Abstract
Advances in sequencing technology have significantly advanced the landscape of developmental biology research. The dissection of genetic networks in model and nonmodel organisms has been greatly enhanced with high-throughput sequencing technologies. RNA-seq has revolutionized the ability to perform developmental biology research in organisms without a published genome sequence. Here, we describe a protocol for developmental biologists to perform RNA-seq on dissected tissue or whole embryos. We start with the isolation of RNA and generation of sequencing libraries. We further show how to interpret and analyze the large amount of sequencing data that is generated in RNA-seq. We explore the abilities to examine differential expression, gene duplication, transcript assembly, alternative splicing and SNP discovery. For the purposes of this article, we use Xenopus laevis as the model organism to discuss uses of RNA-seq in an organism without a fully annotated genome sequence.
Keywords: RNA-seq, differential expression, Xenopus
1. Introduction
1.1 RNA-seq as a powerful tool in developmental biology
Over the past few decades, several advances have been made in dissecting the molecular pathways that govern basic developmental biology. Forward genetic screens in model organisms have been useful in identifying components of signaling pathways and transcription factors involved in processes ranging from the early patterning of embryos to the proper specification and differentiation of organs that are crucial to various stages of development [1–4]. These screens have been extremely valuable, not only in identifying the functional requirements of genes, but also in showing that no prior knowledge of a genes’ molecular function is needed to order genes in a genetic pathway [5]. These phenotype-based screens have provided a directed method to identify genes and assign them functions. This information can then be better utilized in addressing how these genes organize into networks during development.
Gene network dissection has been accelerated with many collaborative efforts undertaken to sequence the genomes and transcriptomes not only of humans, but also of the model organisms, their subspecies, and additional emerging non-model organisms used in developmental biology research [6–12]. Annotation of these genomes has allowed researchers to take reverse genetic approaches to specifically knock down genes of interest and assay for phenotypes to determine gene functions using directed mutagenesis, RNA interference (RNAi), or morpholino (MO) knockdown [13–15]. These approaches have been successfully implemented to further refine our ability to order genes with conserved function in molecular networks.
The molecular basis of genetic networks was first addressed at a genome-wide level with the use of microarrays [16–19]. Microarrays allowed researchers to assay for differential gene expression during development or as a result of genetic manipulation. A disadvantage of microarrays is that they require prior knowledge of cDNA library sequences and use genome annotation data to refine their models. For this reason, microarray platform design requires constant refinement as gene models improve. This includes updating previous versions of microarrays and therefore unnecessary expense. Furthermore, although microarrays have been used extensively for some model organisms in developmental biology research, the percentage of total genes represented on microarrays for organisms with poorly annotated genomes can be low. This is especially true in light of the recent advances in the number of emerging model organisms that are being exploited to study developmental biology and evolution [20]. Moreover, arrays are limited by cDNAs or genes that have been previously described and cannot capture differences in gene models that arise. They are further limited in that they sometimes do not provide insight into small RNAs, alternative splicing, differential 3’ untranslated region (UTR) patterns, or differences in expression of transcripts due to SNPs. As these new model organisms come to the forefront of developmental biology research, alternative methods have been developed to study global gene expression.
Massive parallel high-throughput sequencing has largely superseded microarray technology for most applications, but especially for transcriptomics, as it can be used to address all the limitations highlighted here. High-throughput sequencing of RNA, or RNA-seq, has become a popular method for identifying and quantifying the full set of transcripts in cells or tissues and for differential expression analysis [21–23]. This method does not require prior knowledge of the genome sequence or lists of known genes. For genomes that have been partly or fully sequenced, RNA-seq reads can be aligned with genomic sequences to perform differential expression analysis. The added advantage of RNA-seq in these organisms is that, as genome annotation improves, RNA-seq reads can be re-analyzed with the updated gene annotations, an option that is not available with microarrays. Moreover, data obtained from RNA-seq can help in refining existing gene and alternative splicing models.
For emerging model organisms with incomplete or no genome sequence, RNA-seq provides two major advantages to microarrays. First, the workflow of RNA-seq can be universally applied to these organisms, whereas microarrays would need to be designed specifically for each species, a potentially costly endeavor. Second, RNA-seq data can be used to generate de novo gene lists, which can subsequently be used to analyze differential expression [24]. This requires little to no information regarding the genome prior to analysis and avoids the pitfalls of microarrays for poorly annotated genomes, thereby providing more avenues to studies in model organisms, an example being Xenopus.
The western African clawed frog, Xenopus, has been used extensively as a model organism in vertebrate developmental biology. Xenopus species develop externally, allowing for various manipulations to study fertilization, induction, and cell movements. Xenopus are nearly transparent at tadpole stages, allowing for dynamic visualization of tissue and organ development in live animals. Tissues, such as the developing heart, can be easily dissected from the embryo and allowed to develop in culture. Antisense oligonucleotides, or morpholinos (MOs, Genetools, LLC), have been used successfully to disrupt gene function in Xenopus. These properties have made Xenopus an excellent model organism for the study of cardiac development [25].
Two Xenopus species, X. laevis and X. tropicalis, are widely utilized in cellular and developmental biology. The genome of the diploid X. tropicalis was recently published [26], making it an ideal model organism for genetic studies in the frog. X. laevis embryos and frogs are larger than those of X. tropicalis and are, therefore, preferentially exploited for experimental biology techniques. The X. laevis genome underwent tetraploidization 40 million years ago, and thus, classical genetic analysis was less feasible in this species [27]. Despite this hurdle, an X. laevis genome sequencing project is underway, and the availability of extensive expressed sequence tag (EST) sequences on Xenbase [28] allow for functional analysis of genetic pathways. Moreover, targeted mutations can be generated in Xenopus using TALE nuclease (TALENs) or zinc finger nucleases (ZFNs) [29–31]. These advances, combined with their suitability for experimental biology, make Xenopus a tractable model to dissect gene function in organ development. Here, we describe some examples of the many uses of RNA-seq technology, which we have applied in advancing our understanding of X. laevis cardiac development. Furthermore, these studies provide a basis to study the evolution of gene function as a result of gene duplication and polymorphisms.
The methods outlined here should provide a framework for those working not only with X. laevis, but with any organism, particularly those that lack a published genome sequence.
2. Preparation of total RNA
In this review, we describe the use of Xenopus embryos for RNA-seq experiments, including the isolation of RNA from whole embryos and dissected tissues. These approaches should be transferrable to the organism/tissue of interest. Caution should be taken to avoid introducing RNases into the RNA preparation protocol. RNase-free tips and tubes should be used along with gloves to minimize RNases in materials and handling.
2.1 In vitro fertilization of X. laevis embryos
To obtain sufficient numbers of developmental stage-matched embryos we fertilize embryos in vitro. Here we describe the fertilization method used to obtain X. laevis embryos. This method can be coupled with the injection of MOs at the 1-cell stage to deplete gene function in the entire embryo or in individual blastomeres fated to give rise to specific tissue or cell types [32]. In addition, microinjection of mRNA into early-stage embryos can provide gene overexpression or dominant-negative effects [33, 34]. These methods are well established in Xenopus and have been useful alternatives in the absence of gene knockout tools to study gene function. Recent advances in Xenopus mutagenesis, through forward genetic screening and the use of ZFNs or TALENs to mutate genes of interest, will further aid in functional analysis of genes in Xenopus. Careful planning should be used to determine when embryos will reach proper stage of development for expression studies. Embryos can also be collected at different stages for differential expression studies.
2.1.1 In vitro fertilization materials and reagents
Human Chorionic Gonadotropin, hCG (Chorulon), 1000U/ml stock in PBS, stored at 4°C
1X Marc’s Modified Ringer’s solution (MMR) (0.1 M NaCl, 2.0 mM KCl, 1 mM MgSO4, 2 mM CaCl2, 5 mM HEPES (pH 7.4), 0.1 mM EDTA)
Testes Buffer - Leibovitz's L-15 medium (Invitrogen) supplemented with 10% fetal calf serum and 50 µg/ml gentamycin
Forceps
0.1X Magnesium-buffered saline (MBS) (8.8 mM NaCl, 0.1 mM KCl, 0.1 mM MgSO4, 0.5 mM HEPES (pH 7.8), 0.25 mM NaHCO3, 0.7 mM CaCl2)
2% L-cysteine hydrochloride monohydrate (Sigma), pH 8.0 made fresh daily in ddH2O
2.1.2 In vitro fertilization procedure
Inject adult X. laevis females with 500U hCG 17–18 hours prior to egg collection. Allow adults to recover in frog system water at 16°C after priming injection. We prime at least two females to increase the chances of obtaining high quality eggs.
Transfer females to 1 × MMR and allow to lay eggs undisturbed.
While females are laying eggs, euthanize an X. laevis male. Dissect both testes and store in Testes Buffer. Testes can be stored in this solution for up to one week.
Collect 500–1000 eggs on a petri dish and remove excess buffer with transfer pipette.
Coarsely macerate one-third of a testis. Collect pieces of testis with forceps and paint surface of eggs in petri dish.
After 5 minutes, flood eggs on plate with 0.1X MBS. Allow to recover for 20 minutes.
Remove excess 0.1X MBS and flood fertilized eggs with cysteine solution to remove jelly coats from eggs. Incubate until embryos pack together (approximately 3–5 minutes), then wash 4–5 times with 0.1X MBS.
Culture embryos in 0.1X MBS at desired temperature. At this point, embryos can be injected with morpholinos or mRNA as described elsewhere [32, 35–37].
2.2 Total RNA isolation
Once embryos have reached the desired stage of development, they must be processed to collect total RNA. A minimum of 1 µg total RNA is recommended to obtain enough material for RNA-seq using this protocol. When performing RNA-seq on dissected tissue, the number of explants required to reach the minimal RNA desired will need to be determined. For example, we have found that 100 dissected hearts are required for stage 37 X. laevis embryos to obtain 1 µg of total RNA, while a minimum of 50 hearts are required for stage 45 embryos. It is also critical from this point forward to avoid introduction of RNases. Sterile RNase-free water or DEPC-treated water should be used to make solutions needed for RNA extraction and subsequent procedures.
2.2.1 RNA isolation materials and reagents
0.5X MBS supplemented with 50 µg/ml BSA
Trizol reagent (Invitrogen)
Chloroform
Isopropyl alcohol
Ethanol (70%)
Forceps (for dissection)
RNase-free tubes and pipette tips
RNase-free ddH2O
RQ1 DNase and 10X RQ1 DNase buffer (Promega)
Bench-top centrifuge
RNeasy Mini Kit (Qiagen)
Qubit fluorometer and RNA Assay Kit (Invitrogen)
2.2.2 RNA isolation procedure
Collect 10 embryos in 1 ml Trizol. Alternatively, if collecting RNA from tissue, dissect embryos in 0.5X MBS with 50 µg/ml BSA supplement to aid dissection and separation of tissues. Collect dissected tissue in Trizol (25 explants per 500 µl Trizol). Flash freeze embryos/dissected tissue in Trizol in liquid nitrogen and store at −80°C. This step will enhance the homogenization process during Trizol extraction.
Thaw tubes at room temperature. Homogenize embryos/tissue with pipet, vortex briefly and incubate at room temperature for 5 minutes.
Centrifuge lysate at 22,000 × g for 1 minute to precipitate insoluble material. Transfer supernatant to a new tube.
Add 200 µl chloroform per ml initial Trizol volume. Vortex and incubate at room temperature for 3 minutes followed by centrifugation at 12,000 × g for 15 minutes at 4°C.
Transfer aqueous layer to a new tube and add 500 µl isopropyl alcohol per ml of Trizol used.
Incubate 10 minutes at room temperature. Precipitate RNA by centrifugation at 12,000 × g for 10 minutes at 4°C.
Wash pellet twice with 70% ethanol and centrifuge at 8,000 × g for 5 minutes between washes.
Air-dry pellet and resuspend in 25 µl DEPC-treated water. Add 2 µl RQ1 DNase and 3 µl 10X DNase buffer. Incubate at 37°C for 30 minutes.
Purify total RNA using the RNeasy Mini Kit, according to manufacturer’s instructions.
Quantify purified product using a Qubit fluorometer and RNA Assay Kit according to manufacturer’s instructions (Invitrogen). Store at −80°C or proceed immediately to mRNA enrichment (section 3.1). Note: This protocol is optimized for a minimum of 1 µg total RNA. If RNA yield is too low, you may consider RNA amplification protocols before proceeding [38, 39]. This may lead to amplification biases that should be taken into account downstream when analyzing RNA-seq data [40, 41].
3. RNA-seq
Once total RNA is purified from tissues or embryos of choice, poly-adenylated mRNA is purified and fragmented before library preparation. RNA-seq kits can be purchased from a number of suppliers (i.e., Illumina, NEB, Clontech and Ambion); however, prior to library preparation it is important to ensure that the provided adaptor sequences are compatible with the high-throughput sequencing machines being used. Here we describe a protocol to generate sequencing libraries for the Illumina Genome Analyzer II (GAII), using the Illumina mRNA Sequencing Sample Preparation Guide, September 2009. This protocol can be used to generate libraries from a minimum of 1 µg total RNA obtained from dissected tissue.
3.1 mRNA purification
mRNA comprises a very small percentage of the total RNA population and is therefore difficult to quantify in a total RNA preparation. Total RNA purified must be of high quality. This can be assessed before proceeding with mRNA enrichment. For example, a small amount of total RNA can be analyzed by running on a denaturing gel and examining the ratio of 28S and 18S ribosomal RNA, which for high quality RNA should be approximately 2.0 [42]. This generally requires a large amount of total RNA, but if RNA yield is precious, samples can be analyzed for example using the Pico RNA kit for the RNA Bioanalyzer 2100 (Agilent), which requires a total RNA concentration of 50–5000 pg/µL in water.
The following protocol enriches for poly-adenylated mRNAs from the total RNA pool using magnetic bead capture. As with preparation of total RNA, great care should be taken to avoid introduction of RNases to the samples.
3.1.1 mRNA purification materials and reagents
1 µg total RNA (minimum)
RNase-free water
1.5-ml pre-lubricated RNase-free tube (Costar, cat. no. 3207)
65°C and 80°C dry heat block
Sera-Mag Oligo [dT] beads (ThermoScientific)
Dynal magnetic strand (Invitrogen)
Bead-binding buffer (BBB; 20 mM Tris-HCl, pH 7.5, 1M LiCl, 2 mM EDTA)
Wash Buffer (WB; 10 mM Tris-HCl, pH 7.5, 0.15 M LiCl, 1 mM EDTA)
10 mM Tris-HCl
200-µl thin-walled PCR tubes
3.1.2 mRNA purification procedure
Dilute 1 µg of total RNA to a final volume of 50 µl in RNase-free water in a 1.5-ml pre-lubricated RNase-free tube.
Heat at 65°C for 5 minutes, and immediately place on ice.
While RNA is heating, prepare Sera-Mag beads (15 µl/sample). For each wash step in this procedure, buffer is added to beads and then the beads are collected by placing on a magnetic stand for 1.5 minutes. To prepare beads, wash twice with 100 µl BBB.
After second wash, resuspend beads in 50 µl BBB and combine with 50 µl of ice-cold RNA. Rotate end-over-end at room temperature for 5 minutes.
Wash beads twice with 200 µl WB and resuspend in 50 µl 10 mM Tris-HCl, pH 7.5. Heat for 80°C for 2 minutes.
Collect beads on magnet, and transfer supernatant, containing mRNA, to a clean tube.
Add 50 µl BBB to the mRNA solution to bring total volume to 100 µl and heat for 5 minutes at 65°C. Immediately place on ice at end of 5 minute incubation.
While RNA is heating, wash beads twice with 200 µI WB.
After second wash, resuspend beads with 100 µl ice-cold mRNA and rotate at room temperature for 5 minutes.
Wash beads twice with 200 µl WB.
After second wash, resuspend beads in 17 µl 10 mM Tris-HCl, and heat at 80°C for 2 minutes, and immediately place on magnet.
Elute mRNA (16 µl) and transfer to a 200-µl thin-walled PCR tube.
3.2 mRNA fragmentation
Once mRNA has been enriched from total RNA pools, it must be fragmented. This ensures a uniform size of fragments when preparing the sequencing library and reduces the risk of fragment size-based amplification bias. We use a chemical fragmentation protocol to quickly and efficiently fragment the mRNA. This step needs to be optimized depending on the tissue source, as mRNA obtained from different species may require shorter or longer fragmentation times. Alternatively, this process can be skipped and cDNA fragments can be digested by DNAseI after generating double-stranded cDNA (section 3.4.2) as described elsewhere [43]. However, this may lead to biases of overrepresented dinucleotide sequences [41]. Optimal size of fragments ranges from 200 to 400 base pairs.
3.2.1 mRNA fragmentation materials and reagents
RNase-free water
10X Fragmentation Buffer (Ambion, AM8740)
Stop Solution (200 mM EDTA pH 8.0)
70°C heat block
3 M NaOAc (pH 5.2)
Glycogen (5 µg/µl)
70% Ethanol
RNase-free 1.5 ml centrifuge tubes
Bench-top centrifuge
3.2.2 mRNA fragmentation procedure
Bring mRNA solution to 18 µl with RNase-free water.
Fragment by adding 2 µl 10× Fragmentation Buffer and heating at 70°C for exactly 4 minutes (The incubation time in this step is dependent on tissue. We have found 4 minutes works well for X. laevis).
Add 2 µl Stop Solution and place sample on ice.
Precipitate mRNA by adding 2 µl 3 M NaOAc (pH 5.2), 2 µl glycogen (5 µg/µl), and 60 µl 100% ethanol and storing at −80°C overnight.
Centrifuge tubes 22,000 g for 25 minutes at 4°C and remove supernatant.
Wash pellet with 300 µl 70% ethanol and air-dry.
Resuspend pellet in 11.1 µl RNase-free water.
Optional: RNA can be run on an agarose gel to check for average fragment size, concentrated at 200–400 bp. If RNA is not fragmented enough, increase reaction time in step 2. If RNA is too fragmented, reduce the time in step 2.
3.3 First strand cDNA synthesis
Once mRNA is purified and fragmented from total RNA pool, cDNA is transcribed from mRNA, followed by second strand cDNA synthesis. The ends of the generated cDNA are repaired and adaptors are ligated to these ends to prepare sequencing libraries.
3.3.1 First strand cDNA synthesis materials and reagents
Random Primers (3 µg/µI Invitrogen)
65°C heat block
5× First Strand Buffer (Invitrogen)
100 mM DTT
25 mM dNTPs
RNase Inhibitor (Enzymatics)
SuperScript II (Invitrogen)
Thermal cycler
3.3.2 First strand cDNA synthesis procedure
Add 1 µl of Random Primers (3 µg/µl) to mRNA. Incubate sample at 65°C for 5 minutes and immediately place on ice.
Make master mix containing 4 µl 5× First Strand Buffer, 2 µl 100 mM DTT, 0.4 µl 25 mM dNTP, and 0.5 µl RNase inhibitor. Add 6.5 µl freshly prepared master mix to the sample and incubate at 25°C for 2 minutes.
Add 1 µl SuperScript II to sample and incubate at 25°C for 10 minutes, 42°C for 50 minutes, 70°C for 15 minutes, and then maintain at 4°C.
3.4 Second strand cDNA synthesis
3.4.1 Second strand cDNA synthesis materials and reagents
5× Second Strand Buffer (Invitrogen)
25 mM dNTP
2 U/µl RNase H (Enzymatics)
5 U/µl DNA Pol I (Enzymatics)
QIAquick PCR Purification Kit (Qiagen)
EB buffer; 10 mM Tris-Cl, pH 8.5 (Qiagen)
3.4.2 Second strand cDNA synthesis procedure
Make master mix of 52.8 µl water, 20 µl 5× Second Strand Buffer (Invitrogen), and 1.2 µI 25 mM dNTP.
Add 74 µl freshly prepared master mix to first strand cDNA samples. Incubate on ice for 5 minutes.
Add 1 µl of 2 U/µl RNase H and 5 µl of 5 U/µl DNA Pol I. Incubate at 16°C for 2.5 hours.
Purify cDNA using PCR purification kit according to manufacturer’s instructions. Elute in 50 µl elution buffer (EB; 10 mM Tris-Cl, pH 8.5).
3 5 Ligation of Adapters
3.5.1 Ligation of Adapters materials and reagents
10× T4 Ligation Buffer (including ATP; New England Biolabs, NEB)
10 mM dNTP mixture
Polynucleotide Kinase (10U/µI; NEB)
Klenow (1U/µI; NEB)
T4 DNA Polymerase (3U/µI; NEB)
PCR Purification Kit and Mini-Elute PCR Purification Columns(Qiagen)
Klenow 3’-5’ Exo- (5U/µI; NEB)
10 mM dATP
10× NEB Buffer 2 (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1mM DTT, pH 7.9)
2× Quick Ligation Buffer (NEB)
- Genomic Adapter Oligo Mix (Illumina cat. no. 10000531)
- Pair-end adapters, these can be used for single-end sequencing as well: 5' P- GATCGGAAGAGCGGTTCAGCAGGAATGCCGAG 5' ACACTCTTTCCCTACACGACGCTCTTCCGATCT
Quick Ligase (NEB, approximately 500 U/µl)
AMPure-XP (SPRI) Beads (Agencourt)
EB Buffer; 10 mM Tris-Cl, pH 8.5
70% and 80% Ethanol
3.5.2 Ligation of Adapters procedure
Blunt cDNA by incubating 16 µl cDNA with 20 µl 10× T4 Ligation Buffer (including ATP), 2 µl 10 mM dNTPs, 1 µl polynucleotide kinase (NEB), 1 µl Klenow (NEB), and 1.2 µl T4 DNA polymerase (NEB) for 30 minutes at 20°C.
Purify blunted fragments on a Qiagen PCR Purification Column and elute in 42 µl EB.
To purified fragments, add 3 µI Klenow 3’–5’ Exo- (NEB), 1 µl dATP, and 5 µl 10× NEB Buffer 2 (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, pH 7.9) for a total volume of 50 µl for 1 hour at 37°C to generate “A” overhangs.
Purify fragments with Qiagen Mini-Elute PCR Purification Columns and elute with 17 µl EB.
Ligate adapters to DNA in a reaction mixture consisting of 16 µl DNA, 20 µl 2× Quick Ligation Buffer, 2 µl 1:10 Genomic Adapter Oligo Mix, and 2 µl Quick Ligase at 23°C for 25 minutes.
Unused adapter oligonucleotides and dimers are removed using AMPure Beads. Add 10 µl EB to reaction sample, bringing the total volume to 50 µl.
Add 45 µl AMpure XP Beads to samples and mix by pipetting up and down. Incubate at room temperature for 10 minutes.
Place tubes in magnetic stand for 5 minutes and carefully remove and discard the supernatant without disturbing the beads.
Rinse the bead pellet twice with 200 µl 80% ethanol. Keep the tubes on the magnet and try not to disturb the beads.
After removing second ethanol wash, briefly spin down tubes and return to magnetic stand. Remove remaining liquid with a small bore pipet.
With lid open, incubate tubes at 37°C, 2 minutes or until cracks begin to appear on the pellet surface.
Add 50 µl EB directly to the pellet and resuspend well, pipetting up and down 20 times. Incubate at room temperature for 2 minutes.
Place on magnet for 2 minutes and save the library-containing supernatant to a new tube.
Repeat steps 7–13, (using 70% ethanol in place of 80% ethanol) to remove adapters and dimers a total of two times.
3.6 PCR Amplification of cDNA libraries
Once adapter oligonucleotides are ligated onto cDNA fragments, PCR is used to amplify these fragments to generate the libraries that will be used for high throughput sequencing. PCR reactions are carried out in duplicate for each library to avoid jackpot effects in which some transcripts may become overrepresented by amplification bias. The number of PCR cycles is dependent on the initial amount of cDNA present in the sample. For example, when starting with larger initial amounts (>100 ng), fewer cycles are required, as less amplification will be needed to reach the desired amount of library. Moreover, increasing the number of PCR cycles can result in log phase amplification of genes very highly expressed in the sample and therefore leading to a bias of these samples and generating additional variation from one sample to the next. This can be observed by increased numbers of duplicated sequences in RNA-seq data sets [44], therefore potentially masking other transcripts that are present in a sample at much lower levels. Verify the DNA amount/concentration required for a specific machine or sequencing platform to minimize amplification cycles needed. Note: we use the AMPure beads for selection of amplified products; however, if cDNA fragment size is not optimal (∼200–400 bp on average), or for cost-reduction, gel extraction of cDNA fragments of the correct size range can be used [43] .
3.6.1 PCR Amplification of cDNA libraries materials and procedure
DNase-free water
5× Phusion Buffer HF (NEB)
10 mM dNTP
25 µM primer Solexa PE.1 (5'AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGA TCT)
25 µM primer Solexa PE.2 (5'CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTT CCGATCT)
Phusion Polymerase (NEB)
AMPure-XP (SPRI) Beads (Agencourt)
EB buffer; 10 mM Tris-Cl, pH 8.5
70% ethanol
Qubit Fluorometer and dsDNA HS Assay Kit (Invitrogen)
Bioanalyzer 2100 HS DNA chip (Agilent)
Bioanalyzer 2100 (Agilent)
3.6.2 PCR Amplification of cDNA libraries procedure
Set up PCR reactions in duplicate for each library.
Make PCR master mix consisting of 24 µl water, 20 µl 5× Phusion Buffer HF (NEB), 3 µl 10 mM dNTP, 1 µl each 25 µM primers Solexa PE.1 and PE.2, and 1 µl Phusion polymerase (NEB).
Add master mix (50 µl) to 50 µl cDNA and divide sample equally between two tubes.
PCR with initial denaturation at 98°C for 30 seconds followed by 16 cycles of 98°C for 10 seconds, 65°C for 30 seconds, and 72°C for 30 seconds, followed by final extension at 72°C for 5 minutes. PCR cycles may be altered to account for differences in starting RNA amounts. Fewer cycles of PCR are recommended to minimize amplification bias of highly expressed genes.
Add 40 µl AMpure XP Beads to each 50 µl PCR sample. Mix by pipetting up and down and incubate at room temperature for 10 minutes.
Place tubes in magnetic stand for 5 minutes and carefully remove and discard the supernatant without disturbing the beads.
Rinse the bead pellet twice with 200 µl 70% ethanol. Keep the tubes on the magnet and try not to disturb the beads.
After removing second ethanol wash, briefly spin down tubes and return to magnetic stand. Remove remaining liquid with a small bore pipet.
With lid open, incubate tubes at 37°C, 2 minutes or until cracks begin to appear on the pellet surface.
Add 15 µl EB directly to the pellet and resuspend well, pipetting up and down 20 times. Incubate at room temperature for 2 minutes.
Place on magnet for 2 minutes and save the library-containing supernatant to a new tube. For each sample, pool the two 15 µl elutions into one 30 µl sample
Quantify purified product using a Qubit Fluorometer and dsDNA HS Assay according to manufacturer’s instructions (Invitrogen). Check specific sequencing machine or platform to determine minimal DNA amount and concentration for submission.
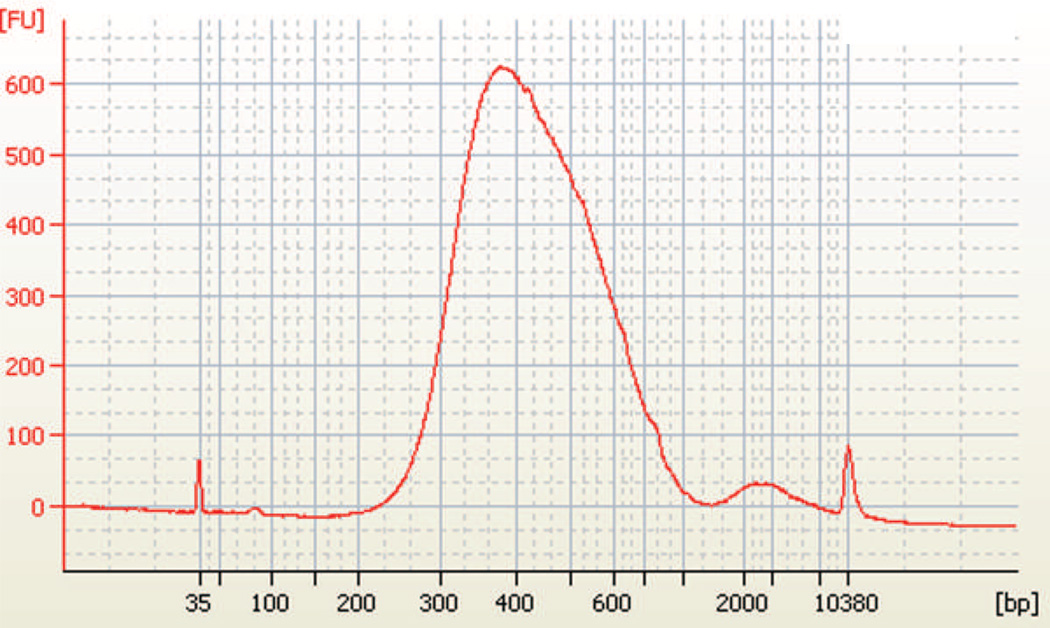
Prior to sequencing, it is usually necessary (consult your high throughput sequencing facility) to check quality of your RNA-seq library by running 2 ng of the sample on a Bioanalyzer 2100 (Agilent) to determine fragment size distribution at a mean length of 300–400 bp. Figure 1 shows an example of an RNA-seq library made from X. laevis stage 37 heart explants. The library contains primarily DNA averaging approximately 400 bp in size. This is consistent with the fragmentation size of RNA described in section 3.2, combined with the size of sequencing adapters. The absence of a peak at 50–100 bp indicates that purification of PCR products and removal of primers and primer-dimers was successful.
Figure 1. Fragment analysis of RNA-seq libraries.
Bioanalyzer trace files for final RNA-seq libraries prepared from stage 37 X. laevis hearts. Graph represents DNA content with amount on y-axis measured by fluorescent unit (FU) and fragment size on the x-axis. Two peaks at 35 and 10380 base pair (bp) correspond to DNA standards used. Note the abundance of DNA in the 300–600 bp range.
4. Sequencing results and data analysis
In this section, we explore the output of high-throughput sequencing, initial quality assessment, and briefly summarize methodology that can be used by biologists to interpret the millions of sequence reads obtained per sample. It should be noted that this knowledge is important to have before undertaking an RNA-seq experiment, as the end-point analysis can largely depend on the methodology used to prepare and run the sequencing libraries. For example, if the goal of an RNA-seq experiment is transcriptome discovery, external RNA controls of known quantity, or spike-ins, may be crucial to determine the sensitivity of RNA-seq to identify rare transcripts in a sample [45]. Alternatively, if analyzing global differential expression among samples, it is necessary to have biological replicates to determine the variability that exists between samples. It may be beneficial to prepare and index libraries from biological replicates so they can be sequenced in bulk on a single lane of a flow cell. Securing the help of a bioinformatics specialist can be quite useful, especially prior to designing the experiment, to outline the goals of the RNA-seq experiment and plan accordingly.
4.1 Sequencing platforms
The protocol outlined here is designed for RNA-seq experiments using the Genome Analyzer II platform from Illumina. We typically use 76 cycles, or 76 base pairs (bp) per read, and single-end sequencing for differential expression studies. Note that as technologies for high-throughput sequencing advance, machines available at sequencing cores may differ greatly. Alternative platforms include the HiSeq2000 and MiSeq (Illumina), Ion Torrent (Invitrogen), and 454 (Roche). These platforms are capable of different size reads (36-, 76-, 50-, and 100-bp, etc. as well as single- or paired-end). Consult with the manufacturers to determine minimum library amount and proper adapter sequences. The choice of sequencing platform and ultimately, the size of reads needed may be dictated by the type of experiment performed, the cost to user, and platform availability.
4.2 Sequencing format and quality analysis
RNA-seq analysis can be complicated if initial read qualities are poor. Prior to sequencing a library, samples are checked for quality of fragment size and concentration to ensure that equal amounts of library are loaded for each sample (see section 3.6.2). Once sequencing is completed, it is important to document a workflow of data handling/analysis, including sequence manipulation, versions of software used to map sequences, and parameters used to determine differential expression. Many of the programs discussed here are continually being updated and therefore parameters used by the programs change frequently. Alternatively, sequence analysis can be performed using Galaxy (http://galaxyproject.org/), a publicly available website preloaded with sequence analysis software. Other commercially available software, such as Partek Genomic Suite or CLC Genomics Workbench, can also be useful to those wishing to avoid using UNIX-based analyses.
4.2.1 General quality assessment
High throughput sequence reads are reported in large text files for each lane of the flow cell. Each sequence read generated by the sequencing platform is assigned a set of quality scores used to determine the reliability of each nucleotide per read. Each sequence read output consists of four lines of text (fastq format, Figure 2), which will vary among sequencers and sequencing facilities. However, the general organization remains the same for a fastq file. Lines 1 and 3 contain information that uniquely identifies each cluster that has been read and the machine that performed the sequencing. Line 2 contains the actual sequence of the read and line 4 contains the quality score assigned to each corresponding nucleotide in line 2. Quality scores are presented in ASCII format, with quality value identities varying depending on the sequencing platform used. FastQC is a powerful Java-based program that can be used to load fastq files generated from high-throughput sequencing and examine various aspects of read quality.
Figure 2. RNA-seq data interpretation and quality analysis.
Overview of fastq file format and example of a single read output for wild type heart sample.
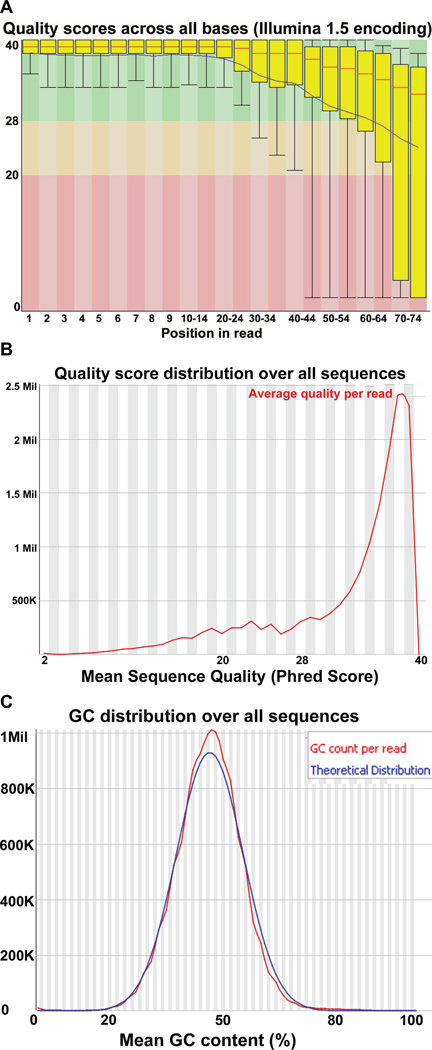
Once fastq files are uploaded to FastQC, parameters of the sequence reads will be displayed. These include: 1) per base quality score, 2) per sequence quality score, 3) sequence length distribution, 4) duplication levels, 5) k-mer profiles (a string of distinct sequences), 6) per_base n content, 7) per base sequence content, 8) per sequence GC content, and 9) per base GC content. Each attribute is taken from the sequence data and compiled into an easy-to-view graphical format (Figure 3 for examples). Careful attention should be given to the quality scores for each base, as lower quality scores may represent erroneous base calls from the sequencing platform. Generally, scores above 20 are acceptable. Also, very high or very low GC content may represent errors in sequencing – we generally see a mean of 50% of GC content per read (Figure 3C) – while duplicated sequences may represent amplification bias or presence of adaptor-dimers formed during library preparation.
Figure 3. Output from fastqc software for RNA-seq data quality analysis.
(A) Quality score per base position for reads. x-axis represents base pair position and y-axis represents interquartile range of quality value (from 0 to 40). Note the overall reduction in quality of scores toward the end of the 76 bp reads. (B) Graphical summary of quality scores shown in (A). (C) G/C distribution over all sequence reads represented on x-axis, with total reads corresponding on y-axis. Overall distribution (red line) versus the theoretical distribution in sample (blue line).
4.2.2 Filtering reads
Low-quality reads can be trimmed using a number of different methods, such as the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), Btrim, Trimmomatic, and ConDeTri [46–48]. Trimming aids in removal of ambiguous reads that may be erroneously identified as SNPs or mutations. Trimming also improves mapping of reads by reducing the number of mismatches to the reference genome or gene list. Sequences commonly overrepresented are those of sequencing adapter primer-dimers remaining in the library. These reads, if present, can easily be removed using the program TagDust [49].The tradeoff of trimming or removing reads is the generation of a reduced number or shorter reads, potentially resulting in less coverage. The choice is generally left to the investigator. If trying to align more reads, reads can be left without filtering and mapping can be altered to allow for more mismatches.
4.3 Mapping sequence reads
Once reads have been checked for quality, and filtered based on preferences determined in section 4.2, these reads are then mapped to the reference genome using programs such as Bowtie, Bowtie2, BWA, and BFAST [50–53]. Because of the file sizes involved (greater than 3–4 GB), mapping analyses can be done by using UNIX commands, for example, on the UNC Research Computing Cluster. This cluster employs a Load Sharing Facility (LSF), which manages jobs on multiple processors and computing nodes, greatly shortening the time needed to perform analysis of RNA-seq data. The LSF can also have modules (programs), including the ones discussed here, pre-installed on the system available for use. Check with your institution or company for access to LSF and for a list of programs available.
4.3.1 Genome/gene reference lists
For sequenced and annotated genomes, a reference list of all annotated genes is usually preloaded by these alignment programs and no further preparation would be required prior to mapping. For species without annotated genomes, a fasta file consisting of gene names and their sequences can be loaded and subsequently formatted by these programs. For example, although the complete X. laevis genome sequence is not yet available, extensive efforts are underway to obtain full-length cDNA libraries (http://xgc.nci.nih.gov/). This allows for the compilation of cDNA sequences to which sequence reads can be mapped to obtain read counts for known genes. For example, all X. laevis cDNA sequences can be downloaded from XenBase [28], and annotated RefSeq sequences can be selected to restrict the analysis to high-quality sequences. As a result, 8,879 cDNA sequences (on May 1, 2011) are used to generate a reference library for mapping, reducing the redundancy of genes and partial transcripts from the larger sets of cDNA sequences available, thereby simplifying the mapping of reads to unique cDNA sequences in subsequent steps. These sequences are available at http://www.marcottelab.org/index.php/Xenopus_reference. To maximize the number of unique genes for mapping, other groups have chosen to align reads to one another to generate de novo transcripts [24]. This method can generate additional X. laevis transcripts compared to the reference list used here (Taejoon Kwon, personal communication) and be more applicable to those attempting to determine the transcriptome of a sample.
The Bowtie2 alignment program can be used to map reads back to the generated reference list. It allows alignments across gaps compared to the reference list, which is useful in the case of SNPs and incomplete cDNA sequences. The program also integrates well with other software packages used to assemble transcripts, such as Cufflinks, or detect splice junctions, such as Tophat [54]. Before Bowtie2 can be used to map reads to the reference list, it must first index the list for mapping. This can be done at the command line of the LSF as follows:
bowtie2-build <ref_list.fa> <base_name>
**base_name refers to name given to indexed library and ref_list is a FASTA file that contains the entire reference library**
4.3.2 Obtaining gene counts
Once a reference library is indexed, Bowtie2 will map reads to this indexed library and report the reads in a Sequence Alignment/Map (.sam) file. The default Bowtie2 usage for this is:
bowtie2 -x index_file –U RNA-seq.fastq -S aligned.sam
***-x refers to reference list that was indexed, -U refers to file name of sequence reads, -S instructs bowtie2 to output data in SAM format with the indicated name.***
To determine the number of reads mapping to each gene, .sam files must first be converted to smaller binary mapping (.bam) files, sorted, then indexed by gene in order to be tabulated to give gene counts using the following commands in Samtools [55]:
samtools view -bt ref_list.txt -o aligned.bam aligned.sam
samtools sort aligned.bam aligned.sorted
samtools index aligned.sorted.bam
samtools idxstats aligned.sorted.bam
The resultant output file generated after these commands are executed will be presented in tab-delimited format with gene names and number of reads mapped to each gene reported.
4.3.3. Alternative alignment parameters
Bowtie2 default parameters assign reads to a single gene in the reference. If a read maps to more than one location in the genome, and the alignment among these regions of the genome is identical, the read is randomly assigned to one of the regions. These parameters can be modified to report all alignments, rather than a single random assignment, or to report user-specified number of alignments. These options should be considered based on the application of RNA-seq experiments. For example, in a species with duplicated genomes, most reads will map to at least two genes.
The X. laevis genome underwent tetraploidization 40 million years ago [27], resulting in duplication of many genes. Gene duplication followed by functional divergence is one mechanism underlying evolution of novel gene function [56]. At least one example of this has been demonstrated in X. laevis, in that alloalleles from two unique, independent loci of hairy2 exhibit different expression patterns and functional requirements [57]. This poses a minor challenge in mapping because duplicated Xenopus genes are quite similar in sequence. Known duplicated genes are present in the cDNA reference library used for mapping. Under default mapping parameters in Bowtie2, reads that map to more than one gene are randomly assigned to one of these genes. These reads are reported in the output file from mapping.
It is unclear what the general contributions of alloalleles of duplicated genes in the Xenopus genome are during development. In the reference list described in section 4.3.1, there are duplicated genes present, corresponding to known alloalleles previously characterized. To determine if any alloallele-specific biases occur in a sample, i.e. one alloallele is more prevalent than the other but is masked by the random mapping, uniquely mapped reads from the .sam files can be filtered out from the Bowtie2 alignment output. This allows one to determine the number of reads that are alloallele-specific in the sample by comparing to mapping of all reads that mapped to the alloallele, including reads that also map to other genes in the reference list. This can be done by selecting all reads that have an “AS” header, but not an “XS” header in the .sam file using the UNIX commands and the bowtie2 output file from section 4.3.2:
grep “AS:” aligned.sam | grep -v “XS:” > unique_alignments.sam
The new file generated here can be processed using Samtools as described in section 4.3.2 to generate gene counts for uniquely mapped reads. These can then be directly compared to the gene counts of all mapped reads from the same sample.
Here, we provide an example of selecting specific parameters to analyze RNA-seq data from stage 37 X. laevis wild-type hearts. The results are summarized in Table 1 and show genes known to be expressed in the heart and to have characterized alloalleles: bmp2; gata4, gata5, gata6; nkx2–5, foxc1, and myl3 [28, 58–62]. We compared the relative amounts of alloalleles present in the sample using all mapped reads (initial bowtie2 output file) or uniquely mapped reads (filtered bowtie2 output file). Using these criteria, we see slight differences in these ratios for some genes. However, the ratios still indicate whether one alloallele is more abundant than the other in the sequencing sample (Table 1). Similar studies could further elucidate the potential divergent expression of alloalleles in different tissues, or duplicate genes in other organisms.
Table 1.
Read counts and ratios of alloallele-specific reads from RNAseq of X. laevis hearts.
| “A” alloallele |
gene length (bp) |
total reads mapping |
unique reads mapping |
“B” alloallele |
gene length |
total reads mapping |
unique reads mapping |
A/B ratio (total) |
A/B ratio (unique) |
|---|---|---|---|---|---|---|---|---|---|
| bmp2|NM_0 01085884 |
2630 | 1281 | 740 | bmp2|NM_ 001101666 |
1992 | 159 | 68 | 8.06 | 10.88 |
| foxc1|NM_0 01088214 |
2400 | 992 | 376 | foxc1|NM_ 001096377 |
2378 | 736 | 279 | 1.35 | 1.35 |
| gata4|NM_0 01090629 |
1626 | 1219 | 568 | gata4|NM_ 001091886 |
1639 | 1770 | 942 | 0.69 | 0.60 |
| gata5|NM_0 01086362 |
2095 | 2764 | 1912 | gata5|NM_ 001088493 |
2050 | 3823 | 2677 | 0.72 | 0.71 |
| gata6|NM_0 01087983 |
4183 | 4555 | 3447 | gata6|NM_ 001090256 |
3720 | 4821 | 3237 | 0.94 | 1.06 |
| myl3|NM_00 1089148 |
1189 | 3383 | 1756 | myl3|NM_0 01089150 |
855 | 15106 | 7763 | 0.22 | 0.23 |
| nkx2–5|NM_ 001086721 |
2506 | 6175 | 4568 | nkx2– 5|NM_ 001172192 |
903 | 1115 | 280 | 5.54 | 16.31 |
4.4 Differential expression analysis
The end goal of an RNA-seq for many developmental biologists is in most cases to identify and quantify differential gene expression between two or more samples. In this section, we describe the methodology used to identify differential expression and identify putative pathways or cellular functions. Although proper statistical methods can be used to identify classes of differential expression, it is important to also use alternative methods of validation. We briefly discuss such methods here.
4.4.1 Normalization methods and statistical analysis
Biological replicates allow the researcher to use correlation between experiments and conduct statistical analysis to determine the significance of differential expression [63, 64]. To properly determine differential expression between samples, normalization must be performed to account for unequal sampling (unequal number of reads among samples). There are a number of methods used for normalization of read counts between samples. These methods have been developed in the absence of truly invariant genes among these different sample types. Spike-in controls of known quantities can be used to mimic the invariant genes and properly compare two or more samples. Alternatively, if spike-in controls are not used, the trimmed mean of the M value (TMM) and an upper-quartile normalization method (RPKM) are popular methods to normalize RNA-seq reads across samples [65, 66].
A number of statistical programs have been designed for R that can be used to analyze differential expression. These include edgeR, DEGseq and DESeq [67–69].
4.4.2 Gene ontology (GO) analysis
When performing differential expression studies, it is often desirable to examine the global differences between two samples. Once a list of differentially expressed genes is determined, this list can be loaded, along with a list of all genes examined for differential expression, into gene ontology software. For example, the program GOrilla identifies GO terms overrepresented in the differentially expressed genes compared to the total number of genes tested [70]. GOrilla examines GO term enrichment in three aspects: cellular process, function and component.
4.4.3 Verification of differential expression
Proper statistical methods and additional biological replicates may limit the number of genes falsely identified as differentially expressed in an RNA-seq experiment, but additional validation is often necessary to eliminate the possibility of improper conclusions. One manner in which RNA-seq results can be validated is by performing quantitative reverse transcription-PCR (qRT-PCR) [71, 72]. PCR primers can be designed against the differential expressed genes and results can be validated on a gene-by-gene basis.
In addition to examining levels of differential gene expression, other methods of validation provide visual verification of RNA-seq results. Standard protocols for in situ hybridization and antibody staining exist in Xenopus that allow researchers to examine spatial-temporal expression of genes and their proteins [73, 74]. This information can be crucial to understanding differential expression. For example, organs such as the heart are comprised of different cell types. RNA-seq or qRT-PCR may identify transcripts present in the heart, but not other organs. Examining spatial expression could give further resolution as to the transcript localization to specific chambers or cell types, as shown recently with differentially expressed genes in tcf21-depleted embryos [75]. This information, together with quantification, will help elucidate gene networks that lead to proper specification and differentiation of the multitude of cell types generated during development.
4.5 Additional RNA-seq applications
The amount of information obtained from an RNA-seq experiment greatly exceeds the uses of the microarray. In addition to differential expression analyses and studies of gene duplication, RNA-seq data can be applied to examine transcript structure and evolution as well as building gene models.
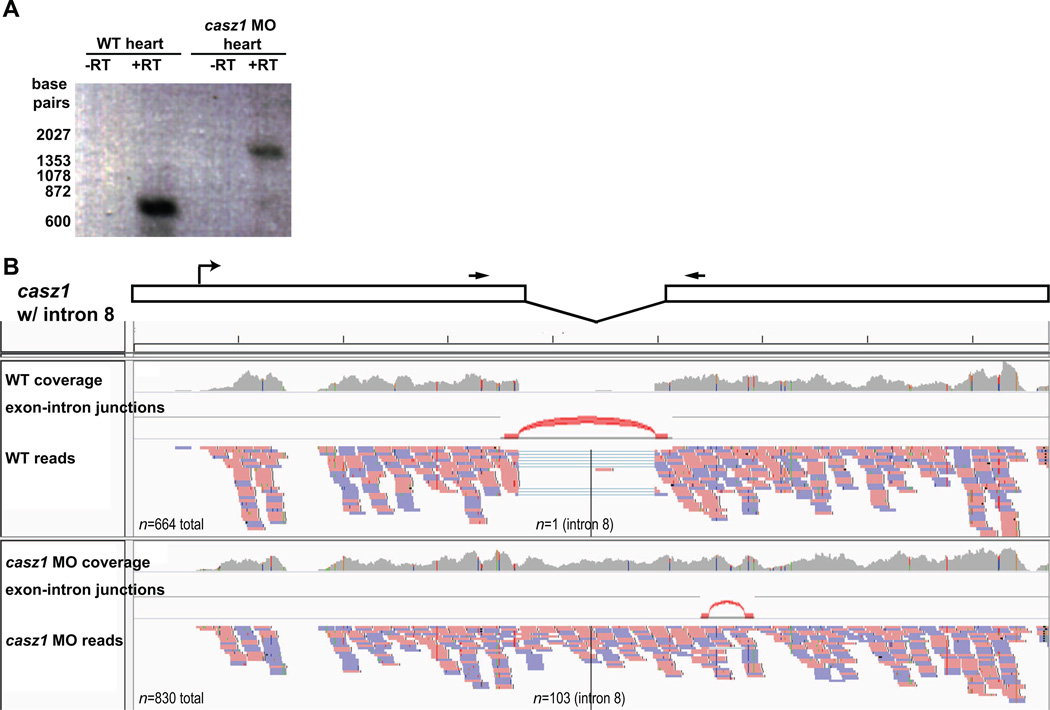
An additional advantage of RNA-seq is the ability to directly test the efficacy of a MO. For example, we have previously shown that casz1 morpholinos (MOs) efficiently block proper splicing of casz1 intron 8, thereby introducing extraneous sequence, and causing a frameshift in the rest of the open reading frame and a premature stop in translation [76]. This can be seen in the results of RT-PCR performed on mRNA derived from hearts dissected from casz1 MO embryos (Figure 4A). To examine these effects with RNAseq data, genomic sequence must be used for the reference sequence for mapping. We sequenced the entirety of casz1 intron 8 from the RT-PCR product and included this sequence between exons 8 and 9 in a .fasta file consisting of the casz1 coding sequence. For simplicity, this sequence was used as the reference file for mapping RNA-seq reads from wild-type (WT) and casz1 MO hearts. Subsequently, after we sequenced this intron, the draft of the X. laevis genome was made publicly available on Xenbase. Therefore, genomic sequences containing intronic sequences can be extracted from Xenbase to generate .fasta format reference sequences for the gene(s) of interest. To determine the effects of splice-blocking by MO using RNA-seq, we use the program Tophat (version 2.0.5) [77]:
tophat -o /WThea xCasz1_w_intron8.fa /WT_read_file.txt
tophat -o /MOhea xCasz1_w_intron8.fa /MO_read_file.txt
***The “-o” option generates an output folder to where the Tophat results are placed.***
Figure 4. Determining morpholino (MO) efficacy by RNA-seq.
(A) RT-PCR of cDNA generated from wild type and casz1 MO hearts shows aberrant splicing of casz1 intron 8 in MO-treated embryos. (B) Modified IGV viewer screenshot with schematic of casz1 mRNA (boxes) with inclusion of intron 8. Arrows indicate primers used for PCR shown in (A). Overall read coverage and individual reads mapping to casz1 mRNA in wild type and casz1 MO hearts are shown below schematic. Exon-intron boundaries predicted by the program Tophat based on read coverage are displayed on genome browser.
Tophat integrates Bowtie2 alignments of RNA-seq reads to the reference genome while also giving information of splice junctions within transcripts. In the output folder for the Tophat alignments, these junctions are represented in .bed files, which can be viewed, together with the individual reads that map to the reference sequence (.bam files that are generated by Tophat using Bowtie2 and Samtools), using IGV viewer [78] or uploaded to a genome browser (when the genome has been annotated).
As shown in Figure 4B, we were able to identify ten reads in wild-type (WT) hearts in which intron 8 had been spliced out, and only a single read that mapped to intron 8. In contrast, casz1 MO hearts had no reads with intron 8 spliced out, but over 100 reads that mapped to intron 8, suggesting that proper splicing was not occurring in MO hearts. Moreover, aberrant splicing was observed downstream of intron 8 in the MO heart (Figure 4B), presumably a secondary result of the first splice-blocking events. Taken together, these results suggest that RNA-seq can be used to not only determine MO efficiency, but also the full consequences of MOs on RNA splicing.
Another option for RNA-seq users is to assemble transcripts de novo. This is especially useful for organisms that have neither a sequenced genome nor an extensive list of ESTs like the one used to generate the Xenopus reference list described here. The program Cufflinks accepts RNA-seq reads and assembles them into a parsimonious set of transcripts [79]. Moreover, Tophat and Cufflinks results can be integrated with statistical software such as CummeRbund to examine differential expression from this newly assembled set of transcripts [54].
Finally, RNA-seq is becoming a popular method in single nucleotide polymorphism (SNP) detection. The abundance of reads obtained in an RNA-seq experiment, together with the quality scores obtained for each base pair in these reads (see section 4.2) allow for the rapid detection of SNPs. Detection of SNPs in transcripts will help identify conserved and rapidly evolving portions of genes and their protein products, giving insight to the structure and function of these proteins. SNPs can be detected using the publicly available program Maq [80], or commercially available programs such as Partek Genomic Suite or CLC Genomics Workbench.
5. Concluding remarks
The advances made in sequencing technology during the last decade have revolutionized our ability to construct gene expression profiles. Here we have described methodology covering new ways to probe the transcriptomes of the model organisms of choice, regardless of genome annotation. We have provided avenues to study differential gene expression and evolution of gene families as discussed with gene duplication, splicing and SNPs. These methods will accelerate the progress being made in understanding gene networks required for proper development, without restricting researchers to organisms with sequenced genomes.
Acknowledgements
We thank Taejoon Kwon, Jason Lieb and Hemant Kelkar for critical discussions and their tutelage in high throughput sequencing data management and analysis. This work is supported by grants to F.L.C from NIH/NHLBI (RO1 DE018825 and RO1 HL089641). N.M.A. is supported by an AHA Postdoctoral Fellowship 11POST6320005.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Russell WL, Kelly EM, Hunsicker PR, Bangham JW, Maddux SC, Phipps EL. Proc Natl Acad Sci U S A. 1979;76:5818–5819. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nusslein-Volhard C, Wieschaus E. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 3.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avery L, Wasserman S. Trends Genet. 1992;8:312–316. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 7.Mouse Genome Sequencing C, Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, L Kolbe D, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 8.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. C. International Human Genome Sequencing, Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 9.Ce.S. Consortium. Science. 1998;282:2012–2018. [Google Scholar]
- 10.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, L Gabor G, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Shtrong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 11.Drosophila 12 Genomes C, Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, Bernardo de Carvalho A, Caspi A, Castrezana S, Celniker SE, Chang JL, Chapple C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia AC, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, Jeck WR, Johnson J, Jones CD, Jordan WC, Karpen GH, Kataoka E, Keightley PD, Kheradpour P, Kirkness EF, Koerich LB, Kristiansen K, Kudrna D, Kulathinal RJ, Kumar S, Kwok R, tnder E, Langley CH, Lapoint R, Lazzaro BP, Lee SJ, Levesque L, Li R, Lin CF, Lin MF, Lindblad-To LaTuopart K, Long M, Low L, Lozovsky E, Lu J, Luo M, Machado CA, Makalowski W, Marzo M, Matsuda M, Matzkin L, McAllister B, McBride CS, McKernan B, McKernan K, Mendez-Lago M, Minx P, Mollenhauer MU, Montooth K, Mount SM, Mu X, Myers E, Negre B, Newfeld S, Nielsen R, Noor MA, O'Grady P, Pachter L, Papaceit M, Parisi MJ, Parisi M, Parts L, Pedersen JS, Pesole G, Phillippy AM, Ponting CP, Pop M, Porcelli D, Powell JR, Prohaska S, Pruitt K, Puig M, Quesneville H, Ram KR, Rand D, Rasmussen MD, Reed LK, Reenan R, Reily A, Remington KA, Rieger TT, Ritchie MG, Robin C, Rogers YH, Rohde C, Rozas J, Rubenfield MJ, Ruiz A, Russo S, Salzberg SL, Sanchez-Gracia A, Saranga DJ, Sato H, Schaeffer SW, Schatz MC, Schlenke T, Schwartz R, Segarra C, Singh RS, Sirot L, Sirota M, Sisneros NB, Smith CD, Smith TF, Spieth J, Stage DE, Stark A, Stephan W, Strausberg RL, Strempel S, Sturgill D, Sutton G, Sutton GG, Tao W, Teichmann S, Tobari YN, Tomimura Y, Tsolas JM, Valente VL, Venter E, Venter JC, Vicario S, Vieira FG, Vilella AJ, Villasante A, Walenz B, Wang J, Wasserman M, Watts T, Wilson D, Wilson RK, Wing RA, Wolfner MF, Wong A, Wong GK, Wu CI, Wu G, Yamamoto D, Yang HP, Yang SP, Yorke JA, Yoshida K, Zdobnov E, Zhang P, Zhang Y, Zimin AV, Baldwin J, Abdouelleil A, Abdulkadir J, Abebe A, Abera B, Abreu J, Acer SC, Aftuck L, Alexander A, An P, Anderson E, Anderson S, Arachi H, Azer M, Bachantsang P, Barry A, Bayul T, Berlin A, Bessette D, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Bourzgui I, Brown A, Cahill P, Channer S, Cheshatsang Y, Chuda L, Citroen M, Collymore A, Cooke P, Costello M, D'Aco K, Daza R, De Haan G, DeGray S, DeMaso C, Dhargay N, Dooley K, Dooley E, Doricent M, Dorje P, Dorjee K, Dupes A, Elong R, Falk J, Farina A, Faro S, Ferguson D, Fisher S, Foley CD, Franke A, Friedrich D, Gadbois L, Gearin G, Gearin CR, Giannoukos G, Goode T, Graham J, Grandbois E, Grewal S, Gyaltsen K, Hafez N, Hagos B, Hall J, Henson C, Hollinger A, Honan T, Huard MD, Hughes L, Hurhula B, Husby ME, Kamat A, Kanga B, Kashin S, Khazanovich D, Kisner P, Lance K, Lara M, Lee W, Lennon N, Letendre F, LeVine R, Lipovsky A, Liu X, Liu J, Liu S, Lokyitsang T, Lokyitsang Y, Lubonja R, Lui A, MacDonald P, Magnisalis V, Maru K, Matthews C, McCusker W, McDonough S, Mehta T, Meldrim J, Meneus L, Mihai O, Mihalev A, Mihova T, Mittelman R, Mlenga V, Montmayeur A, Mulrain L, Navidi A, Naylor J, Negash T, Nguyen T, Nguyen N, Nicol R, Norbu C, Norbu N, Novod N, O'Neill B, Osman S, Markiewicz E, L Oyono O, Patti C, Phunkhang P, Pierre F, Priest M, Raghuraman S, Rege F, Reyes R, Rise C, Rogov P, Ross K, Ryan E, Settipalli S, Shea T, Sherpa N, Shi L, Shih D, Sparrow T, Spaulding J, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Strader C, Tesfaye S, Thomson T, Thoulutsang Y, Thoulutsang D, Topham K, Topping I, Tsamla T, Vassiliev H, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Young G, Yu Q, Zembek L, Zhong D, Zimmer A, Zwirko Z, Jaffe DB, Alvarez P, Brockman W, Butler J, Chin C, Gnerre S, Grabherr M, Kleber M, Mauceli E, MacCallum I. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 12.Cahais V, Gayral P, Tsagkogeorga G, Melo-Ferreira J, Ballenghien M, Weinert L, Chiari Y, Belkhir K, Ranwez V, Galtier N. Molecular ecology resources. 2012;12:834–845. doi: 10.1111/j.1755-0998.2012.03148.x. [DOI] [PubMed] [Google Scholar]
- 13.Moulton JD, Jiang S. Molecules. 2009;14:1304–1323. doi: 10.3390/molecules14031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melton DW. Bioessays. 1994;16:633–638. doi: 10.1002/bies.950160907. [DOI] [PubMed] [Google Scholar]
- 15.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 16.Reinke V. Nat Genet. 2002;(32 Suppl):541–546. doi: 10.1038/ng1039. [DOI] [PubMed] [Google Scholar]
- 17.Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, Stennard FA, Wise N, Schaft D, Wolstein O, Furtado MB, Shiratori H, Chien KR, Hamada H, Black BL, Saga Y, Robertson EJ, Buckingham ME, Harvey RP. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald MJ, Rosbash M. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 19.Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL. Curr Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- 20.Emerging model organisms : a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2009. [Google Scholar]
- 21.Wang Z, Gerstein M, Snyder M. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 23.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaltenbrun E, Tandon P, Amin NM, Showell C, Waldron L, Conlon F. Journal of Birth Defects Research Part A: Clinical and Molecular Teratology. doi: 10.1002/bdra.20793. (in review). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellsten U, Khokha MK, Grammer TC, Harland RM, Richardson P, Rokhsar DS. BMC Biol. 2007;5:31. doi: 10.1186/1741-7007-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowes JB, Snyder KA, Segerdell E, Gibb R, Jarabek C, Noumen E, Pollet N, Vize PD. Nucleic Acids Res. 2008;36:D761–D767. doi: 10.1093/nar/gkm826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young JJ, Cherone JM, Doyon Y, Ankoudinova I, Faraji FM, Lee AH, Ngo C, Guschin DY, Paschon DE, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Harland RM, Zeitler B. Proc Natl Acad Sci U S A. 2011;108:7052–7057. doi: 10.1073/pnas.1102030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Cheng CH, Dawid IB, Chen Y, Zhao H. Proc Natl Acad Sci U S A. 2012;109:17484–17489. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakajima K, Nakajima T, Takase M, Yaoita Y. Dev Growth Differ. 2012;54:777–784. doi: 10.1111/dgd.12006. [DOI] [PubMed] [Google Scholar]
- 32.Tandon P, Showell C, Christine K, Conlon FL. Methods Mol Biol. 2012;843:29–46. doi: 10.1007/978-1-61779-523-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amaya E, Musci TJ, Kirschner MW. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- 34.Hopwood ND, Gurdon JB. Nature. 1990;347:197–200. doi: 10.1038/347197a0. [DOI] [PubMed] [Google Scholar]
- 35.Sive HL, Grainger RM, Harland RM. Cold Spring Harb Protoc, 2010. 2010 doi: 10.1101/pdb.ip81. pdb prot5538. [DOI] [PubMed] [Google Scholar]
- 36.Sive HL, Grainger RM, Harland RM. Cold Spring Harb Protoc, 2010. 2010 doi: 10.1101/pdb.ip81. pdb prot5537. [DOI] [PubMed] [Google Scholar]
- 37.Sive HL, Grainger RM, Harland RM. Cold Spring Harb Protoc, 2010. 2010 doi: 10.1101/pdb.ip81. pdb ip81. [DOI] [PubMed] [Google Scholar]
- 38.Lang JE, Magbanua MJ, Scott JH, Makrigiorgos GM, Wang G, Federman S, Esserman LJ, Park JW, Haqq CM. BMC Genomics. 2009;10:326. doi: 10.1186/1471-2164-10-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauss M, Vierlinger K, Weinhaeusel A, Szameit S, Kaserer K, Noehammer C. Virchows Archiv : an international journal of pathology. 2007;451:1019–1029. doi: 10.1007/s00428-007-0522-4. [DOI] [PubMed] [Google Scholar]
- 40.Sengupta S, Ruotti V, Bolin J, Elwell A, Hernandez A, Thomson J, Stewart R. Biotechniques. 2010;49:898–904. doi: 10.2144/000113556. [DOI] [PubMed] [Google Scholar]
- 41.Zheng W, Chung LM, Zhao H. BMC Bioinformatics. 2011;12:290. doi: 10.1186/1471-2105-12-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. BMC molecular biology. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagalakshmi U, Waern K, Snyder M. Curr Protoc Mol Biol, Chapter. 2010;4 doi: 10.1002/0471142727.mb0411s89. Unit 4 11 11–13. [DOI] [PubMed] [Google Scholar]
- 44.Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang L, Schlesinger F, Davis CA, Zhang Y, Li R, Salit M, Gingeras TR, Oliver B. Genome Res. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong Y. Genomics. 2011;98:152–153. doi: 10.1016/j.ygeno.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Smeds L, Kunstner A. PLoS One. 2011;6:e26314. doi: 10.1371/journal.pone.0026314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. Nucleic Acids Res. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lassmann T, Hayashizaki Y, Daub CO. Bioinformatics. 2009;25:2839–2840. doi: 10.1093/bioinformatics/btp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Homer N, Merriman B, Nelson SF. PLoS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, L Rinn J, Pachter L. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. S. Genome Project Data Processing, Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koonin EV. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 57.Murato Y, Nagatomo K, Yamaguti M, Hashimoto C. Dev Genes Evol. 2007;217:665–673. doi: 10.1007/s00427-007-0176-x. [DOI] [PubMed] [Google Scholar]
- 58.Tonissen KF, Drysdale TA, Lints TJ, Harvey RP, Krieg PA. Dev Biol. 1994;162:325–328. doi: 10.1006/dbio.1994.1089. [DOI] [PubMed] [Google Scholar]
- 59.Smith SJ, Ataliotis P, Kotecha S, Towers N, Sparrow DB, Mohun TJ. Dev Dyn. 2005;232:1003–1012. doi: 10.1002/dvdy.20274. [DOI] [PubMed] [Google Scholar]
- 60.Koster M, Dillinger K, Knochel W. Mech Dev. 1998;76:169–173. doi: 10.1016/s0925-4773(98)00123-3. [DOI] [PubMed] [Google Scholar]
- 61.Jiang Y, Evans T. Dev Biol. 1996;174:258–270. doi: 10.1006/dbio.1996.0071. [DOI] [PubMed] [Google Scholar]
- 62.Hemmati-Brivanlou A, Thomsen GH. Dev Genet. 1995;17:78–89. doi: 10.1002/dvg.1020170109. [DOI] [PubMed] [Google Scholar]
- 63.Auer PL, Doerge RW. Genetics. 2010;185:405–416. doi: 10.1534/genetics.110.114983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yendrek CR, Ainsworth EA, Thimmapuram J. BMC research notes. 2012;5:506. doi: 10.1186/1756-0500-5-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bullard JH, Purdom E, Hansen KD, Dudoit S. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson MD, Oshlack A. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, Huber W. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robinson MD, McCarthy DJ, Smyth GK. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L, Feng Z, Wang X, Wang X, Zhang X. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 70.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heid CA, Stevens J, Livak KJ, Williams PM. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 72.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 73.Harland RM. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 74.Lee C, Kieserman E, Gray RS, Park TJ, Wallingford J. CSH protocols. 2008;2008 doi: 10.1101/pdb.prot4957. pdb prot4957. [DOI] [PubMed] [Google Scholar]
- 75.Tandon P, Miteva YV, Kuchenbrod LM, Cristea IM, Conlon FL. Development. 2013 doi: 10.1242/dev.093385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Christine KS, Conlon FL. Dev Cell. 2008;14:616–623. doi: 10.1016/j.devcel.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trapnell C, Pachter L, Salzberg SL. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, Ruan J, Durbin R. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]