschinger K, Wilson IB. Specificity analysis of lectins and antibodies using remodeled glycoproteins. Anal Biochem. 2009;386:133–146. doi: 10.1016/j.ab.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Wuhrer M, Stam JC, van de Geijn FE, Koeleman CA, et al. Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics. 2007;7:4070–4081. doi: 10.1002/pmic.200700289. [DOI] [PubMed] [Google Scholar]
- 28.van de Geijn FE, Wuhrer M, Selman MH, Willemsen SP, et al. Immunoglobulin G galactosylation and sialylation are associated with pregnancy-induced improvement of rheumatoid arthritis and the postpartum flare: results from a large prospective cohort study. Arthritis Res Ther. 2009;11:R193. doi: 10.1186/ar2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruhaak LR, Uh HW, Beekman M, Koeleman CA, et al. Decreased levels of bisecting GlcNAc glycoforms of IgG are associated with human longevity. PLoS ONE. 2010;5:e12566. doi: 10.1371/journal.pone.0012566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pucic M, Knezevic A, Vidic J, Adamczyk B, et al. High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol Cell Proteomics. 2011;10:M111 010090. doi: 10.1074/mcp.M111.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bones J, Mittermayr S, O’Donoghue N, Guttman A, Rudd PM. Ultra performance liquid chromatographic profiling of serum N-glycans for fast and efficient identification of cancer associated alterations in glycosylation. Anal Chem. 2010;82:10208–10215. doi: 10.1021/ac102860w. [DOI] [PubMed] [Google Scholar]
- 32.Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, et al. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17:1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 33.Wada Y, Tajiri M, Ohshima S. Quantitation of saccharide compositions of O-glycans by mass spectrometry of glycopeptides its application to rheumatoid arthritis. J Proteome Res. 2010;9:1367–1373. doi: 10.1021/pr900913k. [DOI] [PubMed] [Google Scholar]
- 34.Bereman MS, Muddiman DC. The effects of abundant plasma protein depletion on global glycan profiling using nanoLC FT-ICR mass spectrometry. Anal Bioanal Chem. 2010;396:1473–1479. doi: 10.1007/s00216-009-3368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boschetti E, Righetti PG. The art of observing rare protein species in proteomes with peptide ligand libraries. Proteomics. 2009;9:1492–1510. doi: 10.1002/pmic.200800389. [DOI] [PubMed] [Google Scholar]
- 36.Huhn C, Ruhaak LR, Wuhrer M, Deelder AM. Hexapeptide library as a universal tool for sample preparation in protein glycosylation analysis. J Proteomics. 2012;75:1515–1528. doi: 10.1016/j.jprot.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 37.Ruhaak LR, Deelder AM, Wuhrer M. Oligosaccharide analysis by graphitized carbon liquid chromatography-mass spectrometry. Anal Bioanal Chem. 2009;394:163–174. doi: 10.1007/s00216-009-2664-5. [DOI] [PubMed] [Google Scholar]
- 38.Wuhrer M. Glycomics using mass spectrometry. Glycoconj J. 2012 doi: 10.1007/s10719-012-9376-3. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu CS, Ninonuevo MR, Clowers BH, Perkins PD, et al. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9:1939–1951. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mechref Y, Hu Y, Garcia A, Zhou S, et al. Defining putative glycan cancer biomarkers by MS. Bioanalysis. 2012;4:2457–2469. doi: 10.4155/bio.12.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hua S, Lebrilla C, An HJ. Application of nano-LC-based glycomics towards biomarker discovery. Bioanalysis. 2011;3:2573–2585. doi: 10.4155/bio.11.263. [DOI] [PubMed] [Google Scholar]
- 42.Kronewitter SR, de Leoz ML, Peacock KS, McBride KR, et al. Human serum processing and analysis methods for rapid and reproducible N-glycan mass profiling. J Proteome Res. 2010;9:4952–4959. doi: 10.1021/pr100202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Packer NH, Lawson MA, Jardine DR, Redmond JW. A general approach to desalting oligosaccharides released from glycoproteins. Glycoconj J. 1998;15:737–747. doi: 10.1023/a:1006983125913. [DOI] [PubMed] [Google Scholar]
- 44.Kronewitter SR, An HJ, de Leoz ML, Lebrilla CB, et al. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics. 2009;9:2986–2994. doi: 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parekh RB, Dwek RA, Sutton BJ, Fernandes DL, et al. Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature. 1985;316:452–457. doi: 10.1038/316452a0. [DOI] [PubMed] [Google Scholar]
- 46.Rudd PM, Guile GR, Kuster B, Harvey DJ, et al. Oligosaccharide sequencing technology. Nature. 1997;388:205–207. doi: 10.1038/40677. [DOI] [PubMed] [Google Scholar]
- 47.Huhn C, Selman MH, Ruhaak LR, Deelder AM, Wuhrer M. IgG glycosylation analysis. Proteomics. 2009;9:882–913. doi: 10.1002/pmic.200800715. [DOI] [PubMed] [Google Scholar]
- 48.Pompach P, Chandler KB, Lan R, Edwards N, Goldman R. Semi-Automated Identification of N-Glycopeptides by Hydrophilic Interaction Chromatography, nano-Reverse-Phase LC-MS/MS, and Glycan Database Search. J Proteome Res. 2012 doi: 10.1021/pr201183w. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolarich D, Weber A, Turecek PL, Schwarz HP, Altmann F. Comprehensive glyco-proteomic analysis of human alpha1-antitrypsin and its charge isoforms. Proteomics. 2006;6:3369–3380. doi: 10.1002/pmic.200500751. [DOI] [PubMed] [Google Scholar]
- 50.Kodar K, Stadlmann J, Klaamas K, Sergeyev B, Kurtenkov O. Immunoglobulin G Fc N-glycan profiling in patients with gastric cancer by LC-ESI-MS: relation to tumor progression and survival. Glycoconj J. 2012;29:57–66. doi: 10.1007/s10719-011-9364-z. [DOI] [PubMed] [Google Scholar]
- 51.Shikata K, Yasuda T, Takeuchi F, Konishi T, et al. Structural changes in the oligosaccharide moiety of human IgG with aging. Glycoconj J. 1998;15:683–689. doi: 10.1023/a:1006936431276. [DOI] [PubMed] [Google Scholar]
- 52.Parekh R, Roitt I, Isenberg D, Dwek R, Rademacher T. Age-related galactosylation of the N-linked oligosaccharides of human serum IgG. J Exp Med. 1988;167:1731–1736. doi: 10.1084/jem.167.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parekh RB, Roitt IM, Isenberg DA, Dwek RA, et al. Galactosylation of IgG associated oligosaccharides: reduction in patients with adult and juvenile onset rheumatoid arthritis and relation to disease activity. Lancet. 1988;1:966–969. doi: 10.1016/s0140-6736(88)91781-3. [DOI] [PubMed] [Google Scholar]
- 54.Selman MHJ, Niks EH, Titulaer MJ, Verschuuren JJGM, et al. IgG Fc N-Glycosylation Changes in Lambed-Eaton Myasthenic Syndrome and Myasthenia Gravis. J Proteome Res. 2011;10:143–152. doi: 10.1021/pr1004373. [DOI] [PubMed] [Google Scholar]
- 55.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 2009;8:226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 56.Walker MR, Lund J, Thompson KM, Jefferis R. Aglycosylation of human IgG1 and IgG3 monoclonal antibodies can eliminate recognition by human cells expressing Fc gamma RI and/or Fc gamma RII receptors. Biochem J. 1989;259:347–353. doi: 10.1042/bj2590347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeitlin L, Pettitt J, Scully C, Bohorova N, et al. Enhanced potency of a fucose-free monoclonal antibody being developed as an Ebola virus immunoprotectant. Proc Natl Acad Sci U S A. 2011;108:20690–20694. doi: 10.1073/pnas.1108360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumpel BM, Rademacher TW, Rook GA, Williams PJ, Wilson IB. Galactosylation of human IgG monoclonal anti-D produced by EBV-transformed B-lymphoblastoid cell lines is dependent on culture method and affects Fc receptor-mediated functional activity. Hum Antibodies Hybridomas. 1994;5:143–151. [PubMed] [Google Scholar]