Abstract
The mature neocortex is a unique six-layered mammalian brain region. It is composed of morphologically and functionally distinct subpopulations of primary projection neurons that form complex circuits across the central nervous system. The precisely-timed generation of projection neurons from neural stem cells governs their differentiation, postmitotic specification, and signaling, and is critical for cognitive and sensorimotor ability. Developmental perturbations to the birthdate, location, and connectivity of neocortical neurons are observed in neurological and psychiatric disorders. These facts are highlighting the importance of the precise spatiotemporal development of the neocortex regulated by intricate transcriptional, but also complex post-transcriptional events. Indeed, mRNA transcripts undergo many post-transcriptional regulatory steps before the production of functional proteins, which specify neocortical neural stem cells and subpopulations of neocortical neurons. Therefore, particular attention is paid to the differential post-transcriptional regulation of key transcripts by RNA-binding proteins, including splicing, localization, stability, and translation. We also present a transcriptome screen of candidate molecules associated with post-transcriptional mRNA processing that are differentially expressed at key developmental time points across neocortical prenatal neurogenesis.
Introduction
The adult neocortex is the central circuit of consciousness, complex cognition, language and the coordination of voluntary motor activity in mammals (Weiler et al., 2008, Lui et al., 2011). Throughout mammalian evolution, the neocortex is the brain region that has exhibited the greatest expansion in mass relative to body weight (i.e., encephalization) (Shultz and Dunbar, 2010). In this way, the neocortex can be thought of as the evolutionary foundation for cognitive advances, including the uniquely human “theory of mind” and language. However, with these advances, human-specific ailments such as schizophrenia, autism spectrum disorders, Parkinson’s disease, Alzheimer’s disease, and Amyotrophic Lateral Sclerosis have also developed (Garey, 2010, Wegiel et al., 2010, Morgen et al., 2011, Ozdinler et al., 2011, Rapoport and Nelson, 2011, Yang et al., 2011). Therefore, understanding the molecular and cellular mechanisms underlying neocortical formation, maintenance, and dysfunction is critical not only for furthering basic neuroscience knowledge of brain development and architecture but also for better understanding neuropsychiatric disorders. In addition, these efforts may improve current therapeutic approaches to these neocortical ailments.
Neocortical function relies on precise interactions among an array of cell types, which can be broadly divided into epithelial cells, glia and neurons. Neocortical neurons belong to two main classes: interneurons and primary projection neurons. Interneurons are inhibitory GABAergic cells that have short processes forming local circuits. Interneurons migrate tangentially into the developing neocortex from the lateral, medial and caudal ganglionic eminences and can be delineated from projection neurons based on their morphology and expression of markers such as parvalbumin, somatostatin, vasoactive intestinal peptide, neuropeptide Y and cholecystokinin (Corbin et al., 2001, Tanaka and Nakajima, 2012, DeFelipe et al., 2013, van den Berghe et al., 2013).
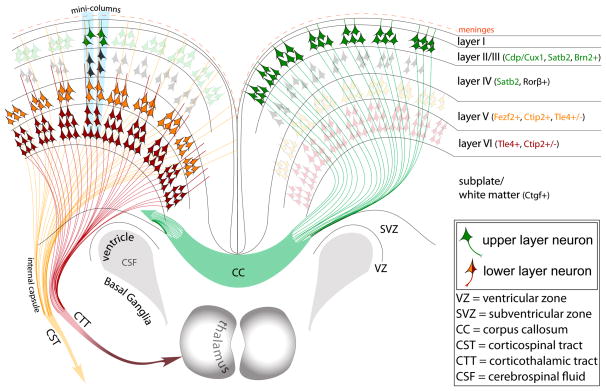
By contrast, primary projection neurons are excitatory glutamatergic cells which carry out the mainstay of the signaling in the neocortex and extend processes over long distances. Importantly, of all of the neurons that populate the neocortex, 75–85% are excitatory projection neurons. The earliest systematic investigation of the neocortex by Santiago Ramón y Cajal revealed that these neurons have characteristic morphological features, including a pyramidal-shaped cell body, many basal dendritic processes, a single apical dendrite oriented toward the pial surface of the neocortex that gives rise to a variable number of oblique branches, and a single axon that usually stems from the base of the cell body or proximal parts of the basal dendrites (Ramón y Cajal S., 1988). Later seminal studies demonstrated histological differences in the density and size of neocortical cell bodies, which define what are now recognized as six distinct layers (Caviness, 1975, Ramón y Cajal S., 1988, Brodmann K., 2006, Hevner, 2006). The target of each projection neuron is related to its position within the six neocortical layers (I–VI) (Figure 1). Lower-layer (V–VI) neurons mainly project subcortically, with axons often terminating in the thalmaus, brain stem and spinal cord, although numerous collaterals for intermediate targets also exist (Floeter and Jones, 1985, Zhang and Deschenes, 1997). Upper-layer (II–IV) neurons exclusively project intracortically, either within the ipsilateral hemisphere or reaching the contralateral hemisphere via the corpus callosum.
Figure 1. A simplified schematic of the postnatal organization and projections of neocortical projection neurons.
The neocortex is highly organized in both horizontal and vertical dimensions. Horizontally, six layers are defined by highly organized subpopulations of glutamatergic projection neurons, which represent approximately 85% of all neocortical neurons. These subpopulations of projection neurons are characterized by specific molecular identities, dendritic morphologies and terminal targets corresponding to each layer. Projection neurons that are born during later stages of prenatal neurogenesis will be predominantly placed in upper layers II–IV (green neurons). These neurons express specific transcription factors like CDP/Cux1, and project solely intracortically forming the corpus callosum that connects the two hemispheres. However, there is also a smaller portion of intracortically projecting neurons placed in lower layers too (not shown). In contrast, earlier born projection neurons will be placed in lower layers V–VI (orange and red neurons). These subopulations will express transcription factors like FEZF2, and will project subcortically to form long range tracts across the central nervous system like the corticothalamic tract (CTT) originating mainly from layer 6, and somewhat from layer 5, or corticospinal tract (CST) originating solely from layer 5. Within the subventricular zone (SVZ) of the corticostriatal junction, adult progenitors are found giving rise to olfactory cortex neurons.
The delineation of the neocortex into six layers arose from neuroanatomical and electrophysiological evidence. More recent work, initially in rodents, has defined subgroups of neurons based on of the expression of transcription factors (TFs) (Molyneaux et al., 2007, Kriegstein and Alvarez-Buylla, 2009). For example, subcortically projecting neurons selectively express Ctip2, Fezf2, and Tle4, whereas intracortically projecting neurons selectively express Cdp/Cux1 and Satb2 (Hevner et al., 2006, Molyneaux et al., 2007). These findings were recently extended to human and non-human primates, with gestational and postnatal investigations showing that the specificity of TF expression in neocortical projection neurons is at least partially conserved across species. As particular TFs correspond to differences in dendritic complexity and axonal projections and, hence, the function of distinct neuronal subpopulations (Kwan et al., 2012b), there are continuing efforts to identify additional markers of neuron subtypes. Moreover, ongoing studies continue to elucidate the molecular and cellular mechanisms underlying TF specification of neocortical neuron subpopulations.
The remaining text of this ForeFront Review will be dedicated to reviewing the current understanding of neocortical development with a focus on neural stem cells, projection neurons, the use of state-of-the-art transcriptome analyses and the emerging field of the role of posttranscriptional processing steps.
Neural Stem Cells in the Developing Neocortex
All functionally-distinct subgroups of neocortical projection neurons are generated prenatally through a highly-orchestrated set of developmental processes. Projection neurons emerge from a pool of neural stem cell progenitors called radial glia (RG) that divide at the ventricular zone (VZ) surface (Figure 2). Lower-layer, subcortically projecting neurons are born first, followed by upper- layer, intracortically projecting neurons. The laminar organization of newborn cells results in the arrangement of distinct columns of functionally related neurons spanning different layers (Mountcastle et al., 1957, Hubel and Wiesel, 1962). According to the radial unit hypothesis (Rakic, 1988), the cytoarchitecture of these columns is the outcome of neuroblasts migrating along basal RG processes from the VZ of the prenatal neocortex. This hypothesis was later confirmed using retroviral green fluorescent protein (GFP) transfection, allowing the tracking of daughter cells from dividing RG (Kornack and Rakic, 1995). Thus, the organization of the mature neocortex arises prenatally, where prenatal neurogenesis is believed to produce all of the diverse subgroups of neocortical projection neurons (Casanova and Trippe, 2006, Kriegstein and Alvarez-Buylla, 2009, Kwan et al., 2012b). Therefore, the basis for advanced neocortical functions during adulthood is largely determined by the complex spatiotemporal control of changes in gene expression early in life, starting from neural stem cells (NSC).
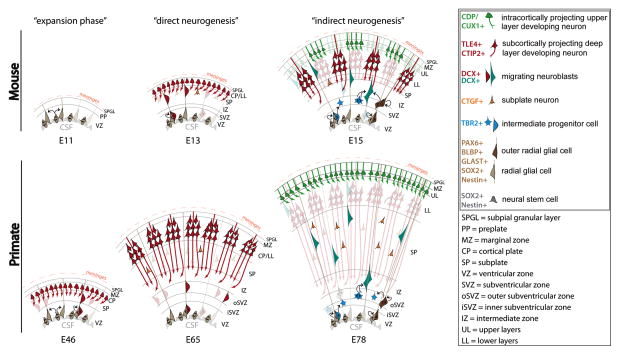
Figure 2. Schematic of distinct stages of neocortical neurogenesis in developing mouse and primate neocortices.
The first “phase” of neocorticogenesis in mouse and primates is characterized by symmetric divisions of neural stem cells called neuroepithelium cells (NEC), which amplifies the number of neocortical progenitors at the ventricular zone (VZ) surface (left panels). This initial phase is accordingly called the “expansion phase”. NECs will then transition into a different lineage of neocortical neural stem cells called radial glia (RG), which divide asymmetrically and first predominantly produce neuronal progeny. This phase was named “direct neurogenesis” (middle panels). As the neurogenic phase progresses, RG continue to undergo a series of asymmetric divisions, but they predominantly produce another progenitor subtype, intermediate progenitor cells (IPCs) and outer radial glia (oRG) (right panels). IPCs and oRG will divide in the subventricular zone (SVZ), which in primates is divided into inner (iSVZ) and outer (oSVZ) portions. Importantly, IPCs terminally divide symmetrically and produce at least two neural progenies. However, oRG will self renew and give rise to neural progenies and IPCs. In this way, both IPCs and oRGs amplify the output of RG, and thus, this later stage of neurogenesis was named “indirect neurogenesis”. These progressive changes in differentiation of neocortical neural stem cells define the birth of distinct subpopulations of projection neurons. Deep layer neurons that will project subcortically into thalamus, brain stem and spinal cord will be born before upper layer intracortically projecting neurons. (Adapted from Angevine and Sidman, 1961, Rakic, 1974, Smart et al., 2002, and Molnár et al., 2006).
As recent molecular and cellular work on NSCs has primarily used mice because of their amenability to genetic manipulation, the embryonic time points mentioned here are specific to mouse neocorticogenesis. NSCs share characteristics of other stem cells, such as self renewal and pluripotency, but have diverse progeny ranging from other types of progenitors to neurons, glia and ependymal cells. However, neocortical NSCs are highly polarized cells with basal and apical processes that span the neocortical wall and attach to tissue surfaces. During the prenatal period, neocortical NSC nuclei undergo a unique process called interkinetic nuclear migration (INM), during which nuclei move toward and away from the lateral ventricle along radial processes. During INM, NSC nuclei are exposed to extracellular cues that may be proliferative, neurogenic, or gliagenic (Taverna and Huttner, 2010, Kosodo, 2012). In this view, INM may influence NSC fate and lead to pseudostratification, a characteristic feature of proliferative region of the neocortex (Taverna and Huttner, 2010). Importantly, INM motions correspond with NSC cycle stages; M-phase and cytokinesis occur when the nucleus approaches the apical epithelial surface, and S-phase and DNA synthesis occur when the nucleus moves away from the ventricle.
The functional significance of INM is advancing rapidly. Recent studies show that disruption of this process is associated with aberrations in NSC cycling and that INM may influence the fate of dividing progenitors (Ueno et al., 2006, Taverna and Huttner, 2010, Yang et al., 2012). Furthermore, species-specific differences in NSC cycle length suggest that INM is involved in evolutionary changes in neocortical neurogenesis (Kornack and Rakic, 1998, Breunig et al., 2011). Finally, abnormalities in INM are implicated in human developmental disorders like lissencephaly (Hatten, 2005). Therefore, regulation of RG cell cycle and INM during neocorticogenesis is important for maintaining the progenitor population and influencing the products of their divisions.
Neural Stem Cell Lineages in the Developing Neocortex
The earliest phase of neocorticogenesis begins with the proliferation of NSCs lining the lateral ventricle of the dorsal telencephalon (Figure 2). As the origin of these cells is epithelial, the earliest lineage of these progenitors is known as neuroepithelial cells (NECs), which are characterized by the expression of Nestin, Prominin-1 (CD133), and ZO-1 (Committee, 1970, Bystron et al., 2008). NECs maintain contact with both the pial (basal) surface of the developing neocortex and the apical epithelial lining of the lateral ventricle via radial processes that progressively elongate throughout neocorticogenesis. NECs of the dorsal telencephalon proliferate prior to embryonic day 10 (E10) and undergo multiple rounds of symmetric division, producing two daughter cells per cycle that expand the pool of NSCs for later neurogenesis (Kriegstein and Alvarez-Buylla, 2009). This phase is known as the “expansion phase” (Galli et al., 2002, Bishop et al., 2003, Noctor et al., 2004), which later transitions into the neurogenic phase of neocortical development. Importantly, only those NECs occupying the dorsal telencephalon region of the lateral ventricle cavity will give rise to later lineages of NSCs that ultimately produce distinct subpopulations of neocortical projection neurons (Sidman et al., 1959).
At approximately E10.5–12, while Nestin is still expressed but expression of CD133 is declining, neocortical NSCs begin to express markers similar to those of glial cells, such as the glutamate aspartate transporter (GLAST) and brain lipid binding protein (BLBP) (Rakic, 2003, Molyneaux et al., 2007, Kriegstein and Alvarez-Buylla, 2009). Although they maintain a polarized morphology, their nomenclature and the products of their division change; RG progenitors of NSC origin begin to undergo asymmetric divisions, producing one daughter self-renewing RG and one postmitotic neuron. In the earliest stages of neurogenesis, the postmitotic product of these divisions predominantly migrates directly into the cortical plate (CP)through an elegantly defined series of four phases (Noctor et al., 2004). Postmitotic neuroblasts derived from RG initially migrate rapidly along radial processes basally into the subventricular zone (SVZ) adjacent to the VZ, where they pause for approximately 24 hours. Progeny then undergo retrograde migration apically toward the VZ and finally turn back toward the CP. As these progeny remain in the CP without further division, this phase of neocorticogenesis is called “direct neurogenesis.” The earliest born neurons project subcortically and occupy the deepest neocortical layer (VI), whereas subsequently born neurons migrate past the deepest layer and into the more superficial layer (Vb). Interestingly, postmigratory CP neurons are positioned in a way that they possibly split a pre-existing matrix structure called the preplate into the basal marginal zone, containing the most specialized layer of Cajal Retzius cells, and the subplate, a monolayer of cells below the CP (Meyer, 2010, Nichols and Olson, 2010) (for review, see (Kostovic I, 1990, Allendoerfer and Shatz, 1994).
After the formation of the deep neocortical layers predominantly through direct neurogenesis, a new phase of “indirect neurogenesis” begins to take place (Figure 2). This occurs in mice around E14.5, when asymmetric divisions of RG at the ventricular surface begin to predominantly produce a specialized cell subtype known as an intermediate progenitor cell (IPC) or basal progenitor cell (Noctor et al., 2004). Some controversy about the contribution of IPCs to neocorticogenesis exists, however, as there is evidence that IPC progeny may contribute to all neocortical layers (Pontious et al., 2008, Kowalczyk et al., 2009) and not just superficial layers. Nevertheless, IPCs migrate away from the proliferative VZ and populate the adjacent SVZ. There, they undergo symmetric divisions giving rise to at least two postmitotic neuroblasts that will become part of the superficial neocortical layers (II–IV) and project intracortically (Kriegstein and Alvarez-Buylla, 2009). In this manner, IPCs serve to amplify the output of a single RG.
IPCs can be distinguished from RG not only by their position and division type and final product but also by their morphology and molecular identity (Pontious et al., 2008). IPCs have a multipolar morphology and maintain no connection with either the pial or epithelial surface. Furthermore, transcriptional programming of the two stem cell populations is mutually exclusive; RG express Sox2 and Pax6, whereas IPCs express Tbr2, which is essential for IPC formation and maintenance (Englund et al., 2005, Pontious et al., 2008, Sessa et al., 2008).
After the formation of the most superficial neocortical layers (II/III), neurogenesis ceases and the SVZ becomes less populated, reducing in size around E18 (Knoblich, 2008, Kriegstein and Alvarez-Buylla, 2009). Final terminal neurogenic divisions take place at the VZ surface, where RG divide symmetrically and produce postmitotic neurons, thereby reducing the available pool of progenitors.
In addition to neurons, other neocortical cell types include microglia, astrocytes, oligodendrocytes, and endothelial cells. Many neocortical astrocytes, oligodendrocytes and endothelial cells arise from the lineage of precursors aligning VZ as primary neurons, although at later stages of neocorticogenesis (Mission JP, 1991, Sun et al., 2005, Li et al., 2012). Thus, when neurogenesis ceases, the remaining RG give rise to astrocytes and oligodendrocyte precursors, and ependymal cells, which is beyond the scope of the current review.
Are RG in the Developing Neocortex Homogenous?
Historically, RG have been thought to be relatively homogenous in nature and to respond to temporal cues in the generation of distinct subpopulations of projection neurons (Molyneaux et al., 2007). However, recent evidence suggests that the situation is more complex. A morphologically similar class of NSCs in the VZ, short neural precursors (SNPs), has been found to contribute to neocorticogenesis in a similar manner as RG (Gal et al., 2006). These cells have a short basal process of variable length that further shortens during mitotic division. Numbers of SNPs and RG are equivalent except at E14.5 during the “direct” to “indirect” neurogenesis transitioning time point, when SNPs outnumber RG. Although SNPs are morphologically distinct from RG, they appear to follow similar patterns of cell cycling and generation of postmitotic progeny.
More recent correlative evidence points toward a subpopulation of RG-like cells sporadically distributed across the VZ that express Cux2 mRNA (Franco et al., 2012). It is possible that these cells are identical to SNPs; although, subsequent work is required to further unravel the morphological and molecular signatures of different NSC subpopulations. Cux2-expressing RG may predetermine their neuronal subtype cell-autonomously during asymmetric division at the ventricular surface as early as E10.5 (Franco et al., 2012). Importantly, Cux2 is also expressed in the SVZ, where IPCs predominately give rise to upper-layer neurons (Zimmer et al., 2004). Using a transgenic reporter mouse with FLEx (Flip-Exclusion) technology, morphologically similar RG were distinguished by Cux2 expression though dTomato (Cux2-negative) or GFP (Cux2-positive) labeling (Franco et al., 2012). Cux2-positive cells generated upper-layer neurons in both in vivo and in vitro conditions and were more likely to re-enter the cell cycle, whereas Cux2-negative cells were more likely to terminally divide in symmetric fashion. These findings indicate that cell fate is programmed into the transcriptomes of the neocortical progenitor pool very early in neurogenesis. Future loss- and gain-of- function studies in which Cux2 is directly manipulated may determine whether this molecule is necessary for narrowing RG fate and further identify possible overlaps and discrepancies between Cux2-positive RG and short NSCs. Thus in the RG populated VZ, there may be either a single type of progenitor that progressively differentiates or there is a co-existence of multiple progenitor subtypes (Franco and Müller, 2013). However, in either case, progenitors must respond promptly to spatiotemporally regulated extracellular cues, as summarized below.
Similarities and Differences between Human and Mouse Neocortical NSCs
Although the mouse neocortex does not fully reflect the remarkable complexity of the folded human neocortex (see (www.brainmuseum.org, 2012), the basic molecular mechanisms of neocorticogenesis in mice have been confirmed in humans (Bayatti et al., 2008), including the spatiotemporal specification of the six neocortical layers (Hevner, 2007, Fertuzinhos et al., 2009, Koopmans et al., 2010, Zhu et al., 2010, Saito et al., 2011, Huang et al., 2012, Kwan et al., 2012a). In humans, however, the molecular and cellular processes of brain development are more complex and the proliferative regions are proportionally larger than in mice (Figure 2). Generation of the human neocortex takes place over the entire course of gestation, with neurogenic divisions starting around gestational week 9–11 (Rakic P, 1968, Sidman RL, 1973, Zecevic et al., 2005, Fish et al., 2008, Lui et al., 2011, Malik et al., 2013). During later stages of neurogenesis, the SVZ of humans and non- human primates is significantly increased in thickness compared with other developing zones/layers and compared with mice (Cheung et al., 2010).
Another proliferative region outside the SVZ was recently discovered in humans (Hansen et al., 2010) and subsequently described in both carnivores and rodents (Fietz et al., 2010, Hansen et al., 2010, Wang et al., 2011). This region, called the outer subventricular zone (oSVZ), is populated by RG-like neurogenic NSCs called outer radial glia (oRG) (Figure 2). oRG are found basally to Tbr2- positive cells in the SVZ, express Pax6 and Sox2, undergo several cell cycles, and maintain a basal process to the pial surface. TFs or other markers specific to oRGs and the specific postmitotic neural progeny that oRG contribute to the developing neocortex remain to be identified. Existing evidence, however, suggests that the oSVZ may be the primary region of proliferative expansion corresponding to the evolutionary advancement of neocortical size and function.
Sophisticated in vitro techniques now enable the modeling of remarkable steps of human cortical development in culture. This system uses human induced pluripotent stem cells (hiPSCs) and has been successful in mimicking the progression of neocorticogenesis—from NECs to RG and the subsequent generation of deep- and upper-layer neurons (Mariani et al., 2012, Espuny-Camacho et al., 2013). Initial studies using this technique demonstrate the ability of cultured hiPSCs exposed to distinct extracellular cues to aggregate in a sphere-like structure with a central cavity. RG-like stem cells line the inner opening, whereas postmitotic progeny expressing cortical TFs are found more superficially. Importantly, the sequential birth of subpopulations of projection neurons is also preserved. Remarkably, these in vitro subpopulations of human projection neurons can be transplanted into a mouse neocortical slice culture, where they become electrophysiologically active and integrate into functional circuits. This new finding could begin to bridge a gap between in vitro and in vivo work and aid the translation of mouse-based research to humans.
Molecular and Cellular Mechanisms of Neocortical NSC Differentiation
During CNS development, the formation of the telencephalon, which contains the neocortex, is induced by a dynamic interplay of multiple intrinsic and extrinsic cues (Rallu et al., 2002). The progressive differentiation of neocortical NSCs and the transition from the expansion of a mostly homogenous population of NEC precursors to the specification of RG and ultimately specialized neuronal subpopulations occurs within a narrow time frame (Shen et al., 2006, Okano and Temple, 2009, Seuntjens et al., 2009, Siegenthaler et al., 2009). Therefore, neocortical NSCs appear to have intricate intrinsically programmed molecular systems that dictate differentiation while responding to extrinsic cues (Shen et al., 2006, Okano and Temple, 2009, Seuntjens et al., 2009, Siegenthaler et al., 2009).
Intrinsic Mechanisms Regulating Neocortical NSCs
Elegant in vitro analyses, first in mice and later in humans, indicated that timed developmental mechanisms are intrinsic to neocortical NSCs (Mariani et al., 2012, Shi et al., 2012, Espuny-Camacho et al., 2013). A seminal study using mouse neocortical NSC lineages showed that the sequential generation of cultured neurons mimics the in vivo temporal order (Shen et al., 2006); as each cortical layer arises, cultured NSCs lose their potency and become restricted in their generation of different neurons. Similarly, cultured human NSCs first generate early neuron subtypes followed by later neuron subtypes (Mariani et al., 2012, Shi et al., 2012, Espuny-Camacho et al., 2013).
At perhaps the deepest intrinsic level, an open chromatin structure influences pluripotency and differentiation of ES cells (Hajkova et al., 2008, Gaspar-Maia et al., 2009). As neocortical neurogenesis progresses, the chromatin structure of DNA in RGs becomes more condensed, with an increase in the High Mobility Group A (HMGA) protein as neurogenic stages progress (Kishi et al., 2012). This protein group is associated with modulating chromatin structure and accessibility to transcription factors through DNA cross-linking (Vogel B, 2011). When MGA proteins were silenced in neural progenitor cells (i.e., NSCs) in vitro and in vivo, reduced levels of these proteins led to more differentiated states of transfected cells. This conclusion was supported by a greater proportion of cells expressing Beta III tubulin or exhibiting a loss of cell cycling. Over-expression of HMGA proteins produced the opposite outcome, with cells more likely to express proliferative markers or to incorporate the S-phase-labeling thymidine analog EdU. These findings indicate that the intrinsic mechanism of chromatin remodeling, which is clearly at work in other stem cell types, also influences neurogenic phases in NSCs.
Open chromatin states likely increase the accessibility of TFs to genomic regions, where they play a role in determining NSC fate. Further, there is also evidence of direct TF influence over the chromatin state in development (Magklara et al., 2011, Guo et al., 2012). The neocortex occupies the dorsal part of the telencephalon and is characterized by specific expression of several TFs in NSCs, such as Empty spiracles homologue 2 (Emx2), Paired box 6 (Pax6), and Forkhead box G1 (Foxg1) (Muzio et al., 2002, Hanashima et al., 2004). These TFs prevent the expansion of ventral and medial neurogenic regions of the telencephalon, which correspond to future basal ganglia and hippocampi, respectively.
A subgroup of TFs, Forkhead box (Fox) TFs, which are mainly described as transcriptional repressors, have been studied in the context of neural stem cell maintenance (Rousso et al., 2012). Over-expression of Foxp2 or Foxp4 has redundant effects, with either protein sufficient to promote differentiation. Conversely, Foxp4 knockout (KO) mice show a lower number of differentiated neurons at early developmental stages and a greater proportion of cells positive for Ki67, an indicator of cell cycling. Mechanistically, this study implicates Foxp2/4 in the disruption of adherens junctions, which are critical for the maintenance of stem proliferative fates (Stepniak et al., 2009). Within these junctions, a host of proteins have been found to be responsible for junction maintenance, including N- Cadherins (Kadowaki et al., 2007, Rasin et al., 2007, Bultje et al., 2009, Stepniak et al., 2009). Foxp4 specifically down-regulates N-Cadherin mRNA without affecting other factors in the junctions. These data suggest an intricate interplay of intrinsic molecules in progressive NSC differentiation.
Finally, T-brain gene-2 (TBR2) TF constitutive depletion is lethal. However, conditional forebrain silencing of Tbr2 revealed its function in IPC formation and maintenance (Englund et al., 2005, Sessa et al., 2008).). Intriguingly, early Tbr2 depletion resulted in reduced production of all neocortical layers, suggesting that indeed Tbr2 contributes to lower layer, subcortically projecting neurons as well as those of upper layers (Sessa et al., 2008). Strikingly, Tbr2 overexpression resulted in ectopic SVZ regions within the RG niche of the VZ. These findings determined Tbr2 is a key intrinsic molecule for identity and proliferation of IPCs.
Collectively, this brief overview on TF functions in NSCs clearly indicates the significance of complex intrinsic regulation of NSC proliferation and differentiation (for additional reviews see (Hevner, 2007, Molyneaux et al., 2007)). However, intrinsic pathways can be regulated by timed extrinsic cues, as follows.
Notch as an Extrinsic Regulator of Neocortical NSCs
Perhaps the best studied example of extrinsic influence over intrinsic gene expression is notch signaling. Notch signaling takes place through a receptor-ligand relationship involving the surface contact of two cells (Artavanis-Tsakonas et al., 1999, Yoon and Gaiano, 2005, Louvi and Artavanis- Tsakonas, 2006, Kopan and Ilagan, 2009, Ables et al., 2011). Binding of ligand components, such as Jagged or Delta proteins, to notch receptors (subtypes notch1–4) causes a γ-secretase cleavage of the intracellular notch receptor domain (ICN). ICN translocates to the nucleus, where it interacts with one of several TFs. The activated protein displaces a repressor complex varying by cell type, thereby activating transcription (Cau and Blader, 2009, Latasa et al., 2009). The result of this process is, with few exceptions, to promote a stem cell fate and repress differentiation (Gaiano et al., 2000).
Although the main molecular players of the notch signaling system are found in the neocortex, evolution has conserved the pathway while modifying some aspects of its regulation. A recent neocortex-specific example concerns the Numb and NumbL proteins, which polarize RG through maintenance of Cadherin-based adherens junctions at the epithelial surface (Rasin et al., 2007). Through a putative association with Numb/NumbL proteins, mPar3, a conserved protein that is asymmetrically distributed during RG division, serves to enhance notch signaling (Bultje et al., 2009). Furthermore, notch signaling in the proliferative VZ/SVZ is distributed such that tight spatial regulation of RG is required for access to the signal (Del Bene et al., 2008, Sessa et al., 2008, Sessa et al., 2010). In particular, there is an apical enrichment of Notch signaling, which promotes NSC renewal (Buchman and Tsai, 2008, Del Bene et al., 2008). These examples strongly implicate INM in this signaling cascade, with RG progenitors contacting distinct members of notch pathway when they are closest to or either furthest away from the VZ surface.
Also, Tbr2-positive IPCs may express the notch ligand Jagged and thereby influence RG potency. Depletion of Tbr2 not only ablates IPC populations in the SVZ due to Notch interactions between IPCs and RG but also results in an early depletion of RG progenitors due to premature differentiation (Mizutani et al., 2007). Further investigations of this phenomenon revealed that mind bomb-1 is expressed in a subset of IPCs and newly born postmitotic neuroblasts, where it promotes endocytosis of ubiquitinated Notch ligands and mediates the fate choices of RG during both symmetric and asymmetric divisions (Koo et al., 2005, Yoon et al., 2008). Interestingly, findings in zebrafish indicate that Par3 selectively distributes mind bomb-1 to the self-renewing cell of an asymmetric pair (Dong et al., 2012). In this manner, neurogenesis is regulated in part by the selective spatial expression of a basally positioned, IPC-specific Notch signal modifier.
Of note, however, some degree of lack of conservation in notch signaling appears to exist among species. In zebrafish, Par3 in the neural tube is asymmetrically localized to the neural-fated progenitor, and loss of Par3 function results in a significant increase in symmetric divisions that generate two progenitors (Alexandre et al., 2010). In mouse neocortical neurogenesis, however, over- expression of mPar3 increases the number of progenitor-producing symmetrical, progenitor producing divisions, where RNA interference more often drove symmetrical neuron generating divisions (Bultje et al., 2009).
Other Extrinsic Cues Regulating Neocortical NSCs
Of the many extracellular influences on neocorticogenesis, perhaps one of the most striking recent examples is the role of the trophic factor Fgf10 in the NEC-to-RG transition (Sahara and O’Leary, 2009). In Fgf10−/− mice, a marker of RG (BLBP) was diminished at E11.5, 12.5, and 13.5, indicating a late shift in the transition from the expansion phase of symmetric NSC division to the appearance of mature neurogenic RG. Subsequent experiments support this finding, showing an increased thickness of the rostral cortex during the postnatal period, whereas caudal regions were unaffected. Together, these results indicate that Fgf10 is a key mediator of regionally selective early NSC differentiation in the developing neocortex.
Extrinsic regulatory cues also can originate from outside the cortex and brain. In one example, loss of the meninges, sheets covering the developing neocortical wall, reduced the production of both neurons and IPCs, indicating less asymmetric divisions (Siegenthaler et al., 2009). Several subsequent elegant approaches revealed that retinoic acid is a powerful extrinsic cue derived from meninges driving proper NSC differentiation from symmetric to asymmetric divisions. For example, in utero retinoic acid treatment rescued the effect of depleting meninges on asymmetric divisions. Thus, these findings indicate that meninges can provide extracellular cues for the neocortical NEC to RG transition.
Interestingly, immature neurons may also send extrinsic cues that provide feedback and maintain differentiation of NSCs. For example, conditional deletion of a transcription factor Sip1 (also known as Zfhx1b) in young neurons regulates the production of subcortically projecting deep-layer neurons and intracortically projecting upper-layer neurons in a non-cell-autonomous manner (Seuntjens et al., 2009). Specifically, Sip1 deletion in early born neurons destined to reside in deep layers induced the premature production of upper-layer neurons and even glial precursors. In this way, Sip1 regulates the timing of cell fate switches during neurogenesis and the total number of neocortical projection neurons.
There is also evidence of the extrinsic influence of maternal endocrine signaling on neocorticogenesis. Maternal thyroid hormone (MTH) in pregnant dams induces profound changes in the maintenance and cycling of cortical progenitors and affects cortical thickness in developing rat embryos (Mohan et al., 2012). Pups from MTH-deficient dams had Pax6-deficient neocortices at E14, although this early deficit was corrected by E18. However, levels of Tbr2, which are indicative of upper-layer-generating IPCs, progressively declined in pups from hormone-deficient dams. This effect was only partially rescued by exogenous supplementation of MTH, indicating a coordination of other relevant factors. Nonetheless, this striking case confirms the importance of extrinsic factors to neocortical neurogenesis, even when they are generated outside the developing neocortex.
Collectively, these findings provide a platform of multiple converging extracellular factors on the intrinsic fate choices of NSCs in the developing neocortex. However, there is an additional intricate set of steps to ultimately define distinct subpopulations of neocortical projection neurons, as follows.
Postmitotic Differentiation and Specification of Subpopulations of Neocortical Projection Neurons
After NSC progeny commit to a postmitotic fate, nascent neuroblasts migrate along the basal radial processes of RG (for review, see (Casanova and Trippe, 2006, Rakic P, 2007, Metin et al., 2008, Molnar Z, 2012)). The diversity of neocortical projection neurons suggested the importance of accurately timed intrinsic programming in postmitotic differentiation. Indeed, precisely timed changes in functional gene expression must occur for the progenies of RG division to produce the hundreds of distinct subtypes of neurons and glia that populate the mature neocortex and contribute to its proper function (Molyneaux et al., 2007). Generally, neocortical layer VI will predominantly project to thalamus via corticothalamic axons. Layer Vb will predominantly project to the brain stem and spinal cord via corticobulbar and corticospinal tracts, respectively. Superficial layers will project intracortically and to a smaller extent into superficial parts of the striatum. There is growing evidence of unique regulation at the DNA level by specific neuron subtypes.
A recent meta-analysis, together with a loss-of-function of coup-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 2 (Ctip2) mutant, showed that corticospinal motor neurons (CSMNs) and callosal projection neurons (CPNs) express several similar but mutually exclusive factors (Arlotta et al., 2005). Expression patterns vary across neocorticogenesis, with TF expression becoming exclusive to each group of cells as they begin to differentiate. When Ctip2 is silenced, the subcortically projecting subset of layer Vb neurons fail to differentiate, indicating that this factor is necessary for their proper formation. When Ctip2 is repressed by the DNA-binding protein special AT-rich sequence-binding protein 2 (Satb2), which is required for the generation of CPNs, later-born upper-layer neurons differentiate into a separate subset of projection neurons (Alcamo et al., 2008, Britanova et al., 2008).
In addition, several groups simultaneously discovered the role of FEZ family zinc finger 2 (Fezf2) TF in CSMN axonal projections (Chen et al., 2005a, Chen et al., 2005b, Molyneaux et al., 2005). Fezf2 is both necessary and sufficient for proper formation of subcortical projections. Developmental silencing of Fezf2 prevented corticospinal tract axons reaching the spinal cord. In contrast, overexpression of Fezf2 in upper layer neurons results in ectopic subcortical projections (Chen et al., 2005b, Chen et al., 2008). In addition, Fezf2 can alter upper layer specification into early postnatal life (Rouaux and Arlotta, 2013), and is sufficient to alter the fate of progenitors from the basal telencephalon when overexpressed (Rouaux and Arlotta, 2010). In addition, Fezf2 expression in lower-layer neurons drives the expression of the lower-layer TF Tbr1. Collectively, these findings indicate Fezf2 as potent regulator of deep layer neuron projections and specification.
Just as the axonal projections of CPN and CSMNs are regulated by Satb2, Ctip2 and Fezf2 TFs, Cux1 (CDP) and Cux2 are important for the formation of dendritic trees in these upper-layer neurons (Cubelos et al., 2010). The lack of Cux1 and Cux2 results in fewer dendritic branches, smaller post-synaptic densities, and reduced excitatory post-synaptic currents, which are all indicators of differentiation failure. Similarly, Fezf2 expression in lower-layer neurons is necessary for normal dendritic architecture of layer 5 projection neurons (Chen et al., 2005b)
These findings were recently extended by an elegant study using double-mutant Fezf2, Ctip2, or Satb2 mice (Srinivasan et al., 2012). Using a beta galactosidase (LacZ) labeling system, the McConnell group discovered networking of TFs in mutual repression and derepression that ultimately determine postmitotic fates of projection neurons in the developing neocortex. Briefly, upper layer neurons with a conditional EMX1 promoter-driven deletion of Satb2 projected ectopically to subcortical structures, but double knockout of Ctip2 and Satb2 lead only to a partial restoration of LacZ-positive intracortical axon projections. In mice with a EMX-Cre driven deletion of Fezf2, subcortical Fezf2-placental alkaline phosphatase (PLAP)-labeled axons were reduced, as expected. However, double knockout of Fezf2 and Satb2 does not restore this loss. Importantly, Ctip2 is downregulated in Fezf2 mutants, but restored in Fezf2/Satb2 double mutants, indicating that Fezf2 represses Satb2 expression, which in turn represses Ctip2 expression.
Corticothalamic projections are lost when Tbr1 expression is developmentally ablated. In Fezf2 mutants, the loss of corticospinal projections is paralleled by an increase in corticothalamic innervation and an expansion of Tbr1 expression (Hevner et al., 2002, McKenna et al., 2011). In addition, Tbr1 represses Fezf2 expression in layer 6 to restrict axons to corticothalamic tract (Han et al., 2011). Beside these roles in the formation of subcortical projections, Tbr1 overexpression was found to rescue intracortical projections in Satb2 mutants. In a conditional EMX1-Cre driven deletion of Satb2, Tbr1 overexpression at E15.5 was sufficient to rescue the intracortical callosal projections of transfected neurons. This, taken together with previous findings that Tbr1−/− mice show abnormalities in callosal connectivity, suggests that Tbr1 has an early role in specifying layer VI neurons, but also plays a role in establishing the connectivity of superficial layers (Hevner et al., 2001).
Reports of nascent neurons with bifurcated axons—one putatively bound for an intracortical target and the other bound for a subcortical target (Garcez et al., 2007)— are further evidence of fate repression. These migrating neuroblasts are found in the intermediate zone (IZ) and become CPNs during early neocorticogenesis. A later study found that some neuroblasts migrating through the IZ co-express Ctip2 and Satb2, which is preserved in later postnatal stages (Lickiss et al., 2012). However, although a large population of bifurcated cells was identified through retrograde DiI labeling, these cells were never found to co-express Ctip2 and Satb2. Findings such as these convey a theme in neocortical development—that substantial decisions in postmitotic differentiation occur via both the promotion of specific cell fates and the active inhibition of alternative fates.
In humans, much less has been demonstrated about differentiation and specification of pyramidal neurons. However, subpopulation and layer specific markers identified in mouse neocortex are reproducible in human neocortices. Even though the number of molecular identity markers for human projection neurons is increasing, there is a need for significant work to fill gaps (Hevner, 2007). Substantially more is known on dendritic differentiation in some prefrontal neocortical regions. Initially it was found that during the perinatal period projection neurons of the human prefrontal cortex have a phase of rapid dendritic growth (Mrzljak L, 1992). This was recently extended by findings in newborn to 91 year old specimens. Here, layer 3 neurons showed a biphasic pattern of growth during early postnatal life, with about a year of stagnation in growth. Layer 5 neurons reached stable adult values sooner (Petanjek et al., 2008). These findings, along with the even more complex developmental pace of synaptogenesis in human neocortices (Petanjek et al., 2011), suggest differential molecular mechanisms behind dendritogenesis and perhaps human specific neurological and psychiatric diseases where neocortical circuits are disrupted.
Clearly, unique proteins mediate the differentiation of NSCs and the postmitotic specification of distinct subpopulations of neocortical projection neurons. Although these discoveries have begun to unravel the complexity of transcriptional control in the developing neocortex, the binary nature of this regulation still does not completely explain the subtle differences among neocortical neuronal subtypes. It is unequivocal that these TFs regulate numerous targets and work in concert with many other factors to hone and specify their functional genetic readout. As we will describe further, recent state-of-art global screens have begun to reveal the transcriptomic effects of these developmental regulators.
Neocortical Transcriptomics in Mice
From the scale of whole neocortices to single cells, transcriptome diversity is increasingly being investigated. Transcriptomics of the entire developing neocortex, however, are complicated by the discrete regions and subpopulations of cells contained therein. Therefore, transcriptomics are being used as a tool to investigate subcompartments and even single cell types in the developing and mature neocortex with great specificity, providing a transcriptional “signature” of regions and cellular subtypes. Greater precision of cellular subtype segregation, however, will allow even more specific conclusions to be drawn. For example, RG of the VZ, IPCs of the SVZ, and subpopulations of laminarly-organized neurons are distinct in morphology, TF identity and likely transcriptome, but have been challenging to separate for individual analysis. However, findings from transcriptome screens of the whole neocortex are typically confirmed with quantitative reverse transcription polymerase chain reaction (RT-PCR), in situ hybridization analyses and/or immunohistochemistry, which ultimately reveals the cells expressing the gene of interest.
To this end, a recent study sought to investigate laminar-specific transcriptomic signatures of the developing neocortex was performed using laser capture microdissection to isolate discrete neocortical regions from embryonic mice (Ayoub et al., 2011, Fietz et al., 2012). E18-P7 neocortices were microdissected into VZ, SVZ/IZ, and CP; RNA was harvested; and deep RNA sequencing (RNASeq) was performed. The findings confirmed expression of subregion-specific transcripts and implicated a host of newly-identified and differentially expressed candidates. Interestingly, splicing was found to play a major role in the diversity of subregion transcriptomes, with splicing levels between regions often not agreeing with overall total transcript levels for a given gene. Of genes with two or more splice variants, 15.7% were differentially expressed in the CP, 11.8% in the VZ, and 12.8% in the SVZ. For example, Mfge8 has two variants expressed in the neocortex at E14.5 showing variant 1 selectively enriched in the VZ. These and other findings suggest that post-transcriptional trait-like splicing is a major contributor to neocortical complexity (Black, 2000, Grabowski and Black, 2001, von Holst et al., 2007).
Transcriptomic analysis of distinct subpopulations of projection neurons was recently achieved by elegant retrograde labeling of CSMNs, corticotectal neurons, and CPNs. Labeled neurons were isolated from the neocortex using fluorescent activated cell sorting (FACS) coupled to transcriptomic analysis (Molyneaux et al., 2009). This technique revealed numerous differences in the transcriptomes of distinct subpopulations of projection neurons that were subsequently confirmed by in situ hybridization and immunohistochemistry, including Ctip2. Comparisons of transcriptomes of upper versus lower neocortical layers also yielded discovery of differentially expressed genes, including Fezf2 (Chen et al., 2005b). As neocortical development has been studied extensively in mice, there is already a large body of work in this species that characterizes the compartmentalization and enrichment of transcripts specific to different regions (Molyneaux et al., 2007, http://www.genepaint.org, 2012, Kwan et al., 2012b, Science, 2012). In addition, large scale in situ profiling has been performed on developing mice, with results for many genes of interest publicly available at several websites, including GenePaint (http://www.genepaint.org/), Allen Brain Atlas (http://www.brain-map.org/), and Eurexpress (http://www.eurexpress.org/ee/), some of which were used in recent publications and as part of this review (Yi et al., 2010, Shim et al., 2012).
This type of investigation is beginning to delineate the transcriptional signatures of mouse neocortical subregions, layers, and proliferative versus non-proliferative compartments (Han et al., 2009, Ayoub et al., 2011, Belgard et al., 2011, Fietz et al., 2012). Analyses have also extended to pharmacological and fluorescent reporter transgenic animal models of disease states such as Attention deficit hyperactivity disorder, MDMA use, Alzheimer’s disease, and maternal neglect (Bordner et al., 2011, Fernandez-Castillo et al., 2012, Kim et al., 2012, gensat.org, 2013, Lempp et al., 2013). These studies also demonstrate that the transcriptome is plastic (Peter et al., 2012). Importantly, efforts are also being made to synthesize neocortical transcriptome data from mice, non- human primates, and humans in both normal and adverse prenatal states (e.g., fetal alcohol exposure) (Wang et al., 2010b, Hashimoto-Torii et al., 2011, Kojima et al., 2013).
Neocortical Transcriptomics in Humans
Transcriptomes of brain subregions are also increasingly being used to describe a region’s genetic signature in humans. The transcriptional load of one subregion or condition can be compared against another to determine the specificity or enrichment of transcript complement. These techniques allow for the quantitative differentiation of regions in developing or evolutionarily disparate brains (Bernard et al., 2012).
Using exon-array screen technology on neocortical regions of the mid-gestational human brain, it was demonstrated that not only the expression of distinct transcripts varies between regions, but that they are differentially spliced or expressed asymmetrically between hemispheres (Johnson et al., 2009). Differentially spliced variants were in many cases selectively expressed in (e.g., LIMK2a, CPVex5–6, ROBO1b, and ANRKD32b) or absent from (e.g., NTRK2b, LIMK2b, CPVLex2, ROBO1a, and ANKRD32a) the neocortex. Many of the transcripts that are differentially expressed or spliced have known roles in neocortical development, specification of neuronal subtypes, and axonal outgrowth, and were associated with neurodevelopmental disorders such as autism.
In another study, the human brain transcriptome was investigated at 15 time points across the lifespan, from 5 weeks post-conception to 82 years of age (Kang et al., 2011). Brains from both males and females were subject to Affymetrix Human Exon array to examine genes that were differentially expressed and spliced between sexes and across development. These findings extended previous work and created a partial spatiotemporal map of the human transcriptome. Interestingly, many of the genes profiled showed differential exon inclusion in the neocortex either temporally (88.7%) or spatiotemporally (28.9%). Most differential splicing occurred during embryonic development, indicating that much of this precise genetic control occurs at posttranscriptional level during the specification of primary neurons, RG cycling, and neuroblast migration.
In a study of 269 subjects ranging from before birth to over 70 years of age, next-generation sequencing technology revealed that rates of transcriptional change were high during the prenatal period and taper off in the first half-year of postnatal life, reaching a roughly steady state level by the second decade of life. This steady state persists for some time—until approximately the fifth decade of life—when transcriptional changes begin to progressively increase. Further investigation, coupled with several protein-level analyses, has recapitulated this pattern of splicing changes across age in two brain regions—the hippocampus and the neocortex (Mazin et al., 2013). Future analyses of this kind can aid in the creation of a transcriptomic “signature” of psychiatric disorders, which could facilitate the advancement of translational research.
Human prefrontal neocortex (PFC) complexity, generated at least in part through transcriptional diversity, is a hallmark of humans. Transcriptional signatures of the human prefrontal cortex (PFC) are reproducible across ages and ethnicities (Colantuoni et al., 2011). Specificities in PFC transcriptomes are of special interest given the role of this brain region in higher cognitive function and its involvement in human-specific disorders (Goldman-Rakic, 2002, Diamond, 2011, Arnsten et al., 2012). Indeed, recent work shows differences in the PFC transcriptome of humans, chimpanzees, and macaques, whereas evolutionarily older brain structures (e.g., hippocampus and caudate) show conserved transcriptional profiles. Furthermore, several genes are enriched only in humans, such as CLOCK, which is implicated in psychiatric disease and circadian rhythms, and FOXP2, which is implicated in language (Feuk et al., 2006). Interestingly, more than 200 genes are differentially expressed in the mouse and human prefrontal cortex (PFC), (Lai et al., 2001, Feuk et al., 2006, Vernes et al., 2008). Although differences in gene expression have clear consequences for functional gene output, these findings suggest that intricate differences in post-transcriptional splicing may be a key mechanism through which evolution has honed neocortical development and function.
The laminar specific transcriptome screens in mouse were recently extended to the human developing neocortical wall (Fietz et al., 2012). Interestingly, this screen did not find significantly different transcriptome in the oSVZ, which may be due to presence of both RG and IPCs in both regions. However, in mice the SVZ and cortical plate were more similar in transcriptome signature than to VZ. In contrast, human developing neocortices have more similar oSVZ and iSVZ than the developing cortical plate. This study also pinpointed significance of differentially expressed distinct mRNAs that encode the extracellular matrix proteins.
An additional broad set of screening work known as the ENCODE Project Consortium characterizes functional genetic elements in multiple cell types (Gerstein, 2012). These studies profile genomic regions of transcription factor association, chromatin states, and histone modifications combined with DNA and RNA sequencing to detail the properties of transcriptional activity. This study has generated data that may be instructive for further investigation of neuronal circuits. In addition, genomic analysis at the single-cell level has been refined by isolating cells from the human PFC and caudate nucleus by FACS. Researchers used high-fidelity multiple displacement amplification methods and successfully generated material for whole genome sequencing from single cells sufficient (Evrony et al., 2012). This “single-cell fingerprinting” approach revealed that retrotransposon insertion rates were low in human neural cells of cortex and caudate nucleus and unlikely to account for cell heterogeneity in these regions. Retrotransposition is a remarkable process were transcribed pieces of the DNA are ultimately re-integrated into genome. These pieces of DNA that will be transcribed and then re-integrated are called retrotransposons, of which the best studied example is Long interspersed element-1 (LINE1 or L1) (Thomas et al., 2012). This L1 retrotransposon was shown to be active specifically in neuronal progenitor cells (NPCs) in vitro and in vivo (Muotri et al., 2005). In particular, Sox2 repressed L1 transcription. Levels of Sox2 are decreased in postmitotic progenies, the L1 expression is increased in them and inserts preferentially into genes encoding neuronal mRNAs. These data suggested that retrotransposon elements contribute to the development of nervous system, which due to the dynamics of retrotransposition may or may not be masked in adults. Indeed, in the adult human cortex and caudate, “single-cell fingerprinting” did not support L1 as a major source of neuronal diversity in adult cortex and caudate (Evrony et al., 2012). Nevertheless, the “single-cell fingerprinting” this approach was further used in a patient with hemimegalencephaly to map the mosaic AKT mutation in a lineage of cortically derived cells (Evrony et al., 2012). Results showed that a subpopulation of both neuronal and non-neuronal cells carried this mutation, suggesting that its etiology is within an early multi-potent precursor. These analyses have changed the field and allowed for highly specific diagnostic tools with clinical utility.
Single-cell analysis can also be extended to the transcriptome level (Hashimshony et al., 2012). The highly scalable “CEL-seq” system assigns each isolated cell’s RNA a 5′ barcode and Illumina adaptor during reverse transcription, and then cDNAs from multiple cells are pooled for in vitro transcription and sequencing via a modified Illumina assay. This system was successfully applied to single cells isolated from C. elegans embryos as a proof of principle. The implications for using this technology to dissect the transcriptional character of neocortical circuits are profound. Although these are powerful tools to analyze the neocortex at the genomic and transcriptional levels, the field awaits a proteome-level analysis of the neocortex.
Post-Transcriptional Processing
As the transcriptome of NSCs and postmitotic neuronal subgroups comes into sharper focus, greater attention must be paid to the functional protein expression levels of these transcripts. Given that there are often disconnects reported between the transcriptome and proteome, the transcript complement of cell subtypes must be interpreted as a “first step” in the segregation of neocortical cell subtypes (Chang and Stanford, 2008, Taniguchi et al., 2010, Day et al., 2011). Indeed, the transcriptome level of resolution cannot directly be interpreted as functional genetic makeup. Therefore, correlating transcriptional data with protein levels and the understanding of regulatory posttranscriptional steps will allow researchers to determine the cellular potential for rapidly translating and increasing protein content. Given the highly polarized, rapidly differentiating and functionally-specific cells of the developing neocortex, these regulatory processes may occur disproportionately to those of other brain regions.
Regulation at the post-transcriptional level may fill some of the gaps in the understanding of neocorticogenesis and rapid changes in functional gene expression. The traditional notion that DNA encodes RNA, which in turn encodes protein, assumes a passive role of mRNA in translation. It is now appreciated that mRNA itself is heavily regulated and can be targeted post-transcriptionally at many levels. After mRNA is transcribed, it can be subject to alternative splicing, sequestered or exported from the nucleus, transported throughout the cell, and/or selectively degraded or translated (Keene, 2007) (Figure 3 and graphical abstract).
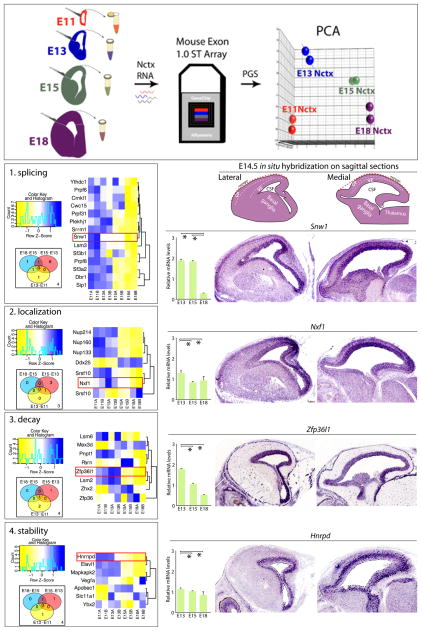
Figure 3. Transcriptome analysis of molecules involved in post-transcriptional mRNA processing steps within the developing mouse neocortex.
(top) Neocortex (Nctx) from embryonic days 11 (E11), E13, E15, and E18 was dissected, and RNA was isolated then assayed using Mouse Exon 1.0 ST Arrays, and analyzed using Partek Genome Suite (PGS) and R/Bioconductor. Principal component analysis (PCA) revealed clustering among replicates and distinct differentiation among developmental stages. (Left) Gene ontology (GO) analysis of whole developing mouse neocortices during key steps of the neurogenesis for distinct steps of post-transcriptional mRNA processing: splicing (1), localization (2), decay (3), and stability (4). GO analysis is presented as heatmap (blue = higher expression; yellow = lower expression; normalized by gene) and corresponding Venn analyses. Heatmaps include all genes on the Affymetrix Mouse Exon array annotated with the listed GO term, and Venn diagrams depict numbers of genes having significant contrasts between adjacent time points. (Right) qRT-PCR of whole developing neocortices for sample genes annotated with red boxes on the heatmaps. Corresponding in situ hybridization of lateral (middle panels) and medial (right panels) sagittal neocortical sections of E14.5 neocortices were obtained from www.genepaint.com. Remarkably, besides temporally distinct expression levels, the post- transcriptional regulatory elements also show restricted enrichment in different compartments of developing neocortices. For example, expression of the decay regulator Zfp36l1 decreased during neurogenesis, while its expression is highly enriched in the VZ where RG reside. In contrast, expression of a splicing regulator, Snw1, dramatically decreased at E18 when neurogenesis ceases, but is enriched at E14.5 in both progenitor characterized compartment VZ and postmitotic compartment CP. All qRT-PCR values were normalized to four housekeeping genes Gapdh, Mrps6, Rps13, and Rps18, and then scaled to average. * p < 0.05.
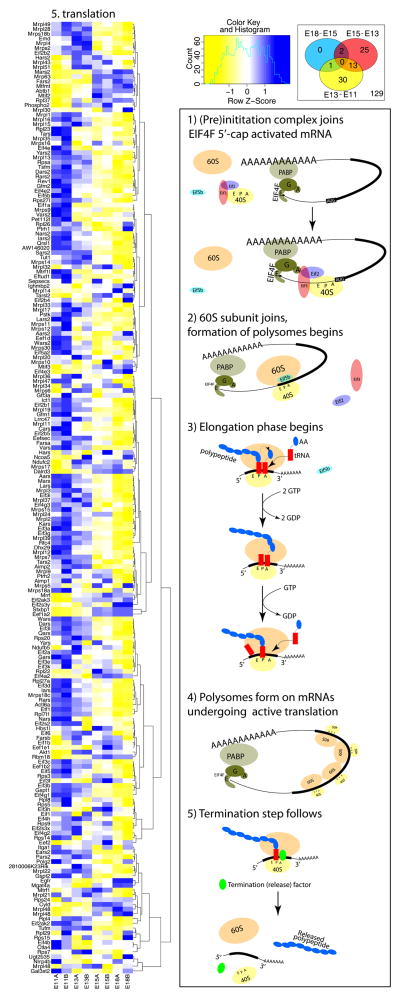
The mechanism of canonical translation is well described. The pathway starts with initiation, when an mRNA is activated via binding to the 5′ untranslated region (UTR) of the EIF4F eukaryotic initiation cap complex, composed of EIF4E, EIF4G, and EIF4A. The activated mRNA is then joined by the 43S pre-initiation complex, consisting of the small 40S ribosomal subunit and a ternary complex of eukaryotic initiation factor 2 (EIF2)-GTP-tRNAMet, which screens for the initiation AUG codon. In the late stages of the translation initiation/pre-elongation steps, the ternary complex is removed from the small 40S subunit, and the 60S ribosomal subunit is recruited to start formation of the actively translating 80S ribosomal polysomes. This is followed by eukaryotic elongation factor 2 (EEF2)- dependent elongation, which is critical in the post-initiation phase for progression from A to P to E sites after the 40S and 60S subunits form the 80S ribosomal polysomes. Ultimately, termination and ribosomal recycling occurs (Figure 4) (Jackson et al., 2010, Kong and Lasko, 2012).
Figure 4. Transcriptome analysis of developing neocortices during prenatal neurogenesis for mRNA translation reveals numerous mRNA clusters showing dynamic expression patterns.
(Right) Canonical process of translation and points of regulation. 5′-cap activated mRNA carries EIF4F complex (EIF4A+EIF4G+EIF4E) bound to the 5′ untranslated region (UTR) (step 1, top right animation). EIF4G from the complex is associating with PABP bound to the 3′ UTR making the active mRNA into a loop (step 2). On these mRNAs the preinitation complex (eIF2–40S ribosome-Eif3) joins the 5′UTR and screens for the start codon. Then the 60S ribosomal subunit joins the 40S to form the 80S ribosomal monosome. This step is partially regulated by Eif5b, while Eif2 and EIf3 are removed from the 40S (step 3). This initiation phase then transitions into the elongation phase when active translation is being governed by polysome assembly and polypeptide elongation (step 4). Once the polypeptide is finalized by reaching the stop codon, the termination step dissociates the 80S ribosome back into 40S and 60S subunits (step 5, bottom right animation). Each step of translation is regulated by distinct molecules, as shown in the next figure.
One well-studied example of post-transcriptional regulation is microRNA (miRNA) antisense silencing of mRNA translation. Broadly, miRNAs are a class of non-protein-coding RNA that function through translational repression of mRNA targets (Bartel, 2004). Typically, they are approximately 21 nucleotides in length in their mature form, and although they have similarities to other non-coding RNAs, they are distinct in target, synthesis, and their location in the genome (Bartel, 2004, He and Hannon, 2004, Bartel, 2009). The importance of post-transcriptional regulation in the neocortex was illustrated in a study in which gene encoding Dicer, the enzyme responsible for the maturation of both miRNAs and siRNAs, was floxed, and Cre was driven by the Emx promoter, which begins expression around E9.5 and is specific to the dorsal telencephalon (De Pietri Tonelli et al., 2008). Conditional Dicer KO embryos show a striking loss of almost all upper-layer neurons, which are preferentially born after E14.5. These neurons, however, are derived from a homogenous population of NSCs at the ventricular surface, indicating that intrinsic molecular programming achieved through the expression of miRNAs sharpens the fidelity of gene expression to allow for a properly formed and functional neocortex. As the upper layers of the neocortex are the newest in an evolutionarily sense (Cubelos et al., 2010), it is fascinating that Dicer as post-transcriptional regulator may play a central role in the formation of layers associated with higher-level cognitive function. In this way, post- transcriptional regulation can be seen as directly necessary for the advancement of cortical function and the generation of upper layers.
Recently developed methods have aided in addressing the gap between transcriptome and proteome via ribosomal profiling on Bacterial Artificial Chromosomes (BAC) (Gong et al., 2002, Gong et al., 2003, Yang et al., 2006, Gong et al., 2010). Given that the genomic sequence is very long and contains regulatory sequences such as promoter and enhancer regions that are located many kilobases away from the poly(A) tail, traditional transfection techniques cannot always accurately address the amount of expression of individual transcripts. BACs offer the advantage of being able to drive cell subtype-specific expression of ribosomal proteins while containing a GFP tag. The GFP tag can then be purified to isolate the mid-translation profile of transcripts with cell types (TRAP) (Heiman et al., 2008, Dougherty et al., 2010, Gong et al., 2010, Dougherty et al., 2012). This technique has successfully revealed expression patterns of several genes (Head et al., 2007, Heiman et al., 2008). In combination with transcriptomic profiling of cell subtypes, this method can allow the functional genetic output of cell subtypes to be weighed against their transcriptional profile to assess their potential verses realized genetic complement. In developmental studies, this technique can be used to discriminate the TRAP of stem and newly postmitotic cells. This information can then be instructive for comparison against disease states or to extrapolate evolutionary characteristics or region-specific profiles.
Post-Transcriptional Regulatory Elements are Differentially Expressed in Space and Time Across Neocortical Neurogenesis
Post-transcriptional regulation is clearly one of the key players in generating neocortical cellular diversity and function. However, studies investigating the regulation of translation, the final step of post-transcriptional regulation, are only in their infancy. Nevertheless, as early as the 1960s, researchers already found clusters of ribosomes in neuronal soma, dendrites, and dendritic spines (Bodian, 1965, Giuditta A, 1977, Giuditta A, 1980, Steward O, 1982, Giuditta A, 1991, Koenig and Martin, 1996). Follow-up studies confirmed these active sites of translation (polysomes) in distant neuronal processes (Giuditta A, 1991, Koenig and Martin, 1996). These findings suggest the existence of localized protein synthesis and multiple post-transcriptional regulatory steps during neuronal development. miRNAs may be functional in this role as they are co-transported with target mRNA and compartmentalized in the cell, regulating translation at the synapse (Pichardo-Casas et al., 2012). However, there is no current screen focused on post-transcriptional processing elements and their significance may be overlooked.
Therefore, we proceeded to demonstrate changes in genes associated with post- transcriptional regulation during murine neurogenesis. We performed a microarray analysis of RNA harvested from whole neocortices at four key developmental time points: E11 was chosen to outline expression during the onset of neurogenesis; E13 to investigate direct neurogenic processes giving rise to predominantly lower layer subcortically projecting neurons; E15 to outline the predominant shift from birth of subcortically to intracortically projecting neurons; and E18 to investigate the final stages of neocortical neurogenesis (Figure 3). Analysis of transcriptomics was performed using GeneChip Mouse Exon 1.0 ST Array (Affymetrix; n=2 per developmental stage or experimental condition) coupled with bioinformatics in BioConductor/R using the oligo and limma packages (GK., 2005, Carvalho and Irizarry, 2010). In R, analysis was performed first with the oligo package to interpret exon data using the “core” transcript annotations with highest confidence and then with the limma package to identify significant differences between groups using an F-test. Our initial analysis focused on transcripts associated with the post-transcriptional analysis steps of mRNA localization, degradation, stability and distinct steps of translation.
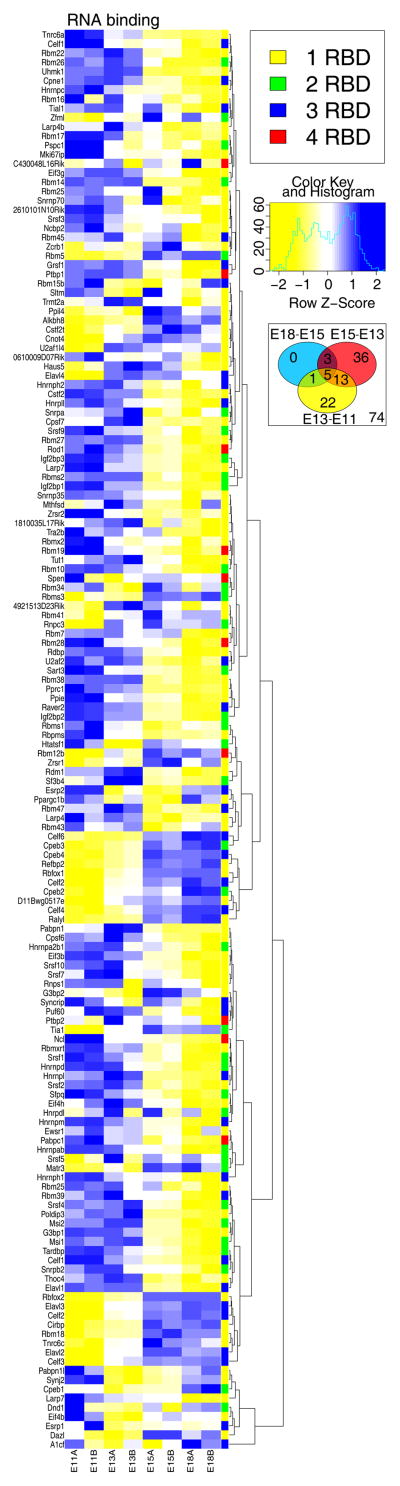
The colors in the heatmap (Figures 3–7) represent RMA scaled gene summary data for the listed gene symbols. The patterns of colors depict changes in expression of the functional groups of genes shown. For example, genes were selected by their annotation with gene ontology biological function codes as follows: splicing (GO:0000398), mRNA localization (GO:0006406), mRNA decay (GO:0006402; also named mRNA breakdown), mRNA stability (GO:0048255), mRNA translation (GO0006412), translation initiation (GO:0006413), translation elongation (GO:0006414), translation termination (GO:0006415), and RNA binding proteins (GO:0003723). For assignment of particular RBD and their number, such as 1 to 4 RRM and/or KH domains, each RBP was manually assessed for RBP domain using UNIPROT (http://www.uniprot.org) and a heatmap for each subset was produced as above.
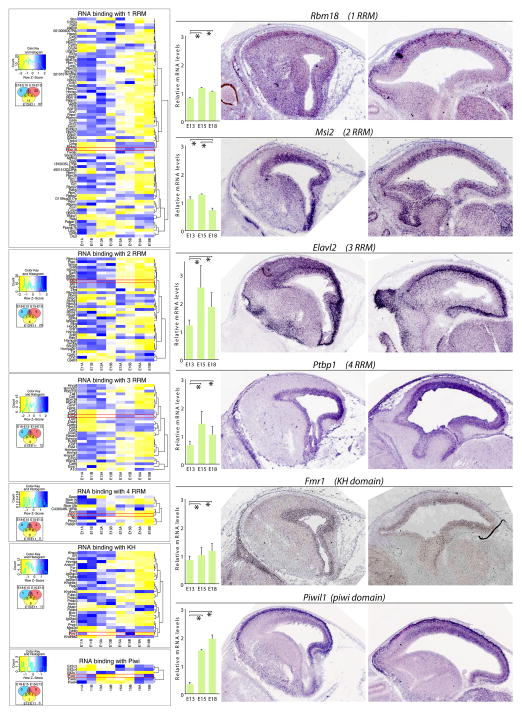
Figure 7. Transcriptome analysis of the developing mouse neocortex for RBPs with RRM, KH or piwi domains.
(Left) RBPs with distinct RBD signatures show differences in their number and temporal expression. For example, RBPs with 1 RRMs are the most numerous group. Corresponding Venn diagrams are provided below the heatmap key. (Right) qRT-PCR and in situ hybridization from www.genepaint.com revealed dynamic spatiotemporal expression of distinct RBPs. One example for each RBP group from the heatmap on the left is presented. Ptbp1, which is characterized by 4 RRM domains is highly enriched in the VZ, while Rbm18 characterized by 1 RRM seems to be in postmitotic neurons of CP, and some signal is detected in the migratory zone between VZ and CP. Piwil1 shows high signal in the VZ and CP, but is somewhat decreased in the anterolateral VZ. All qRT-PCR values were again normalized to four housekeeping genes Gapdh, Mrps6, Rps13, and Rps18, then scaled to average. * p < 0.05.
Notably, for each step of the post-transcriptional processing and RBP subset we found at least one candidate regulatory member to be differentially expressed throughout neocortical neurogenesis. In addition, the greatest number of transcripts associated with post-transcriptional processing that were differentially expressed across neocorticogenesis are involved in translational control (Figure 4). This is not surprising, as mRNA translation is a complex regulatory point that is composed of several tightly controlled steps. Remarkably, when analyzed for different steps of translation (Figure 5), we again found differentially expressed mRNAs important for initiation, elongation and termination. These findings suggest rapid and precise spatiotemporal control of functional protein expression during progressive neocorticogenesis, particularly at the level of translation.
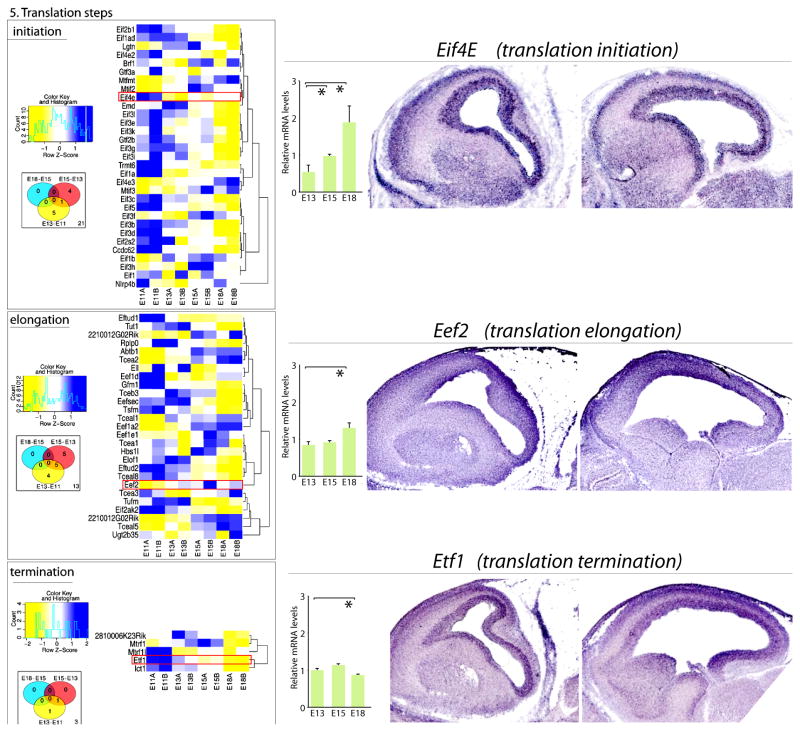
Figure 5. Transcriptome analysis of the developing mouse neocortex for mRNAs encoding regulators of distinct steps of mRNA translation.
(Left) Remarkably, even mRNAs encoding regulators of mRNA translation show dynamic changes in their expression during neurogenesis. GO analysis is again presented as heatmap (blue = higher expression; yellow = lower expression, normalized by gene) together with corresponding Venn analyses. (Right) qRT-PCR of whole developing neocortices for genes corresponds to relative gene expression in the heatmaps (red boxes). Corresponding in situ hybridization of lateral and medial sagittal neocortical sections of E14.5 neocortices were obtained from www.genepaint.com. Interestingly, initiation factor EIf4E and termination factor, Etf1, are both enriched in the VZ and CP, suggesting highly dynamic regulation of these two regulatory steps in RG progenitors and postmitotic differentiating neurons. All qRT-PCR values were normalized to four housekeeping genes (Gapdh, Mrps6, Rps13, and Rps18), and then scaled to average. * p < 0.05.
The array analysis was followed by qRT-PCR for one gene from each subgroup. Stable housekeeping reference genes are critical for credible qRT-PCR results. However, housekeeping genes for developmental neocortices have not been previously detailed in depth and therefore, we tested several previously used housekeeping genes (Gapdh, Pgk1, Rps18, and Rns18) and several new candidates that were unchanged in our across-development microarray analysis of neocortex (Rps13, Rps6kb1, Mrps6, and Pdcl2). The qRT-PCR results using these probes from E13, E15, and E18 neocortices were analyzed using Biogazelle qbasePLUS2 software to determine which housekeeping genes and how many of them should be optimally used, as described (Biogazelle; Zwijnaarde, Belgium) (Pinto F, 2012). Based on this approach, we found that optimally at least four housekeeping genes must be used together to accurately determine the dynamics of mRNA expression levels across neocortical development: Mrps6, Rps13, Rps18, and Gapdh. Thus, the subsequent developmental qRT-PCR results were normalized to these four reference targets per stage and then to E13 to obtain relative mRNA levels across development. A change of p < 0.05 was considered significant using one-way analysis of variance (ANOVA). Finally, the expression sites of these mRNAs were assessed using on-line in situ hybridization databases Euroexpress and Genepaint (http://www.genepaint.org, 2012, www.euroexpress.org, 2012).
The follow up qRT-PCR of whole developing neocortices corroborated array data that post- transcriptional regulatory elements present dynamic changes in their expression levels across neocortical neurogenesis. For example, the mRNA decay regulator, the expression of Zinc finger protein 36, C3H type-like 1 (Zfp36l1) decreased as neurogenesis progressed (Figure 3). Even more remarkable is its spatially restricted enriched expression in the VZ where RG cell bodies are residing, suggesting its role in neocortical progenitors (Figure 3). Interestingly, even mRNA translation regulators showed enriched expression in distinct compartments of developing neocortices. For example, mRNAs encoding initiation factor Eif4E and termination factor Etf1 are enriched in VC and CP, suggesting dynamic control of distinct steps of mRNA translation during neocortical development in these two compartments.
RNA Binding Proteins in Neocorticogenesis
The study of perhaps the most dynamic and ubiquitous of all post-transcriptional regulators, RNA binding proteins (RBPs), has only recently commenced in the context of neocorticogenesis. These proteins are prime candidates for post-transcriptional regulation given that they rapidly influence all steps: the stabilization, degradation, transport, splicing, and translation of mRNA cues (Keene, 2007) (graphical abstract). Thus, they represent a unique regulatory interface between transcriptional programming and functional protein expression and affect virtually every level of RNA processing. Therefore, the dynamic activity of RBPs may be of great importance for the rapid and specific gene expression events that are disproportionately required by the developing neocortex.
Binding of target RNA occurs at an RNA-binding (RBD) domain of which there are almost 40 subtypes known to date (Lunde et al., 2007). RBPs have one or several RBD, with greater numbers associated with increased specificity of RNA binding. RNA-recognition motifs (RRMs) are the most common example of RBDs in eukaryotes. RRMs provide both RNA and protein binding capacity. RRM can be situated that this single type of RBD is responsible for the specific binding of many RNA subtypes (Lunde et al., 2007, Clery et al., 2008). For example, the third RRM of the drosophila RBP, ELAV, is multifunctional and aids in both protein-protein and RNA-protein interactions, thereby influencing splicing events (Toba and White, 2008). In addition to RRMs, there are many other types of RBDs, such as RNP K homology (KH) and piwi domains. The unique quaternary structure of RBPs may allow for the specialized presentation of distinct RBDs. In this manner, RBP-RNA binding can be modified by many of the factors known to modify proteins post-translationally. The molecular mechanisms of these interactions, however, are not well elucidated.
In many cases, RBPs bind co-transcriptionally in the nucleus and begin forming ribonucleoprotein (RNP) complexes varying in structure and composition that ultimately mediate RNA fate (Kishore et al., 2010). RBP-RNA binding occurs through a specific or semi-conserved sequence, by secondary structure, or both. An early study showed that a sequence of 3′ UTR was necessary for localization of Bicoid RNA in Drosophila oocytes (Macdonald and Struhl, 1988). Further studies showed that within this region there are several stem loop structures required for sequential and increasingly specific localization of the transcript (Ferrandon et al., 1997). Sequence mutations of the secondary structures maintaining the stem loop were sufficient to preserve mRNA localization. Also, a cis-acting element within the 3′ UTR of Bicoid was required for RNA dimerization of the transcript itself, and this too was necessary for transcript transport. Therefore, even within a single transcript, there are multiple variations of RBP binding activities that may influence grouping in disparate RNPs.
RBP Roles in Neocorticogenesis
RBP activity is critical to the developing and mature brain. Some of the earliest studies of mRNA localization show that β-actin contains a “zip-code” cis-element located in the 3′ UTR that is responsible for the dendritic localization of the message (Kislauskis et al., 1994). Later research showed that this element preferentially exists in the β-actin 3′ UTR, whereas the γ-actin 3′ region lacks the essential zip code (Willis et al., 2011). This and subsequent work solidified the view that an RBP Zipcode Binding Protein 1 (ZBP1) acts to bind this cis element and that of GAP-43, transporting these messages to the outgrowing neurites of developing neurons in the central and peripheral nervous system (Donnelly et al., 2011). Interestingly, a phosphorylation-deficient ZBP1 mutant failed to release mRNA cargo in the dendrites and initiate translation of β-actin. Therefore, RBPs are involved in mRNA transport and are affected in a spatially-specific manner by post-translational modifications that ultimately influence translation of mRNA cargo.
During neocortical neurogenesis, mRNA distribution in progenitors is dependent upon Staufen2, an RBP. Staufen2 localizes asymmetrically within dividing RG (Kusek et al., 2012). Loss of Staufen2 function via shRNA both in vitro and in vivo demonstrated role in Pax6+ RG maintenance and Tbr2 suppression. Further in vivo results demonstrated that Staufen2 serves to regulate cell cycle re-entry. In particular, silencing of Staufen2 promoted cell cycle exit and differentiation. Early overexpression of Staufen2 in vivo resulted in aberrant pockets of differentiated heterotopias of variable size and cell type, further suggesting its role in stem-maintenance. Interestingly, Staufen2 binds a subset of mRNAs whose role was associated with asymmetric divisions. These findings indicate that the machinery for segregating mRNA between dividing cells is critical for the maintenance of progenitors and the spatiotemporally appropriate generation of neocortical layers.
Alternatively, bound mRNA can be regulated during and after splicing events by RBPs as a regulatory step between transcription and translation. One example in the neocortex is Magoh, which binds at the exon junction complex of target mRNAs (Silver et al., 2010). In this study, the Magohmos2 allele was found to cause a putative frame shift in the Magoh protein, resulting in a truncated protein. This factor is homologous between mice and humans at 100% of its amino acids. Importantly, haploinsufficiency in MagohMos2/+ neocortices lead to a significant decrease in cortical mass, mimicking microcephalic neuropathology. Further investigations found proper numbers of Pax6- positive RG progenitors and a significant reduction in numbers of IPCs. As a result, the neocortex was disorganized and lacking many lower- and upper-layer markers, with lower numbers of Tbr2- positive IPC progenitors. Also, Tuj1- and DCX-positive postmitotic neurons are generated at an increased rate early in neurogenesis, indicating premature cell cycle exit and differentiation. This microcephalic phenotype demonstrates the role of an RBP in neocortex formation, and future studies will parse out the downstream transcripts involved.
Although the Magohmos2 allele mediates the proliferation and generation of neurons from IPCs in the cortex, another RBP, Nova2, modulates an alternatively-spliced isoform of Dab1—an adapter of the Reelin pathway associated with proper placement of neocortical projection neurons (Sheldon et al., 1997, Yano et al., 2010). Nova2 binds Dab1.7bc in the cortex and cerebellum and selectively controls splicing of the Dab1.7bc isoform. Loss of Nova2 function prevents the selective exclusion of this exon, and the anatomical consequences are similar to the Reeler phenotype, with disorganized and ectopic cortical layers, especially those formed by later-born cohorts of neurons. In utero electroporation of wild-type Dab1 rescues the effects of Dab1.7bc expression in Nova2 KO mice. Furthermore, acute over-expression of Dab1.7bc via in utero electroporation also causes cortical disorganization when assessed at E14.5. Interestingly Nova, another member of this protein family, has a function apart from its splicing role in the nucleus. That is, it appears to be involved in translocating GIRK2 mRNA to the dendrites of spinal cord neurons and inhibitor synapses, thereby putatively regulating differentiation or plasticity (Racca et al., 2010). In this manner, the post- transcriptional event of alternative splicing, which is modulated by an RBP, is sufficient to regulate neuronal migration and differentiation in the neocortex.
In addition, splicing has been directly implicated in the self-renewal versus differentiation choice of RG in a polypyrimidine-tract-binding (PTB) protein-2 (Ptbp2) loss-of-function study (Licatalosi et al., 2012). PTBs are RBPs that play an important role in alternative splicing functions (Sawicka K, 2008). Ptbp2 is heavily expressed during the embryonic phases of neurogenesis and greatly reduced after P7. Using the recently generated HITS-CLIP technique (discussed in detail later), researchers found that targets of Ptbp2, including Depdc5, Sphkap, Erbb4, Dzip1, Ank3, Braf, Ppp3cb, and the basal progenitor maintenance factor, Numb, are more likely to be alternatively spliced in Ptbp−/− mice. Functionally, this failure to inhibit splicing results in the premature and aberrant differentiation of RG into DCX-positive neuroblasts. Failure to exclude the alternative third exon of Numb is coincident with this developmental abnormality. Furthermore, INM is perturbed, with BrdU- positive S-phase cells found ectopically at the epithelial surface of the VZ, whereas phospho-histone 3-positive M-phase RG divide aberrantly, away from the LV.
RBPs Characterized by KH, piwi and RRM are Differentially Expressed during Neocortical Neurogenesis
Our microarray analysis coupled to bioinformatics of developing neocortices revealed a large number of RBPs that have dynamic expression changes across neocortical neurogenesis (Figure 6). Interestingly, several clusters of RBPs are highly expressed during “direct neurogenesis,” while others are highly expressed during “indirect neurogenesis.” These data suggest the influence of RBPs across neocorticogenesis and a high level of complexity in their combinatorial roles. Dividing the RBPs based on their domain type, we found that that the most numerous subgroups of RBPs in developing neocortices are characterized by RRM domain, followed by KH (Figure 7). Interestingly, within the RRM subgroup the number of RRMs per molecule range from 1 to 4 for all but RNA binding motif protein 19 (Rbm19) which has 6 RRMs. Each of the subgroups contain members which either decrease or increase in their expression across neocortical neurogenesis suggesting their temporally distinct roles. Furthermore, assessment of available online in situ hybridizations of developing neocortices at E14.5 from www.genepaint.com and/or www.euroexpress.org revealed also spatial specificity in their expression (Figure 7). These findings suggest the dynamic spatiotemporal regulation of RBP’s as well as their function. Further, these data also point to their selective spatiotemporal control of neocortical post-transcriptional processing steps. Taken together previous findings, our screen suggests a significant role of posttranscriptional regulatory elements in neocortical development, and possibly evolution.
Figure 6. Transcriptome analysis of the developing mouse neocortex for RNA binding proteins (RBPs).
(left) GO analysis for RBPs revealed their substantial enrichment in developing neocortices, again with a predominant switch in expression levels occurring around E15. Each RBP has one or more RNA binding domain (RBDs) like KH, piwi or an RNA recognition motif (RRM). Total number of RBDs per RBP are presented with the color key (upper right corner).
Fragile X Mental Retardation Protein
Perhaps the best described example of the functional involvement of an RBP characterized by KH domain in brain development is Fragile X mental retardation protein (FMRP). In the clinic, FMRP mutations result in Fragile X mental retardation, an autism spectrum disorder, and are the most common monogenic cause of autism. The gene Fragile X Mental retardation 1 (FMR1) was identified by positional cloning (Verkerk et al., 1991), whereas encoded FMRP was recognized as an RBP somewhat later (Ashley CT Jr, 1993). Indeed, many of the genes disrupted in autism are associated with FMRP (Iossifov et al., 2012). The FMR1 RBP mutation involves increasing numbers of hereditary CGG repeats in the 5′ UTR of the FMR1 transcript encoding FMRP (Verheij et al., 1993). This may ultimately occur because the promoter region of FMR1 lies within this locus (Khalil et al., 2008). Functionally, FMRP is responsible for the mass exodus of nascent mRNA transcripts from the nucleus and preferential translocation to the dendrites, involving as much as 4% of total mRNA (Santoro et al., 2012). These recent studies show that the lack of FMRP results in underdeveloped synapses and dendritic spines. New research demonstrates that FMRP may be a downstream effector of mTORC1 and may mediate synaptogenesis and dendritic spine maturation through control of GluR and PSD-95 mRNA translation (Liu-Yesucevitz et al., 2011). Remarkably, wild-type offspring nurtured by Fmr1 mutant dams develop hyperactivity, a common trait of autism mouse models, implicating FMRP as a maternal environmental factor that disrupts neurological development (Zupan and Toth, 2012).
At the cellular level, compared with wild-type mouse cortical neurospheres, Fmr1-deficient E13 and P6 neurospheres generate three-fold more neurons and 15% fewer glia cells (Caldwell et al., 2001, Castrén et al., 2005). Neurospheres from humans with Fragile X syndrome (FXS) human embryos showed higher five-fold increase in neurons and a 70% decrease in glial cells. This shift is also found in vivo, where knockdown of Fmr1 by in utero electroporation of shRNA increases IPC production at the expense of RG (Saffary and Xie, 2011).
In the neocortex specifically, FMRP has been implicated in evolutionary differences between mice and humans in the expression of NOS1, a gene associated with synaptogenesis and schizophrenia (Kwan et al., 2012a). FMRP selectively binds NOS1 mRNA in the mid-gestational human neocortex, whereas the homologous mouse protein does not effectively bind Nos1. Furthermore, in Fragile X mental retardation, there is a significant decrease in human NOS1 protein levels, whereas protein levels are preserved in FMRP KO mice. These findings indicate that there is a species-specific role of FMRP in neocortical development. In addition to offering direct evidence linking FMRP and NOS1 expression to FXS, these findings demonstrate a difference in post- transcriptional regulation of NOS1 and suggest that FMRP may have a critical species- and region- specific role in the translation and function of this molecule. A recent study extends these findings by showing that FMRP is necessary for RG self-renewal by suppressing differentiation into TBR2- expressing IPCs (Saffary and Xie, 2011).
Recent examination of the FMRP mechanism has elucidated its role in the translation of bound targets through regulation at the elongation step (Darnell et al., 2011). In particular, the direct targets of FMRP have been identified through in vivo crosslinking-IP (CLIP). CLIP crosslinks protein and RNA molecules that are in direct contact. The subsequent IP can be subject to sequencing analysis (HITS-CLIP) to determine broad RBP-mRNA interactions (Darnell, 2010). Using this methodology, FMRP was found to bind a subset of mRNA targets, most often within their coding regions and frequently to those that are loaded with ribosomes and stalled in translation. Deletion of FMRP using multiple techniques, however, does not restore bound targets to active translation as assessed by sucrose gradient fractionation. Although the specific action of FMRP remains elusive, these findings suggest a dynamic role for this protein, allowing target mRNAs to be loaded with ribosomes and staged for translation while perhaps protecting them from degradation. Many FMRP mRNA targets are also involved in synaptic plasticity and are candidate autism genes, such as mGluR subunits that are altered in FXS (Cruz-Martin et al., 2012). In summary, FMRP is a mass regulator of functional gene expression that appears to be tightly regulated and highly specific. Studies on FMRP shed light on the ability of a single protein to cause multiple types of mental retardation. However, furthering our knowledge of the distinct steps and developmental time windows of FMRP activity are essential for better understanding FMRP-associated disorders and possible pharmacotherapeutics.
Elav RBPs
The Embryonic Lethal Abnormal Vision-Like (ElavL) proteins characterized by 3 RRM domains were identified as autoantibodies of patients with paraneoplastic neurological encephalomyelitis and paraneoplastic sensory neuropathy (Szabo A, 1991, Lövblad KO, 1993, Deschênes-Furry et al., 2006). Mammalian ElavL is homologous to Drosophila Elav, an RBP that is neuron-specific and required for nervous system development (Campos et al., 1987, Robinow and White, 1988, Robinow S, 1988). In mammals, ElavLs are also known as Human Antigen (Hu) family proteins and consist of four members: HuR (Elavl1), HuB (Elavl2), HuC (Elavl3), and HuD (Elavl4). These Hu proteins are mostly neuron-specific with the exception of HuR. They promote the stability and translation of its mRNA targets (Szabo A, 1991, Park et al., 2000).
Perhaps the best studied Hu proteins, HuD, is well conserved across species. HuD and other Elav family members have three RRMs: two at the N-terminus, followed by an intervening “linker” or “hinge” region, and then a C-terminus RRM (Fukao et al., 2009). In general, Elav proteins are known to stabilize transcripts, and HuD specifically does so through binding to AU-rich instability elements at the 3′ UTR, which are the targets of RBPs and exonucleases that preferentially degrade mRNA. HuD stabilization occurs through the competitive inhibition of these factors through binding at the same sequences (Park-Lee et al., 2003). A recent study shows that the first two RRMs have the most efficient RNA binding capacity but that the third RRM is essential for binding the long poly A tail of some neuron-specific transcripts (Bolognani et al., 2010), which preferentially promotes stability and translation of HuD targets. Furthermore, this study identifies members of an RNP that aid the involvement of HuD in translation, including eiF4G, eiF4A, eiF4E, and PAPB. Subsequent analysis using RNAase show that only the eiF4A interaction is protein-dependent and that other members bind to an RNA bridge. This study also demonstrates that the linker region and third RRM of HuD are required for protein-protein interaction and the presence of HuD in translating polysome fractions of a sucrose gradient. These results suggest that HuD functions in neocorticogenesis by selectively binding to translational protein complexes and influencing the translation of target transcripts.
Through transcript stabilization and promoting translation, HuD influences the differentiation of neurons and the proliferation of NSCs. For instance, an assessment of proliferation through in vitro analysis of a constitutive HuD KO mouse revealed a reduction in number of neurospheres, indicating a deficit in proliferation or differentiation of NSCs (Akamatsu et al., 2005). In the behaving mouse, these effects read out to a lack of motor control, evidenced by poor rotorod performance and abnormal paw clasping. In agreement with these results, in vitro studies demonstrate that HuD binds the 3′ UTR of GAP-43, and its over-expression is sufficient to drive the increased stability and expression of GAP-43 and the formation of neurites (Chung et al., 1997, Anderson et al., 2001). Other in vitro studies examining HuD expression in PC12 cells describe an increased expression of HuD in cells that are actively extending neurites. Inhibition of HuD via RNA interference completely stunts neurite outgrowth without affecting existing processes (Dobashi et al., 1998, Aranda-Abreu et al., 1999). As such, HuD is implicated in the initial stages of neurite outgrowth but not the stability of already-formed dendrites. The mechanism of this action appears to be an association of GAP-43 with HuD granules at growth cones that is dependent on translation, although this evidence come from co- localization rather than direct polysome analysis (Smith et al., 2004). Related behavioral studies show that learning tasks, such as the Morris water maze, up-regulate HuD levels and HuD co-localization with Gap-43 in rat hippocampi (Quattrone et al., 2001, Pascale et al., 2004). Thus, HuD is also active in the adult animal, seemingly to modulate plasticity through Gap-43 regulation similar to the mechanism at work in neurite outgrowth. Interestingly, gene ontology studies of HuD binding reveal that approximately 7% of bound targets are mRNAs of other RBPs such as Musashi 2 (Bolognani et al., 2010).
Misregulation of HuD, furthermore, is implicated in a host of diseases. For example, at least two single nucleotide polymorphisms in the ELAVL4 gene locus are associated with the age of onset of Parkinson’s disease (PD), a motor and cognitive disorder, in a clinical study of 1,223 members of 643 families (Noureddine et al., 2005). This study was replicated several years later by genotyping the two previously reported risk alleles for PD and discovering a third minor risk allele, confirming and extending ELAVL4’s link to PD (DeStefano et al., 2008). An interceding study of PD and control patients from the United States, Norway, and Ireland narrowed a possible genetic founder to an Irish population while confirming the locus and identity of the two previously reported risk alleles (rs9675852 and rs3902720) (Haugarvoll et al., 2007). Further investigations implicate HuD, through its role in alternative splicing, to a well-studied lymphoblastic leukemia known as the Philadelphia Syndrome, which is caused by a chromosomal translocation resulting in the BCR-ABL fusion protein (Bellavia et al., 2007, Mullighan et al., 2008). In this disorder, the TF Ikaros is deleted in the vast majority of patients with chronic myelogenous leukemia. HuD functions as an intermediary between notch3 signaling and Ikaros. Notch3 signaling up-regulates HuD and promotes alternative splicing of a non-transcriptionally active form of Ikaros, which disallows its contribution to lymphoid leukemias. HuD is implicated in human malignant neuroblasts due to its abnormal stabilization of the transcript of N-myc pre-RNA during nuclear processing (Cho and Noguchi, 1997, Darina L Lazarovab, 1999). Curiously, HuD mRNA is also increased in the blood of patients with small cell lung cancer (D’Alessandro et al., 2008), and several studies have implicated HuD as a marker for small cell lung cancer at the protein level (King, 1997). More recent in vitro studies using SH-SY5Y cells transfected with either HuD over-expression constructs or antisense RNAi vectors show correlated expression of HuD and N-myc. Furthermore, there is evidence for the loss of an HuD allele that is located on chromosome 1p in patients with neuroblastoma, particularly those with the worst prognoses (Grandinetti et al., 2005).
Clearly, the regulation of HuD expression is critical to the development and steady-state function of biological systems. Furthermore, several studies demonstrate that HuD itself is under heavy translational and post-transcriptional control. For example, HuD is edited by the adenosine deaminase that acts on RNA1 and 2 enzymes (Enstero et al., 2010). RNA editing is a process through which enzymatic activity acts on double-stranded RNA to edit adenosines to inosines, which are interpreted as guanosines by the ribosome during translation. Therefore, the peptide product can be changed. In the previously mentioned study, HuD was found to have five editing sites within its coding region, indicating its variable post-transcriptional regulation. It should also be noted that HuD is a target of alternative splicing, and there is evidence that this is an auto-regulatory event governed by other Hu family proteins. A recent study demonstrated that up-regulation of Hu proteins in HeLa cells promotes the inclusion of HuD exon 6 (Wang et al., 2010a). RNAi against Hu expression, however, decreases the rate of exon6 inclusion. So far, HuD is known to be expressed as four distinct protein isoforms in the mouse (NCBI), each which may have a distinct role in neocorticogenesis.
Other mouse studies implicate HuC/D in steady-state levels of transcripts as well as splicing (Ince-Dunn et al., 2012). Using HITS-CLIP technology to assess splicing and steady-state levels in Elavl3−/− and Elavl4−/− mice, consensus binding was predominately found in the 3′ UTR of mRNA targets. In pre-mRNA targets, however, Elavl3 was often bound and regulating splicing of bound cargo. Furthermore, Elavl3 targets are often involved in synaptic plasticity and signaling. Elavl3/4−/− mice exhibit a 50% loss of cortical glutamate. In the Elavl3−/− single KO, an alternative splicing site is occupied by Elavl3 on the pre-mRNA of the glutaminase enzyme. This enzyme is largely responsible for generating cortical glutamate, and Elav3 controls the generation of two isoforms of this protein. A significant decrease in one isoform (Gls-I) but not the other (Gls-s) may be responsible for this change in excitation. Finally, investigators electrophysiologically detected active seizure patterns in Elavl3 mutant mice. Collectively, members of the Hu family appear to be involved in many complex disorders. Although these disorders are most likely not the outcome of a single causative gene, studies of Hu family proteins may help provide insights in disease mechanisms and potential pharmacotherapeutics.
Musashi
The RBP Musashi is highly conserved across species. Initially discovered in Drosophila, it was found to be required for asymmetric divisions of sensory organ progenitors (Nakamura et al., 1994, Kaneko et al., 2000). In vertebrates, Musashi isoforms Musashi1 and Musashi2 are expressed in both adult and embryonic NSCs. Musashi regulates Numb and p21, a cyclin-dependent kinase (CDK) inhibitor (Imai et al., 2001, Battelli et al., 2006, Nishimoto and Okano, 2010), both which are implicated in neocortical NSC differentiation and cell cycle progression. Musashi appears to maintain stem cell self-renewal and undifferentiated state by disrupting recruitment of the large ribosomal subunit and other factors for polysome assembly. These mechanisms are still not well understood, and more focused research is needed to identify Musashi’s biochemical pathway. Musashi expression levels decrease as differentiation progresses, making it undetectable in postmitotic neurons (Kaneko et al., 2000).
Interestingly, HuD stabilizes Musashi mRNA, and this effect may be critical in the transition from stem cell proliferation to neural differentiation (Ratti et al., 2006). Recent studies show Musashi- Notch signaling to be a key regulator in chronic myeloid leukemia (Griner and Reuther, 2010, Ito et al., 2010). In relation to the CNS, Musashi-1 is ectopically expressed in a large majority of neurons exhibiting neurofibrillary tangles or Pick bodies (Lovell and Markesbery, 2005). Finally, Musashi knockouts show phenotypes similar to FMRP-deficient animals (Sakakibara and Okano, 1997, Sakakibara et al., 2002). Collectively, these findings suggest an important role of Musashi in normal development and the pathogenesis of many neurological and psychiatric disorders.
Discussion and Implications for Future Studies
Human evolutionary advantages of language, complex motor behavior, and advanced cognition can be traced to the morphological and functional expansion of the neocortex. Increased proliferative regions such as the SVZ and oSVZ can explain the dramatic increase in neuronal number, but this alone does not explain the vast functional differences among mammalian neocortices. Other neocortical regions also show evolutionary expansion, one of most important being the subplate, which is vastly expanded in human and non-human primates (Kostovic et al., 1989, Kostovic and Judas, 1998, 2010) (for review and psychiatric implications of this region, see (Kostovic et al., 2011)). Modern techniques, which permit a closer look at the transcriptomic architecture of neocortical compartments across development and species, reveal that the specificity of neocortical regions are dictated at least in part by vastly different transcript complements as well as uniquely expressed splice variants.
Given the relative homology of the mouse and human genomes and their similar number of protein-coding genes, it stands to reason that regulation at the post-transcriptional level may explain the disparity in complexity among mammalian neocortices. Of these post-transcriptional mechanisms, splicing is of particular interest, as it allows an expansion of proteome functionality and a narrowed fidelity without broad DNA-level changes. Investigations of human pre-mRNA demonstrate that 74% of these molecules are subject to alternative splicing (Johnson et al., 2003). A later study showed that these splicing events produce functional isoforms, suggesting that proteins are robust molecules that tolerate excisions and insertions, perhaps allowing the evolution of the proteome (Birzele et al., 2008). It may not be unfounded to extrapolate that evolutionary changes in the neocortex may correlate with greater specificity in the proteome of neocortical compartments.
For the splicing of a target transcript to impact the activity of a cell, it must be translated. At this heavily regulated post-transcriptional step, there is already one species-specific example of transcript regulation by an RBP (Kwan et al., 2012a). However, profiling of the proteome to investigate compartmentalized and species-specific evolutionary changes in the neocortex is not yet complete. This is likely due to the technical difficulties involved in such analyses. The spatiotemporal genetic specificity necessary to generate defined subpopulations of neocortical projection neurons likely involves an interaction of transcript regulation events, such that uniquely spliced variants are transported and rapidly translated based on cellular demand. Subtle alterations in spatiotemporal expression at the post-transcriptional level result in a wide spectrum of neocortex-associated disorders. Clinical applications of post-transcriptional studies are vast, and promising findings already exist. Indeed, future studies could combine the increasingly precise definitions of psychiatric disorders in the Diagnostic and Statistical Manual of Mental Disorders with analyses of transcriptional profiles. Studies employing this approach are already beginning to characterize the transcriptional profiles of schizophrenia, autism, and suicide (Roussos et al., 2012, Sequeira A, 2012, Ziats and Rennert, 2012). Recent work in rats also identifies changes in functional classes of transcripts in neurons that either do or do not “sprout” and regrow connections after a modeled stroke infarct (Li et al., 2010). By comparing animals of different ages, this study also shows that the vast majority of changed transcripts in successfully outgrowing neurons differed depending on age, suggesting treatments that may be tailored for particular age groups. A similar screening method in a study of the effect of maternal exposure to alcohol on mice and humans (Hashimoto-Torii et al., 2011) demonstrates a consistent down-regulation of the proliferative TBR2 transcript and protein as well as postmitotic specification markers, particularly those of upper-layer neurons. A screen of a mouse model of anxiety also confirms transcript changes in the hippocampus and cortex, pointing toward new targets for study (Virok et al., 2011). Furthermore, layer-specific profiling using BAC-TRAP shows translational changes in layer V projection neurons after antidepressant treatment (Schmidt et al., 2012). Finally, acute traumas can alter functions of distinct regulatory members of mRNA translation. For example spinal cord injury alters mRNA binding signature of EIF4E in neocortical neurons contralateral to the hemisection model (Thompson K, 2010), an initiation factor that in our screen came as part of GO:RNA binding. These data suggest that post-spinal cord injury mRNAs for possible regenerative efforts are acutely present in central neurons, but their translation is disrupted. Together, these types of studies demonstrate an increasing trend toward defining brain disorders based on transcript profiles and their posttranscriptional processing.
These types of studies could enhance our understanding of disease mechanisms within varying disease subtypes, leading to more specific and effective treatments. The understanding of the rich post-transcriptional contribution to disease states, however, must proceed to the final functional output of gene expression. The mere presence of a transcript cannot be equated to its proteomic function, and there is new evidence that the initiation and elongation steps of translation are heavily regulated, and perhaps aberrantly in disease (Darnell et al., 2011). The understanding of the metabolism of mRNA may be well served by greater study of RBPs in parallel with their mRNA targets of regulation. Therefore, future studies on the role of RBPs in neocorticogenesis and profiles of the developmental proteome are paramount to understanding the functional genetic complement of a tissue or cell.
Highlights.
historical, developmental and evolutionary perspective on neocortical development
neocortical neural stem cell lineages and subpopulations of projection neurons
impact of post-transcriptional processing on neurodevelopment
screen of post-transcriptional mRNA regulators across neocortical neurogenesis
RNA binding proteins and their roles in neocorticogenesis
Acknowledgments
We extend our gratitude to current and past members of the Rasin laboratory and researchers on Rutgers/RWJMS campuses for discussions and comments. Given the large scope of this review, special thanks are also given to the reviewers for their insightful comments. This work was supported by NIH grants NS064303 (M-R.R.), NS075367 (M-R.R.), DA032984 (R.P.H.), and RWJMS start-up (M-R.R.). Finally, we apologize to the authors of seminal and outstanding work that was not possible to mention here.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Brain Museum. http://brainmuseum.org/
- Ables JL, Breunig JJ, Eisch AJ, Rakic P. Not(ch) just development: Notch signalling in the adult brain. Nat Rev Neurosci. 2011;12:269–283. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, Okano HJ, Sakakibara S, Takano H, Takano T, Takahashi T, Noda T, Okano H. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc Natl Acad Sci U S A. 2005;102:4625–4630. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Alexandre P, Reugels AM, Barker D, Blanc E, Clarke JDW. Neurons derive from the more apical daughter in asymmetric divisions in the zebrafish neural tube. Nat Neurosci. 2010;13:673–679. doi: 10.1038/nn.2547. [DOI] [PubMed] [Google Scholar]
- Allendoerfer KL, Shatz CJ. The subplate, a transient neocortical structure: its role in the development of connections between thalamus and cortex. Annu Rev Neurosci. 1994;17:185–218. doi: 10.1146/annurev.ne.17.030194.001153. [DOI] [PubMed] [Google Scholar]
- Anderson KD, Sengupta J, Morin M, Neve RL, Valenzuela CF, Perrone-Bizzozero NI. Overexpression of HuD accelerates neurite outgrowth and increases GAP-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp Neurol. 2001;168:250–258. doi: 10.1006/exnr.2000.7599. [DOI] [PubMed] [Google Scholar]
- Aranda-Abreu GE, Behar L, Chung S, Furneaux H, Ginzburg I. Embryonic lethal abnormal vision-like RNA- binding proteins regulate neurite outgrowth and tau expression in PC12 cells. J Neurosci. 1999;19:6907–6917. doi: 10.1523/JNEUROSCI.19-16-06907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal Subtype-Specific Genes that Control Corticospinal Motor Neuron Development In Vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Arnsten Amy FT, Wang Min J, Paspalas Constantinos D. Neuromodulation of Thought: Flexibilities and Vulnerabilities in Prefrontal Cortical Network Synapses. Neuron. 2012;76:223–239. doi: 10.1016/j.neuron.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Ashley CT, Jr, WK, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Ayoub AE, Oh S, Xie Y, Leng J, Cotney J, Dominguez MH, Noonan JP, Rakic P. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci U S A. 2011;108:14950–14955. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Molecular and Cellular Neuroscience. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Bayatti N, Sarma S, Shaw C, Eyre JA, Vouyiouklis DA, Lindsay S, Clowry GJ. Progressive loss of PAX6, TBR2, NEUROD and TBR1 mRNA gradients correlates with translocation of EMX2 to the cortical plate during human cortical development. Eur J Neurosci. 2008;28:1449–1456. doi: 10.1111/j.1460-9568.2008.06475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, Garcia-Moreno F, Molnár Z, Margulies EH, Ponting CP. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia D, Mecarozzi M, Campese AF, Grazioli P, Talora C, Frati L, Gulino A, Screpanti I. Notch3 and the Notch3-upregulated RNA-binding protein HuD regulate Ikaros alternative splicing. EMBO J. 2007;26:1670–1680. doi: 10.1038/sj.emboj.7601626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Lubbers LS, Tanis KQ, Luo R, Podtelezhnikov AA, Finney EM, McWhorter MM, Serikawa K, Lemon T, Morgan R, Copeland C, Smith K, Cullen V, Davis-Turak J, Lee CK, Sunkin SM, Loboda AP, Levine DM, Stone DJ, Hawrylycz MJ, Roberts CJ, Jones AR, Geschwind DH, Lein ES. Transcriptional architecture of the primate neocortex. Neuron. 2012;73:1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birzele F, Csaba G, Zimmer R. Alternative splicing and protein structure evolution. Nucleic Acids Res. 2008;36:550–558. doi: 10.1093/nar/gkm1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KM, Garel S, Nakagawa Y, Rubenstein JL, O’Leary DD. Emx1 and Emx2 cooperate to regulate cortical size, lamination, neuronal differentiation, development of cortical efferents, and thalamocortical pathfinding. J Comp Neurol. 2003;457:345–360. doi: 10.1002/cne.10549. [DOI] [PubMed] [Google Scholar]
- Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- Bodian D. SUGGESTIVE RELATIONSHIP OF NERVE CELL RNA WITH SPECIFIC SYNAPTIC SITES. Proc Natl Acad Sci USA. 1965;53:418–425. doi: 10.1073/pnas.53.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2010;38:117–130. doi: 10.1093/nar/gkp863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordner KA, George ED, Carlyle BC, Duque A, Kitchen RR, Lam TT, Colangelo CM, Stone KL, Abbott TB, Mane SM, Nairn AC, Simen AA. Functional genomic and proteomic analysis reveals disruption of myelin- related genes and translation in a mouse model of early life neglect. Frontiers in psychiatry / Frontiers Research Foundation. 2011;2:18. doi: 10.3389/fpsyt.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig Joshua J, Haydar Tarik F, Rakic P. Neural Stem Cells: Historical Perspective and Future Prospects. Neuron. 2011;70:614–625. doi: 10.1016/j.neuron.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, Šestan N, Molnár Z, Tarabykin V. Satb2 Is a Postmitotic Determinant for Upper-Layer Neuron Specification in the Neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Brodmann KGL. Brodmann’s Localisation in the Cerebral Cortex: The Principles of Comparative Localisation in the Cerebral Cortex Based on Cytoarchitectonics: Birkhäuser 2006 [Google Scholar]
- Buchman JJ, Tsai L-H. Putting a Notch in Our Understanding of Nuclear Migration. Cell. 2008;134:912–914. doi: 10.1016/j.cell.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Caldwell MA, He X, Wilkie N, Pollack S, Marshall G, Wafford KA, Svendsen CN. Growth factors regulate the survival and fate of cells derived from human neurospheres. Nat Biotech. 2001;19:475–479. doi: 10.1038/88158. [DOI] [PubMed] [Google Scholar]
- Campos AR, Rosen DR, Robinow SN, White K. Molecular Analysis of the Locus Elav in Drosophila- Melanogaster - a Gene Whose Embryonic Expression Is Neural Specific. Embo Journal. 1987;6:425–431. doi: 10.1002/j.1460-2075.1987.tb04772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova MF, Trippe J., 2nd Regulatory mechanisms of cortical laminar development. Brain Res Rev. 2006;51:72–84. doi: 10.1016/j.brainresrev.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Castrén M, Tervonen T, Kärkkäinen V, Heinonen S, Castrén E, Larsson K, Bakker CE, Oostra BA, Åkerman K. Altered differentiation of neural stem cells in fragile X syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17834–17839. doi: 10.1073/pnas.0508995102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E, Blader P. Notch activity in the nervous system: to switch or not switch? Neural Development. 2009;4:36. doi: 10.1186/1749-8104-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviness VS., Jr Architectonic map of neocortex of the normal mouse. J Comp Neurol. 1975;164:247–263. doi: 10.1002/cne.901640207. [DOI] [PubMed] [Google Scholar]
- Chang WY, Stanford WL. Translational control: a new dimension in embryonic stem cell network analysis. Cell Stem Cell. 2008;2:410–412. doi: 10.1016/j.stem.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci U S A. 2005a;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci U S A. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci U S A. 2005b;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AF, Kondo S, Abdel-Mannan O, Chodroff RA, Sirey TM, Bluy LE, Webber N, DeProto J, Karlen SJ, Krubitzer L, Stolp HB, Saunders NR, Molnar Z. The subventricular zone is the developmental milestone of a 6- layered neocortex: comparisons in metatherian and eutherian mammals. Cereb Cortex. 2010;20:1071–1081. doi: 10.1093/cercor/bhp168. [DOI] [PubMed] [Google Scholar]
- Cho JH, Noguchi M. Expression of HuD (a paraneoplastic encephalomyelitis antigen) mRNA in lung cancer. J Korean Med Sci. 1997;12:305–310. doi: 10.3346/jkms.1997.12.4.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Eckrich M, Perrone-Bizzozero N, Kohn DT, Furneaux H. The Elav-like proteins bind to a conserved regulatory element in the 3′-untranslated region of GAP-43 mRNA. The Journal of biological chemistry. 1997;272:6593–6598. doi: 10.1074/jbc.272.10.6593. [DOI] [PubMed] [Google Scholar]
- Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee TB. Embryonic vertebrate central nervous system: revised terminology. The Boulder Committee. The Anatomical Record. 1970;166:257–261. doi: 10.1002/ar.1091660214. [DOI] [PubMed] [Google Scholar]
- Corbin JG, Nery S, Fishell G. Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nat Neurosci. 2001 doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- Cruz-Martin A, Crespo M, Portera-Cailliau C. Glutamate induces the elongation of early dendritic protrusions via mGluRs in wild type mice, but not in fragile X mice. PLoS ONE. 2012;7:e32446. doi: 10.1371/journal.pone.0032446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Sebastian-Serrano A, Beccari L, Calcagnotto ME, Cisneros E, Kim S, Dopazo A, Alvarez-Dolado M, Redondo JM, Bovolenta P, Walsh CA, Nieto M. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. 2010;66:523–535. doi: 10.1016/j.neuron.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro V, Muscarella LA, Copetti M, Zelante L, Carella M, Vendemiale G. Molecular detection of neuron-specific ELAV-like-positive cells in the peripheral blood of patients with small-cell lung cancer. Cellular oncology: the official journal of the International Society for Cellular Oncology. 2008;30:291–297. doi: 10.3233/CLO-2008-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarovab Darina L, BAS, Biedler June L, Rossa Robert A. HuD, a neuronal-specific RNA-binding protein, is a putative regulator of N-myc pre-mRNA processing/stability in malignant human neuroblasts. Oncogene. 1999;18:2703–2710. doi: 10.1038/sj.onc.1202621. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley interdisciplinary reviews RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RS, McDade KK, Chandran UR, Lisovich A, Conrads TP, Hood BL, Kolli VS, Kirchner D, Litzi T, Maxwell GL. Identifier mapping performance for integrating transcriptomics and proteomics experimental results. BMC Bioinformatics. 2011;12:213. doi: 10.1186/1471-2105-12-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelipe J, Lopez-Cruz PL, Benavides-Piccione R, Bielza C, Larranaga P, Anderson S, Burkhalter A, Cauli B, Fairen A, Feldmeyer D, Fishell G, Fitzpatrick D, Freund TF, Gonzalez-Burgos G, Hestrin S, Hill S, Hof PR, Huang J, Jones EG, Kawaguchi Y, Kisvarday Z, Kubota Y, Lewis DA, Marin O, Markram H, McBain CJ, Meyer HS, Monyer H, Nelson SB, Rockland K, Rossier J, Rubenstein JLR, Rudy B, Scanziani M, Shepherd GM, Sherwood CC, Staiger JF, Tamas G, Thomson A, Wang Y, Yuste R, Ascoli GA. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013;14:202–216. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of Neurogenesis by Interkinetic Nuclear Migration through an Apical-Basal Notch Gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschênes-Furry J, Perrone-Bizzozero N, Jasmin BJ. The RNA-binding protein HuD: a regulator of neuronal differentiation, maintenance and plasticity. BioEssays. 2006;28:822–833. doi: 10.1002/bies.20449. [DOI] [PubMed] [Google Scholar]
- DeStefano AL, Latourelle J, Lew MF, Suchowersky O, Klein C, Golbe LI, Mark MH, Growdon JH, Wooten GF, Watts R, Guttman M, Racette BA, Perlmutter JS, Marlor L, Shill HA, Singer C, Goldwurm S, Pezzoli G, Saint-Hilaire MH, Hendricks AE, Gower A, Williamson S, Nagle MW, Wilk JB, Massood T, Huskey KW, Baker KB, Itin I, Litvan I, Nicholson G, Corbett A, Nance M, Drasby E, Isaacson S, Burn DJ, Chinnery PF, Pramstaller PP, Al-Hinti J, Moller AT, Ostergaard K, Sherman SJ, Roxburgh R, Snow B, Slevin JT, Cambi F, Gusella JF, Myers RH. Replication of association between ELAVL4 and Parkinson disease: the GenePD study. Hum Genet. 2008;124:95–99. doi: 10.1007/s00439-008-0526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond A. Biological and social influences on cognitive control processes dependent on prefrontal cortex. Progress in Brain Research. 2011;189:319–339. doi: 10.1016/B978-0-444-53884-0.00032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobashi Y, Shoji M, Wakata Y, Kameya T. Expression of HuD protein is essential for initial phase of neuronal differentiation in rat pheochromocytoma PC12 cells. Biochem Biophys Res Commun. 1998;244:226–229. doi: 10.1006/bbrc.1998.8247. [DOI] [PubMed] [Google Scholar]
- Dong Z, Yang N, Yeo SY, Chitnis A, Guo S. Intralineage directional Notch signaling regulates self-renewal and differentiation of asymmetrically dividing radial glia. Neuron. 2012;74:65–78. doi: 10.1016/j.neuron.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, Yoon SO, Bassell GJ, Twiss JL. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30:4665–4677. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–4230. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, Zhang J, Feng H, Gong S, Heintz N. Mouse transgenesis in a single locus with independent regulation for multiple fluorophores. PLoS ONE. 2012;7:e40511. doi: 10.1371/journal.pone.0040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enstero M, Akerborg O, Lundin D, Wang B, Furey TS, Ohman M, Lagergren J. A computational screen for site selective A-to-I editing detects novel sites in neuron specific Hu proteins. BMC Bioinformatics. 2010;11:6. doi: 10.1186/1471-2105-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espuny-Camacho I, Michelsen Kimmo A, Gall D, Linaro D, Hasche A, Bonnefont J, Bali C, Orduz D, Bilheu A, Herpoel A, Lambert N, Gaspard N, Péron S, Schiffmann Serge N, Giugliano M, Gaillard A, Vanderhaeghen P. Pyramidal Neurons Derived from Human Pluripotent Stem Cells Integrate Efficiently into Mouse Brain Circuits In Vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, Park PJ, Walsh CA. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Castillo N, Orejarena MJ, Ribases M, Blanco E, Casas M, Robledo P, Maldonado R, Cormand B. Active and passive MDMA (‘ecstasy’) intake induces differential transcriptional changes in the mouse brain. Genes, brain, and behavior. 2012;11:38–51. doi: 10.1111/j.1601-183X.2011.00735.x. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Koch I, Westhof E, Nusslein-Volhard C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3[prime] UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997;16:1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertuzinhos S, Krsnik Ž, Kawasawa YI, Rašin M-R, Kwan KY, Chen J-G, Judaš M, Hayashi M, Šestan N. Selective Depletion of Molecularly Defined Cortical Interneurons in Human Holoprosencephaly with Severe Striatal Hypoplasia. Cerebral Cortex. 2009;19:2196–2207. doi: 10.1093/cercor/bhp009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, Kalervo A, Lipsanen-Nyman M, Skaug J, Nakabayashi K, Finucane B, Hartung D, Innes M, Kerem B, Nowaczyk MJ, Rivlin J, Roberts W, Senman L, Summers A, Szatmari P, Wong V, Vincent JB, Zeesman S, Osborne LR, Cardy JO, Kere J, Scherer SW, Hannula-Jouppi K. Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am J Hum Genet. 2006;79:965–972. doi: 10.1086/508902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J, Wilsch-Brauninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, Huttner WB. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- Fietz SA, Lachmann R, Brandl H, Kircher M, Samusik N, Schröder R, Lakshmanaperumal N, Henry I, Vogt J, Riehn A, Distler W, Nitsch R, Enard W, Pääbo S, Huttner WB. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proceedings of the National Academy of Sciences. 2012;109:11836–11841. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Dehay C, Kennedy H, Huttner WB. Making bigger brains-the evolution of neural-progenitor-cell division. J Cell Sci. 2008;121:2783–2793. doi: 10.1242/jcs.023465. [DOI] [PubMed] [Google Scholar]
- Floeter MK, Jones EG. Transplantation of fetal postmitotic neurons to rat cortex: survival, early pathway choices and long-term projections of outgrowing axons. Brain Res. 1985;354:19–38. doi: 10.1016/0165-3806(85)90065-3. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Gil-Sanz C, Martinez-Garay I, Espinosa A, Harkins-Perry SR, Ramos C, Muller U. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco Santos J, Müller U. Shaping Our Minds: Stem and Progenitor Cell Diversity in the Mammalian Neocortex. Neuron. 2013;77:19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao A, Sasano Y, Imataka H, Inoue K, Sakamoto H, Sonenberg N, Thoma C, Fujiwara T. The ELAV protein HuD stimulates cap-dependent translation in a Poly(A)- and eIF4A-dependent manner. Mol Cell. 2009;36:1007–1017. doi: 10.1016/j.molcel.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli R, Fiocco R, De Filippis L, Muzio L, Gritti A, Mercurio S, Broccoli V, Pellegrini M, Mallamaci A, Vescovi AL. Emx2 regulates the proliferation of stem cells of the adult mammalian central nervous system. Development. 2002;129:1633–1644. doi: 10.1242/dev.129.7.1633. [DOI] [PubMed] [Google Scholar]
- Garcez PP, Henrique NP, Furtado DA, Bolz J, Lent R, Uziel D. Axons of callosal neurons bifurcate transiently at the white matter before consolidating an interhemispheric projection. Eur J Neurosci. 2007;25:1384–1394. doi: 10.1111/j.1460-9568.2007.05387.x. [DOI] [PubMed] [Google Scholar]
- Garey L. When cortical development goes wrong: schizophrenia as a neurodevelopmental disease of microcircuits. J Anat. 2010;217:324–333. doi: 10.1111/j.1469-7580.2010.01231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, Ramalho-Santos M. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- gensat.org. 2013 . The Rockefeller University; New York, NY: 2013. The Gene Expression Nervous System Atlas (GENSAT) Project, NINDS Contracts N01NS02331 & HHSN271200723701C to The Rockefeller University (New York, NY) www.genesat.org. [Google Scholar]
- Gerstein M. Genomics: ENCODE leads the way on big data. Nature. 2012;489:208. doi: 10.1038/489208b. [DOI] [PubMed] [Google Scholar]
- Giuditta ACA, Lazzarini G. Ribosomal RNA in the axoplasm of the squid giant axon. Journal of neurochemistry. 1980;34:1757–1760. doi: 10.1111/j.1471-4159.1980.tb11271.x. [DOI] [PubMed] [Google Scholar]
- Giuditta AME, Perrone Capano C, Langella M, Martin R, Castigli E, Kaplan BB. Active polysomes in the axoplasm of the squid giant axon. Journal of Neuroscience Research. 1991;28:18–28. doi: 10.1002/jnr.490280103. [DOI] [PubMed] [Google Scholar]
- Giuditta AMS, Felsani A, Del Rio A. Factors for protein synthesis in the axoplasm of squid giant axons. Journal of neurochemistry Journal of Neurochemistry. 1977;28:1393–1395. doi: 10.1111/j.1471-4159.1977.tb12339.x. [DOI] [PubMed] [Google Scholar]
- GKS . Limma: linear models for microarray data. In: Gentleman RCV, Dudoit S, Irizarry RA, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Goldman-Rakic P. The “psychic cell” of Ramón y Cajal. Progress in Brain Research. 2002;136:427–434. doi: 10.1016/s0079-6123(02)36035-7. [DOI] [PubMed] [Google Scholar]
- Gong S, Kus L, Heintz N. Rapid bacterial artificial chromosome modification for large-scale mouse transgenesis. Nature protocols. 2010;5:1678–1696. doi: 10.1038/nprot.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- Grandinetti KB, Spengler BA, Biedler JL, Ross RA. Loss of one HuD allele on chromosome [num]1p selects for amplification of the N-myc proto-oncogene in human neuroblastoma cells. Oncogene. 2005;25:706–712. doi: 10.1038/sj.onc.1209095. [DOI] [PubMed] [Google Scholar]
- Griner LN, Reuther GW. Aggressive myeloid leukemia formation is directed by the Musashi 2/Numb pathway. Cancer Biology & Therapy. 2010;10:979–982. doi: 10.4161/cbt.10.10.14010. [DOI] [PubMed] [Google Scholar]
- Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin αpromoter choice. Proceedings of the National Academy of Sciences. 2012;109:21081–21086. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MM, Shin Y, Xu X, Zhu Y, Li M, Sestan N. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci U S A. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wu X, Chung WY, Li T, Nekrutenko A, Altman NS, Chen G, Ma H. Transcriptome of embryonic and neonatal mouse cortex by high-throughput RNA sequencing. Proc Natl Acad Sci U S A. 2009;106:12741–12746. doi: 10.1073/pnas.0902417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 Suppresses Early Cortical Cell Fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Torii K, Kawasawa YI, Kuhn A, Rakic P. Combined transcriptome analysis of fetal human and mouse cerebral cortex exposed to alcohol. Proc Natl Acad Sci U S A. 2011;108:4212–4217. doi: 10.1073/pnas.1100903108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cell Reports. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Hatten ME. LIS-less neurons don’t even make it to the starting gate. The Journal of Cell Biology. 2005;170:867–871. doi: 10.1083/jcb.200506140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugarvoll K, Toft M, Ross OA, Stone JT, Heckman MG, White LR, Lynch T, Gibson JM, Wszolek ZK, Uitti RJ, Aasly JO, Farrer MJ. ELAVL4, PARK10, and the Celts. Movement disorders: official journal of the Movement Disorder Society. 2007;22:585–587. doi: 10.1002/mds.21336. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Head K, Gong S, Joseph S, Wang C, Burkhardt T, Rossi MR, LaDuca J, Matsui S, Vaughan M, Hicks DG, Heintz N, Cowell JK. Defining the expression pattern of the LGI1 gene in BAC transgenic mice. Mamm Genome. 2007;18:328–337. doi: 10.1007/s00335-007-9024-6. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF. From radial glia to pyramidal-projection neuron: transcription factor cascades in cerebral cortex development. Mol Neurobiol. 2006;33:33–50. doi: 10.1385/MN:33:1:033. [DOI] [PubMed] [Google Scholar]
- Hevner RF. Layer-specific markers as probes for neuron type identity in human neocortex and malformations of cortical development. J Neuropath Exp Neur. 2007;66:101–109. doi: 10.1097/nen.0b013e3180301c06. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Miyashita-Lin E, Rubenstein JLR. Cortical and thalamic axon pathfinding defects in Tbr1, Gbx2, and Pax6 mutant mice: Evidence that cortical and thalamic axons interact and guide each other. The Journal of Comparative Neurology. 2002;447:8–17. doi: 10.1002/cne.10219. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y-P, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JLR. Tbr1 Regulates Differentiation of the Preplate and Layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. http://www.genepaint.org (2012) http://www.genepaint.org. Max Planck Institute. [DOI] [PubMed] [Google Scholar]
- Huang H, Jeon T, Sedmak G, Pletikos M, Vasung L, Xu X, Yarowsky P, Richards LJ, Kostovic I, Sestan N, Mori S. Coupling Diffusion Imaging with Histological and Gene Expression Analysis to Examine the Dynamics of Cortical Areas across the Fetal Period of Human Brain Development. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubel DH, Wiesel TN. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. The Journal of physiology. 1962;160:106–154. doi: 10.1113/jphysiol.1962.sp006837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The Neural RNA-Binding Protein Musashi1 Translationally Regulates Mammalian numb Gene Expression by Interacting with Its mRNA. Molecular and Cellular Biology. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince-Dunn G, Okano HJ, Jensen KB, Park WY, Zhong R, Ule J, Mele A, Fak JJ, Yang C, Zhang C, Yoo J, Herre M, Okano H, Noebels JL, Darnell RB. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron. 2012;75:1067–1080. doi: 10.1016/j.neuron.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee Y-h, Narzisi G, Leotta A, Kendall J, Grabowska E, Ma B, Marks S, Rodgers L, Stepansky A, Troge J, Andrews P, Bekritsky M, Pradhan K, Ghiban E, Kramer M, Parla J, Demeter R, Fulton Lucinda L, Fulton Robert S, Magrini Vincent J, Ye K, Darnell Jennifer C, Darnell Robert B, Mardis Elaine R, Wilson Richard K, Schatz Michael C, McCombie WR, Wigler M. De Novo Gene Disruptions in Children on the Autistic Spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, Zhao C, Lagoo A, Gerrard G, Foroni L, Goldman J, Goh H, Kim S-H, Kim D-W, Chuah C, Oehler VG, Radich JP, Jordan CT, Reya T. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J, Castle J, Garrett-Engele P, Kan Z, Loerch P, Armour C, Santos R, Schadt E, Stoughton R, Shoemaker D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;5653 doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and Evolutionary Insights into Human Brain Development through Global Transcriptome Analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki M, Nakamura S, Machon O, Krauss S, Radice GL, Takeichi M. N-cadherin mediates cortical organization in the mouse brain. Dev Biol. 2007;304:22–33. doi: 10.1016/j.ydbio.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, Ogawa Y, Toyama Y, Miyata T, Okano H. Musashi1: An Evolutionally Conserved Marker for CNS Progenitor Cells Including Neural Stem Cells. Developmental Neuroscience. 2000;22:139–153. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, Guennel T, Shin Y, Johnson MB, Krsnik Z, Mayer S, Fertuzinhos S, Umlauf S, Lisgo SN, Vortmeyer A, Weinberger DR, Mane S, Hyde TM, Huttner A, Reimers M, Kleinman JE, Sestan N. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS ONE. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Moon M, Yu SB, Mook-Jung I, Kim JI. RNA-Seq analysis of frontal cortex and cerebellum from 5XFAD mice at early stage of disease pathology. Journal of Alzheimer’s disease: JAD. 2012;29:793–808. doi: 10.3233/JAD-2012-111793. [DOI] [PubMed] [Google Scholar]
- King PH. Differential expression of the neuroendocrine genes Hel-N1 and HuD in small-cell lung carcinoma: evidence for down-regulation of HuD in the variant phenotype. International journal of cancer Journal international du cancer. 1997;74:378–382. doi: 10.1002/(sici)1097-0215(19970822)74:4<378::aid-ijc3>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Kishi Y, Fujii Y, Hirabayashi Y, Gotoh Y. HMGA regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nat Neurosci. 2012;15:1127–1133. doi: 10.1038/nn.3165. [DOI] [PubMed] [Google Scholar]
- Kishore S, Luber S, Zavolan M. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief Funct Genomics. 2010;9:391–404. doi: 10.1093/bfgp/elq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu XC, Singer RH. Sequences Responsible for Intracellular-Localization of Beta-Actin Messenger-Rna Also Affect Cell Phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Koenig E, Martin R. Cortical plaque-like structures identify ribosome-containing domains in the Mauthner cell axon. The Journal of Neuroscience. 1996;16:1400–1411. doi: 10.1523/JNEUROSCI.16-04-01400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T, Higo N, Sato A, Oishi T, Nishimura Y, Yamamoto T, Murata Y, Yoshino-Saito K, Onoe H, Isa T. Functional annotation of genes differentially expressed between primary motor and prefrontal association cortices of macaque brain. Neurochem Res. 2013;38:133–140. doi: 10.1007/s11064-012-0900-4. [DOI] [PubMed] [Google Scholar]
- Kong J, Lasko P. Translational control in cellular and developmental processes. Nat Rev Genet. 2012;13:383–394. doi: 10.1038/nrg3184. [DOI] [PubMed] [Google Scholar]
- Koo B-K, Lim H-S, Song R, Yoon M-J, Yoon K-J, Moon J-S, Kim Y-W, Kwon M-c, Yoo K-W, Kong M-P, Lee J, Chitnis AB, Kim C-H, Kong Y-Y. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- Koopmans PJ, Barth M, Norris DG. Layer-specific BOLD activation in human V1. Hum Brain Mapp. 2010;31:1297–1304. doi: 10.1002/hbm.20936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornack DR, Rakic P. Radial and horizontal deployment of clonally related cells in the primate neocortex: Relationship to distinct mitotic lineages. Neuron. 1995;15:311–321. doi: 10.1016/0896-6273(95)90036-5. [DOI] [PubMed] [Google Scholar]
- Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci U S A. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y. Interkinetic nuclear migration: beyond a hallmark of neurogenesis. Cell Mol Life Sci. 2012;69:2727–2738. doi: 10.1007/s00018-012-0952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostovic I, Judas M. Transient patterns of organization of the human fetal brain. Croat Med J. 1998;39:107–114. [PubMed] [Google Scholar]
- Kostovic I, Judas M. The development of the subplate and thalamocortical connections in the human foetal brain. Acta Paediatr. 2010;99:1119–1127. doi: 10.1111/j.1651-2227.2010.01811.x. [DOI] [PubMed] [Google Scholar]
- Kostovic I, Judas M, Sedmak G. Developmental history of the subplate zone, subplate neurons and interstitial white matter neurons: relevance for schizophrenia. International journal of developmental neuroscience: the official journal of the International Society for Developmental Neuroscience. 2011;29:193–205. doi: 10.1016/j.ijdevneu.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Kostovic I, Lukinovic N, Judas M, Bogdanovic N, Mrzljak L, Zecevic N, Kubat M. Structural basis of the developmental plasticity in the human cerebral cortex: the role of the transient subplate zone. Metab Brain Dis. 1989;4:17–23. doi: 10.1007/BF00999489. [DOI] [PubMed] [Google Scholar]
- Kostovic IRP. Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. Journal of Comparative Neurology. 1990;297:441–470. doi: 10.1002/cne.902970309. [DOI] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, Daza RA, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusek G, Campbell M, Doyle F, Tenenbaum Scott A, Kiebler M, Temple S. Asymmetric Segregation of the Double-Stranded RNA Binding Protein Staufen2 during Mammalian Neural Stem Cell Divisions Promotes Lineage Progression. Cell stem cell. 2012;11:505–516. doi: 10.1016/j.stem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Johnson MB, Dube U, Shim S, Rasin MR, Sousa AM, Fertuzinhos S, Chen JG, Arellano JI, Chan DW, Pletikos M, Vasung L, Rowitch DH, Huang EJ, Schwartz ML, Willemsen R, Oostra BA, Rakic P, Heffer M, Kostovic I, Judas M, Sestan N. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012a;149:899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012b;139:1535–1546. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CSL, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Latasa M, Cisneros E, Frade J. Cell cycle control of Notch signaling and the functional regionalization of the neuroepithelium during vertebrate neurogenesis. International Journal of Develomental Biology. 2009;53:895–908. doi: 10.1387/ijdb.082721ml. [DOI] [PubMed] [Google Scholar]
- Lempp T, Toennes SW, Wunder C, Russe OQ, Moser CV, Kynast KL, Freitag CM, Niederberger E. Altered gene expression in the prefrontal cortex of young rats induced by the ADHD drug atomoxetine. Progress in neuro-psychopharmacology & biological psychiatry. 2013;40:221–228. doi: 10.1016/j.pnpbp.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Li S, Overman JJ, Katsman D, Kozlov SV, Donnelly CJ, Twiss JL, Giger RJ, Coppola G, Geschwind DH, Carmichael ST. An age-related sprouting transcriptome provides molecular control of axonal sprouting after stroke. Nat Neurosci. 2010;13:1496–1504. doi: 10.1038/nn.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Newbern JM, Wu Y, Morgan-Smith M, Zhong J, Charron J, Snider WD. MEK Is a Key Regulator of Gliogenesis in the Developing Brain. Neuron. 2012;75:1035–1050. doi: 10.1016/j.neuron.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickiss T, Cheung AF, Hutchinson CE, Taylor JS, Molnar Z. Examining the relationship between early axon growth and transcription factor expression in the developing cerebral cortex. J Anat. 2012;220:201–211. doi: 10.1111/j.1469-7580.2011.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lövblad KOBJ, Bourdenet S, Burger D, Bernard D, Regli F, Steck AJ. Sensory neuronopathy and small cell lung cancer: antineuronal antibody reacting with neuroblastoma cells. Journal of Neurology. 1993;240:327–332. doi: 10.1007/BF00839961. [DOI] [PubMed] [Google Scholar]
- Lovell MA, Markesbery WR. Ectopic Expression of Musashi-1 in Alzheimer Disease and Pick Disease. Journal of Neuropathology & Experimental Neurology. 2005;64:675–680. doi: 10.1097/01.jnen.0000173891.17176.5b. [DOI] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PM, Struhl G. cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336:595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt Bradley M, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans Zoe A, Kheradpour P, Mountoufaris G, Carey C, Barnea G, Kellis M, Lomvardas S. An Epigenetic Signature for Monoallelic Olfactory Receptor Expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Vinukonda G, Vose LR, Diamond D, Bhimavarapu BBR, Hu F, Zia MT, Hevner R, Zecevic N, Ballabh P. Neurogenesis Continues in the Third Trimester of Pregnancy and Is Suppressed by Premature Birth. The Journal of Neuroscience. 2013;33:411–423. doi: 10.1523/JNEUROSCI.4445-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, Horvath TL, Vaccarino FM. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin P, Xiong J, Liu X, Yan Z, Zhang X, Li M, He L, Somel M, Yuan Y, Phoebe Chen Y-P, Li N, Hu Y, Fu N, Ning Z, Zeng R, Yang H, Chen W, Gelfand M, Khaitovich P. Widespread splicing changes in human brain development and aging. Mol Syst Biol. 2013;9 doi: 10.1038/msb.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JLR, Chen B. Tbr1 and Fezf2 Regulate Alternate Corticofugal Neuronal Identities during Neocortical Development. The Journal of Neuroscience. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metin C, Vallee RB, Rakic P, Bhide PG. Modes and mishaps of neuronal migration in the mammalian brain. J Neurosci. 2008;28:11746–11752. doi: 10.1523/JNEUROSCI.3860-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G. Building a human cortex: the evolutionary differentiation of Cajal-Retzius cells and the cortical hem. J Anat. 2010;217:334–343. doi: 10.1111/j.1469-7580.2010.01266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mission JPTT, Caviness VS., Jr Ontogeny of radial and other astroglial cells in murine cerebral cortex. Glia. 1991;4:138–148. doi: 10.1002/glia.440040205. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Mohan V, Sinha RA, Pathak A, Rastogi L, Kumar P, Pal A, Godbole MM. Maternal thyroid hormone deficiency affects the fetal neocorticogenesis by reducing the proliferating pool, rate of neurogenesis and indirect neurogenesis. Exp Neurol. 2012;237:477–488. doi: 10.1016/j.expneurol.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Molnar ZCG. Cereral Cortical Development in Rodents and Primates. Progress in Brain Research. 2012;195 doi: 10.1016/B978-0-444-53860-4.00003-9. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, Macklis JD. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29:12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Morgen K, Sammer G, Weber L, Aslan B, Muller C, Bachmann GF, Sandmann D, Oechsner M, Vaitl D, Kaps M, Reuter I. Structural brain abnormalities in patients with Parkinson disease: a comparative voxel-based analysis using T1-weighted MR imaging and magnetization transfer imaging. AJNR American journal of neuroradiology. 2011;32:2080–2086. doi: 10.3174/ajnr.A2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountcastle VB, Davies PW, Berman AL. Response properties of neurons of cat’s somatic sensory cortex to peripheral stimuli. J Neurophysiol. 1957;20:374–407. doi: 10.1152/jn.1957.20.4.374. [DOI] [PubMed] [Google Scholar]
- Mrzljak LUH, Kostovic I, van Eden CG. Prenatal development of neurons in the human prefrontal cortex. II. A quantitative Golgi study. Journal of Comparative Neurology. 1992;316:485–496. doi: 10.1002/cne.903160408. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui C-H, Relling MV, Shurtleff SA, Downing JR. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Muzio L, DiBenedetto B, Stoykova A, Boncinelli E, Gruss P, Mallamaci A. Conversion of cerebral cortex into basal ganglia in Emx2−/− Pax6Sey/Sey double-mutant mice. Nat Neurosci. 2002;5:737–745. doi: 10.1038/nn892. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- Nichols AJ, Olson EC. Reelin promotes neuronal orientation and dendritogenesis during preplate splitting. Cereb Cortex. 2010;20:2213–2223. doi: 10.1093/cercor/bhp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto Y, Okano H. New insight into cancer therapeutics: Induction of differentiation by regulating the Musashi/Numb/Notch pathway. Cell Res. 2010;20:1083–1085. doi: 10.1038/cr.2010.122. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Noureddine MA, Qin XJ, Oliveira SA, Skelly TJ, van der Walt J, Hauser MA, Pericak-Vance MA, Vance JM, Li YJ. Association between the neuron-specific RNA-binding protein ELAVL4 and Parkinson disease. Hum Genet. 2005;117:27–33. doi: 10.1007/s00439-005-1259-2. [DOI] [PubMed] [Google Scholar]
- Okano H, Temple S. Cell types to order: temporal specification of CNS stem cells. Current Opinion in Neurobiology. 2009;19:112–119. doi: 10.1016/j.conb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Ozdinler PH, Benn S, Yamamoto TH, Guzel M, Brown RH, Jr, Macklis JD. Corticospinal motor neurons and related subcerebral projection neurons undergo early and specific neurodegeneration in hSOD1G(9)(3)A transgenic ALS mice. J Neurosci. 2011;31:4166–4177. doi: 10.1523/JNEUROSCI.4184-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park-Lee S, Kim S, Laird-Offringa IA. Characterization of the interaction between neuronal RNA-binding protein HuD and AU-rich RNA. The Journal of biological chemistry. 2003;278:39801–39808. doi: 10.1074/jbc.M307105200. [DOI] [PubMed] [Google Scholar]
- Park S, Myszka DG, Yu M, Littler SJ, Laird-Offringa IA. HuD RNA recognition motifs play distinct roles in the formation of a stable complex with AU-rich RNA. Mol Cell Biol. 2000;20:4765–4772. doi: 10.1128/mcb.20.13.4765-4772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale A, Gusev PA, Amadio M, Dottorini T, Govoni S, Alkon DL, Quattrone A. Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory. Proc Natl Acad Sci U S A. 2004;101:1217–1222. doi: 10.1073/pnas.0307674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Judaš M, Kostović I, Uylings HBM. Lifespan Alterations of Basal Dendritic Trees of Pyramidal Neurons in the Human Prefrontal Cortex: A Layer-Specific Pattern. Cerebral Cortex. 2008;18:915–929. doi: 10.1093/cercor/bhm124. [DOI] [PubMed] [Google Scholar]
- Petanjek Z, Judaš M, Šimić G, Rašin MR, Uylings HBM, Rakic P, Kostović I. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proceedings of the National Academy of Sciences. 2011;108:13281–13286. doi: 10.1073/pnas.1105108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Scheuch H, Burkard TR, Tinter J, Wernle T, Rumpel S. Induction of immediate early genes in the mouse auditory cortex after auditory cued fear conditioning to complex sounds. Genes, brain, and behavior. 2012;11:314–324. doi: 10.1111/j.1601-183X.2011.00761.x. [DOI] [PubMed] [Google Scholar]
- Pichardo-Casas I, Goff LA, Swerdel MR, Athie A, Davila J, Ramos-Brossier M, Lapid-Volosin M, Friedman WJ, Hart RP, Vaca L. Expression profiling of synaptic microRNAs from the adult rat brain identifies regional differences and seizure-induced dynamic modulation. Brain Research. 2012;1436:20–33. doi: 10.1016/j.brainres.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto FPC, Ferreira D, Moradas-Ferreira P, Tamagnini P. Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0034983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- Quattrone A, Pascale A, Nogues X, Zhao W, Gusev P, Pacini A, Alkon DL. Posttranscriptional regulation of gene expression in learning by the neuronal ELAV-like mRNA-stabilizing proteins. Proc Natl Acad Sci U S A. 2001;98:11668–11673. doi: 10.1073/pnas.191388398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racca C, Gardiol A, Eom T, Ule J, Triller A, Darnell RB. The Neuronal Splicing Factor Nova Co-Localizes with Target RNAs in the Dendrite. Front Neural Circuits. 2010;4:5. doi: 10.3389/neuro.04.005.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Rakic P. Elusive radial glial cells: Historical and evolutionary perspective. Glia. 2003;43:19–32. doi: 10.1002/glia.10244. [DOI] [PubMed] [Google Scholar]
- Rakic PH-TK, Sarkisian MR. Genetic determinants of neuronal migration in the cerebral cortex. Novartis Found Symp. 2007;288:45–53. doi: 10.1002/9780470994030.ch4. [DOI] [PubMed] [Google Scholar]
- Rakic P., SR Autoradiographic study of supravital DNA synthesis in fetal human brain. Journal of Neuropathology & Experimental Neurology. 1968;27:139–140. [PubMed] [Google Scholar]
- Rallu M, Corbin JG, Fishell G. Parsing the prosencephalon. Nat Rev Neurosci. 2002;3:943–951. doi: 10.1038/nrn989. [DOI] [PubMed] [Google Scholar]
- Ramón y Cajal SDJ, Jones EG. Cajal on the cerebral cortex: an annotated translation of the complete writings. Oxford University Press; 1988. [Google Scholar]
- Rapoport SI, Nelson PT. Biomarkers and evolution in Alzheimer disease. Prog Neurobiol. 2011;95:510–513. doi: 10.1016/j.pneurobio.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, Sestan N. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Ratti A, Fallini C, Cova L, Fantozzi R, Calzarossa C, Zennaro E, Pascale A, Quattrone A, Silani V. A role for the ELAV RNA-binding proteins in neural stem cells: stabilization of Msi1 mRNA. Journal of Cell Science. 2006;119:1442–1452. doi: 10.1242/jcs.02852. [DOI] [PubMed] [Google Scholar]
- Robinow SCA, Yao KM, White K. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science. 1988;242:1570–1572. doi: 10.1126/science.3144044. [DOI] [PubMed] [Google Scholar]
- Robinow S, White K. The Locus Elav of Drosophila-Melanogaster Is Expressed in Neurons at All Developmental Stages. Developmental Biology. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- Rouaux C, Arlotta P. Fezf2 directs the differentiation of corticofugal neurons from striatal progenitors in vivo. Nat Neurosci. 2010;13:1345–1347. doi: 10.1038/nn.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaux C, Arlotta P. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat Cell Biol. 2013;15:214–221. doi: 10.1038/ncb2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso DL, Pearson CA, Gaber ZB, Miquelajauregui A, Li S, Portera-Cailliau C, Morrisey EE, Novitch BG. Foxp- mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron. 2012;74:314–330. doi: 10.1016/j.neuron.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Katsel P, Davis KL, Siever LJ, Haroutunian V. A System-Level Transcriptomic Analysis of Schizophrenia Using Postmortem Brain Tissue Samples. Arch Gen Psychiatry. 2012;69:1–11. doi: 10.1001/archgenpsychiatry.2012.704. [DOI] [PubMed] [Google Scholar]
- Saffary R, Xie Z. FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J Neurosci. 2011;31:1427–1439. doi: 10.1523/JNEUROSCI.4854-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara S, O’Leary DD. Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron. 2009;63:48–62. doi: 10.1016/j.neuron.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Hanai S, Takashima S, Nakagawa E, Okazaki S, Inoue T, Miyata R, Hoshino K, Akashi T, Sasaki M, Goto Y, Hayashi M, Itoh M. Neocortical layer formation of human developing brains and lissencephalies: consideration of layer-specific marker expression. Cereb Cortex. 2011;21:588–596. doi: 10.1093/cercor/bhq125. [DOI] [PubMed] [Google Scholar]
- Sakakibara S-i, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T, Okano H. RNA-binding protein Musashi family: Roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proceedings of the National Academy of Sciences. 2002;99:15194–15199. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara S-i, Okano H. Expression of Neural RNA-Binding Proteins in the Postnatal CNS: Implications of Their Roles in Neuronal and Glial Cell Development. The Journal of Neuroscience. 1997;17:8300–8312. doi: 10.1523/JNEUROSCI.17-21-08300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annual review of pathology. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- Sawicka KBM, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochemical Society Transactions. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- Schmidt EF, Warner-Schmidt JL, Otopalik BG, Pickett SB, Greengard P, Heintz N. Identification of the cortical neurons that mediate antidepressant responses. Cell. 2012;149:1152–1163. doi: 10.1016/j.cell.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Science AIfB. Allen Brain Atlas. 2012 http://www.brain-map.org/
- Sequeira AML, Walsh DM, Cartagena PM, Choudary P, Li J, Schatzberg AF, Watson SJ, Akil H, Myers RM, Jones EG, Bunney WE, Vawter MP. Gene expression changes in the prefrontal cortex, anterior cingulate cortex and nucleus accumbens of mood disorders subjects that committed suicide. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0035367. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Colasante G, Nini A, Klein WH, Broccoli V. Tbr2-positive intermediate (basal) neuronal progenitors safeguard cerebral cortex expansion by controlling amplification of pallial glutamatergic neurons and attraction of subpallial GABAergic interneurons. Genes Dev. 2010;24:1816–1826. doi: 10.1101/gad.575410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave K-A, Huylebroeck D, Tarabykin V. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci. 2009;12:1373–1380. doi: 10.1038/nn.2409. [DOI] [PubMed] [Google Scholar]
- Sheldon M, Rice DS, D’Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell BW, Cooper JA, Goldowitz D, Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, Robinson HPC, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15:477–486. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, Sestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz S, Dunbar RI. Species differences in executive function correlate with hippocampus volume and neocortex ratio across nonhuman primates. J Comp Psychol. 2010;124:252–260. doi: 10.1037/a0018894. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Miale IL, Feder N. Cell proliferation and migration in the primitive ependymal zone: an autoradiographic study of histogenesis in the nervous system. Exp Neurol. 1959;1:322–333. doi: 10.1016/0014-4886(59)90024-x. [DOI] [PubMed] [Google Scholar]
- Sidman RLRP. Neuronal migration, with special reference to developing human brain: a review. Brain Research. 1973;62:1–35. doi: 10.1016/0006-8993(73)90617-3. [DOI] [PubMed] [Google Scholar]
- Siegenthaler JA, Ashique AM, Zarbalis K, Patterson KP, Hecht JH, Kane MA, Folias AE, Choe Y, May SR, Kume T, Napoli JL, Peterson AS, Pleasure SJ. Retinoic Acid from the Meninges Regulates Cortical Neuron Generation. Cell. 2009;139:597–609. doi: 10.1016/j.cell.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ, Liaw HJ, Myung K, Walsh CA, Gaiano N, Pavan WJ. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci. 2010;13:551–558. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Afroz R, Bassell GJ, Furneaux HM, Perrone-Bizzozero NI, Burry RW. GAP-43 mRNA in growth cones is associated with HuD and ribosomes. J Neurobiol. 2004;61:222–235. doi: 10.1002/neu.20038. [DOI] [PubMed] [Google Scholar]
- Srinivasan K, Leone DP, Bateson RK, Dobreva G, Kohwi Y, Kohwi-Shigematsu T, Grosschedl R, McConnell SK. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc Natl Acad Sci U S A. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepniak E, Radice GL, Vasioukhin V. Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb Perspect Biol. 2009;1:a002949. doi: 10.1101/cshperspect.a002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward OLW. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. Journal of Neuroscience. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Goderie SK, Temple S. Asymmetric Distribution of EGFR Receptor during Mitosis Generates Diverse CNS Progenitor Cells. Neuron. 2005;45:873–886. doi: 10.1016/j.neuron.2005.01.045. [DOI] [PubMed] [Google Scholar]
- Szabo ADJ, Manley G, Rosenfeld M, Wong E, Henson J, Posner JB, Furneaux HM. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- Tanaka DH, Nakajima K. Migratory pathways of GABAergic interneurons when they enter the neocortex. European Journal of Neuroscience. 2012;35:1655–1660. doi: 10.1111/j.1460-9568.2012.08111.x. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E, Huttner WB. Neural Progenitor Nuclei IN Motion. Neuron. 2010;67:906–914. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Thomas CA, Paquola ACM, Muotri AR. LINE-1 Retrotransposition in the Nervous System. Annual Review of Cell and Developmental Biology. 2012;28:555–573. doi: 10.1146/annurev-cellbio-101011-155822. [DOI] [PubMed] [Google Scholar]
- Thompson KDV, Dubey A, Crockett DP, Rasin MR. Acute adaptive responses of central sensorimotor neurons after spinal cord injury. Transl Neurosci. 2010;1:268–278. [Google Scholar]
- Toba G, White K. The third RNA recognition motif of Drosophila ELAV protein has a role in multimerization. Nucleic Acids Research. 2008;36:1390–1399. doi: 10.1093/nar/gkm1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Katayama K, Yamauchi H, Nakayama H, Doi K. Cell cycle progression is required for nuclear migration of neural progenitor cells. Brain Res. 2006;1088:57–67. doi: 10.1016/j.brainres.2006.03.042. [DOI] [PubMed] [Google Scholar]
- van den Berghe V, Stappers E, Vandesande B, Dimidschstein J, Kroes R, Francis A, Conidi A, Lesage F, Dries R, Cazzola S, Berx G, Kessaris N, Vanderhaeghen P, van Ijcken W, Grosveld Frank G, Goossens S, Haigh Jody J, Fishell G, Goffinet A, Aerts S, Huylebroeck D, Seuntjens E. Directed Migration of Cortical Interneurons Depends on the Cell-Autonomous Action of Sip1. Neuron. 2013;77:70–82. doi: 10.1016/j.neuron.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Verheij C, Bakker CE, de Graaff E, Keulemans J, Willemsen R, Verkerk AJ, Galjaard H, Reuser AJ, Hoogeveen AT, Oostra BA. Characterization and localization of the FMR-1 gene product associated with fragile X syndrome. Nature. 1993;363:722–724. doi: 10.1038/363722a0. [DOI] [PubMed] [Google Scholar]
- Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu Y-H, Kuhl DPA, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang F, Eussen BE, van Ommen G-JB, Blonden LAJ, Riggins GJ, Chastain JL, Kunst CB, Galjaard H, Thomas Caskey C, Nelson DL, Oostra BA, Warren ST. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcon M, Oliver PL, Davies KE, Geschwind DH, Monaco AP, Fisher SE. A functional genetic link between distinct developmental language disorders. The New England journal of medicine. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virok DP, Kis Z, Szegedi V, Juhasz G, Zvara A, Jr, Muller G, Levay G, Harsing LG, Rajko R, Penke B, Janka Z, Janaky T, Puskas LG. Functional changes in transcriptomes of the prefrontal cortex and hippocampus in a mouse model of anxiety. Pharmacological reports: PR. 2011;63:348–361. doi: 10.1016/s1734-1140(11)70501-1. [DOI] [PubMed] [Google Scholar]
- Vogel BLA, Sauer M, Hock R. Cross-linking of DNA through HMGA1 suggests a DNA scaffold. Nucleic Acids Res. 2011;39:7124–7133. doi: 10.1093/nar/gkr396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Holst A, Egbers U, Prochiantz A, Faissner A. Neural stem/progenitor cells express 20 tenascin C isoforms that are differentially regulated by Pax6. The Journal of biological chemistry. 2007;282:9172–9181. doi: 10.1074/jbc.M608067200. [DOI] [PubMed] [Google Scholar]
- Wang H, Molfenter J, Zhu H, Lou H. Promotion of exon 6 inclusion in HuD pre-mRNA by Hu protein family members. Nucleic Acids Res. 2010a;38:3760–3770. doi: 10.1093/nar/gkq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WZ, Hoerder-Suabedissen A, Oeschger FM, Bayatti N, Ip BK, Lindsay S, Supramaniam V, Srinivasan L, Rutherford M, Mollgard K, Clowry GJ, Molnar Z. Subplate in the developing cortex of mouse and human. J Anat. 2010b;217:368–380. doi: 10.1111/j.1469-7580.2010.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci. 2011;14:555–561. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel J, Kuchna I, Nowicki K, Imaki H, Marchi E, Ma SY, Chauhan A, Chauhan V, Bobrowicz TW, de Leon M, Louis LA, Cohen IL, London E, Brown WT, Wisniewski T. The neuropathology of autism: defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 2010;119:755–770. doi: 10.1007/s00401-010-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler N, Wood L, Yu J, Solla SA, Shepherd GM. Top-down laminar organization of the excitatory network in motor cortex. Nat Neurosci. 2008;11:360–366. doi: 10.1038/nn2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, Xu M, Donnelly CJ, Tep C, Kendall M, Erenstheyn M, English AW, Schanen NC, Kirn-Safran CB, Yoon SO, Bassell GJ, Twiss JL. Axonal Localization of transgene mRNA in mature PNS and CNS neurons. J Neurosci. 2011;31:14481–14487. doi: 10.1523/JNEUROSCI.2950-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- www.brainmuseum.org (2012) www.brainmuseum.org. In: Brain Museum brainmuseum.org.
- www.euroexpress.org (2012) www.euroexpress.org. www.euroexpress.org: Eurexpress project
- Yang Y, Fung SJ, Rothwell A, Tianmei S, Weickert CS. Increased interstitial white matter neuron density in the dorsolateral prefrontal cortex of people with schizophrenia. Biol Psychiatry. 2011;69:63–70. doi: 10.1016/j.biopsych.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YT, Wang CL, Van Aelst L. DOCK7 interacts with TACC3 to regulate interkinetic nuclear migration and cortical neurogenesis. Nat Neurosci. 2012;15:1201–1210. doi: 10.1038/nn.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Jiang H, Chaichanasakul T, Gong S, Yang XW, Heintz N, Lin S. Modified bacterial artificial chromosomes for zebrafish transgenesis. Methods. 2006;39:183–188. doi: 10.1016/j.ymeth.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Yano M, Hayakawa-Yano Y, Mele A, Darnell RB. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron. 2010;66:848–858. doi: 10.1016/j.neuron.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Barnes AP, Hand R, Polleux F, Ehlers MD. TGF-β Signaling Specifies Axons during Brain Development. Cell. 2010;142:144–157. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- Yoon KJ, Koo BK, Im SK, Jeong HW, Ghim J, Kwon MC, Moon JS, Miyata T, Kong YY. Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron. 2008;58:519–531. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Zecevic N, Chen Y, Filipovic R. Contributions of cortical subventricular zone to the development of the human cerebral cortex. J Comp Neurol. 2005;491:109–122. doi: 10.1002/cne.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZW, Deschenes M. Intracortical axonal projections of lamina VI cells of the primary somatosensory cortex in the rat: a single-cell labeling study. J Neurosci. 1997;17:6365–6379. doi: 10.1523/JNEUROSCI.17-16-06365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Yang Y, Gao J, Tao H, Qu C, Qu J, Chen J. Area dependent expression of ZNF312 in human fetal cerebral cortex. Neurosci Res. 2010;68:73–76. doi: 10.1016/j.neures.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Ziats MN, Rennert OM. Aberrant Expression of Long Noncoding RNAs in Autistic Brain. J Mol Neurosci. 2012:1–5. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
- Zupan B, Toth M. Fmr-1 as an Offspring Genetic and a Maternal Environmental Factor in Neurodevelopmental Disease. In: Denman RB, editor. Modeling Fragile X Syndrome. Vol. 54. Springer; Berlin Heidelberg: 2012. pp. 243–253. [DOI] [PubMed] [Google Scholar]