ABSTRACT
Bacteria release a wide diversity of small bioactive molecules that often correspond to secondary metabolites. Among them, volatile molecules produced under various growth conditions were shown to mediate cross-kingdom interactions with plants, nematodes, and fungi. Although the role of volatile compounds in bacterial biology is not well understood, recent reports indicated that they could play a role in airborne interactions between bacteria and influence antibiotic resistance, biofilm formation, and virulence. In this study, we investigated long-distance effects of 14 previously described Escherichia coli volatile compounds upon the bacteria E. coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Bacillus subtilis. We show that several of these molecules constitute chemical cues influencing growth, adhesion, and motility in exposed bacteria. Moreover, we show that aerial exposure to trimethylamine (TMA), a volatile compound produced in animal intestines and tissues upon biogenic reduction of trimethylamine oxide (TMAO), modifies the antibiotic resistance profiles of all tested Gram-positive and Gram-negative bacteria. We demonstrate that the TMA mode of action is distinct from that previously described for ammonia and results from nonspecific transient alteration of antibiotic uptake due to pH increase in the environment of bacteria aerially exposed to TMA. Our study therefore presents a new way by which volatile compounds can affect community behavior and structure in physically separated bacteria. It further demonstrates that bacterial gases and volatile compounds mediate chemical interactions, triggering functional responses that play an important role in the development of bacterial communities.
IMPORTANCE
Bacteria release many different volatile compounds during food transformation and fermentation. Here we sought to investigate the role of several bacterial volatile molecules released by Escherichia coli during long-distance airborne interactions with other bacteria. While several tested volatiles affect bacterial motility and surface adhesion, we show that aerial exposure to trimethylamine, a molecule produced by E. coli and many other Gram-negative bacteria in animal intestines and infected tissues, also modulates antibiotic resistance in all tested bacteria. We demonstrate that exposure to trimethylamine increases the pH of the growth medium of exposed bacteria, resulting in modifications in antibiotic uptake and transient alteration of antibiotic resistance. Our study therefore presents a new mechanism by which volatile compounds can affect community behavior and structure in physically separated bacteria, and it illustrates how airborne chemical interactions between bacteria contribute to the development of bacterial communities.
INTRODUCTION
The capacity of bacteria to sense and respond to small bioactive metabolites contributes to their ability to adapt and thrive in complex polymicrobial environments (1–3). While the role of soluble molecules has been actively investigated, bacteria also release a wide diversity of volatile compounds corresponding to by-products of metabolic reactions occurring under various growth conditions during food transformation and fermentation (4–6). Bacterial volatiles are perceived as odors, attractive scents, or pollutants by an animal’s olfactory receptors or specialized detection devices, but their role in local and distant microbial interactions has generally been overlooked (7). Nevertheless, volatile bacterial secondary metabolites have recently been shown to influence the growth of exposed plants, fungi, and animals (8–12). Moreover, several reports also showed that bacterial volatiles mediate biological interactions between physically separated bacteria and modulate bacterial behavior and phenotypes, such as stress and antibiotic tolerance, colony morphotypes, community behavior, and virulence (8, 13–19). While these studies indicate that bacterial volatiles have broad biological effects upon the bacteria themselves, characterization of the highly complex blend of volatiles produced by each bacterium and elucidation of their biological roles remain challenging tasks (8).
We previously showed that biogenic gaseous ammonia has profound effects upon the antibiotic resistance profiles of both Gram-negative and Gram-positive bacteria (17). In this study, we wished to expand the repertoire of bacterial volatiles that play a potential role in bacterial physiology. Using a simple experimental system to evaluate volatile interactions between bacteria, we investigated long-distance actions of 14 previously described Escherichia coli volatile compounds on motility, biofilm formation, and antibiotic resistance using E. coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Bacillus subtilis as target bacteria for intra- and interspecies interactions. Among our findings, we show that aerial exposure to trimethylamine (TMA), a volatile molecule produced upon biogenic reduction of trimethylamine oxide (TMAO), led to modification of the antibiotic resistance profile in all tested Gram-negative and Gram-positive bacteria (20–22). Interestingly, the TMA-dependent phenotype and mode of action were distinct from those previously described for ammonia and involving the synthesis of intracellular polyamines. In contrast, we show that TMA activity depends on an alteration in antibiotic flux due to a pH increase in the environment of bacteria exposed to TMA. Our study therefore reveals a new mechanism by which bacteria can display transient increased resistance to antibiotics when exposed to volatile compounds produced by neighboring bacteria. The previously unsuspected phenotypic consequence of exposure to TMA could contribute to altering stress resistance and bacterial community behavior in TMA- and TMAO-rich environments, such as animal intestines and tissues.
RESULTS
Screening for phenotypes induced by E. coli volatile compounds in aerially exposed representative bacteria.
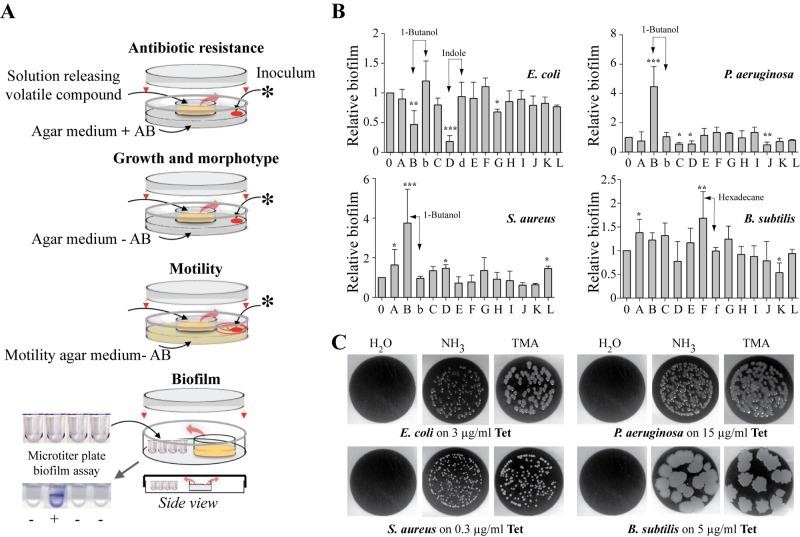
We hypothesized that aerial exposure to E. coli volatile compounds other than ammonia (NH3) could also influence bacterial behavior, including motility, growth, biofilm formation, and antibiotic resistance. To test this, we exposed Gram-positive and Gram-negative bacteria to 14 known and commercially available E. coli volatile compounds emitted from liquid solutions placed in the middle of a previously described 2-petri-dish assay (Fig. 1A and Table 1) (17, 22–25, 26). Exposure of E. coli, P. aeruginosa, S. aureus, and B. subtilis to 14 characterized E. coli volatile compounds showed that, except for 2,3-butanedione and acetaldehyde, none of the remaining 12 tested compounds showed significant growth toxicity and could therefore be used for further phenotypic analyses (Table 1). Whereas none of the 12 nontoxic compounds triggered growth differences on plates, use of 0.3% motility agar plates revealed that aerial exposure to several compounds affected E. coli (1-butanol) and P. aeruginosa (indole, 2-butanone, and acetoin) motility (Table 1). While B. subtilis motility was unaffected by the tested volatiles (Table 1), this phenotype could not be studied with S. aureus, since it is nonmotile (data not shown). We also observed that several volatile compounds, including indole, 1-butanol, and hexadecane, increased or decreased the bacterial ability to form biofilms in microtiter plates. This effect is concentration dependent, since exposure of recipient bacteria to 10-fold dilutions of the corresponding emitting solutions had no effect on biofilm formation and motility (Fig. 1B) (data not shown). Finally, we tested whether exposure to any of the tested volatile compounds could also affect antibiotic resistance, and we showed that, in addition to previously described effects of gaseous ammonia (17), exposure to trimethylamine (TMA) led to increased resistance to tetracycline in all tested Gram-negative and Gram-positive bacteria (Fig. 1C).
FIG 1 .
Phenotypes induced by E. coli volatile compounds in representative bacteria. (A) Variations of the previously described 2-petri-dish assay (17) used to evaluate the impact of volatile compounds on antibiotic resistance, growth and morphotype, motility, and biofilm formation as described in Materials and Methods. *, 20-µl bacterial inoculum. AB, antibiotic. (B) Volatile-mediated modulation of biofilm formation by exposed E. coli MG1655F′, P. aeruginosa PAO1, S. aureus HG001, and B. subtilis 3610 to 12 volatile compounds (listed in Table 1). E. coli graph: B, 1% 1-butanol; b, 0.1% 1-butanol; D, 0.15% indole; d, 0.015% indole. P. aeruginosa graph: B, 1% 1-butanol; b, 0.1% 1-butanol. S. aureus graph: B, 1% 1-butanol; b, 0.1% 1-butanol. B. subtilis graph: F, 1% hexadecane; f, 0.1% hexadecane. The data represent means ± standard deviations (SD) from at least 3 independent experiments in which the OD595 of the nonexposed bacteria was defined as 1. For statistical analysis, P < 0.05 (*), P < 0.01 (**), and P < 0.0001 (***) by one-tailed unpaired Student’s t test in comparison with H2O. (C) Aerial exposure to TMA increases resistance to tetracycline in all tested bacteria. Shown is growth of E. coli MG1655, P. aeruginosa PAO1, S. aureus HG001, and B. subtilis 168 on inhibitory concentrations of tetracycline upon exposure to H2O, NH3 (1%), or TMA (0.5%). Pictures were taken after 24 h of aerial exposure to the volatile source in the 2-petri-dish experimental design presented in panel A.
TABLE 1 .
Phenotypic impact of aerial exposure to E. coli volatile compoundsa
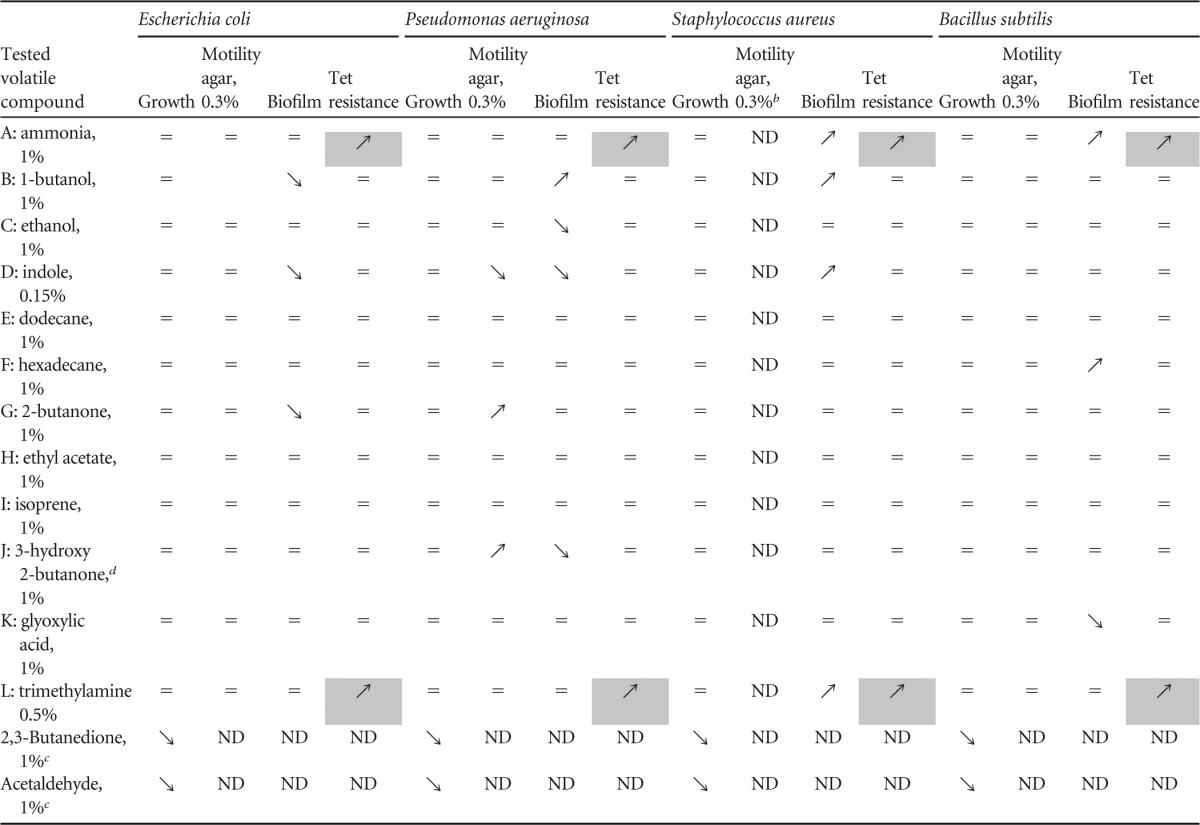
=, not affected; ↘, inhibition; ↗, stimulation; ND, not determined. Shaded cells indicate a phenotype observed with all exposed bacteria.
S. aureus HG001 was not motile on 0.3% motility agar plates.
Both acetaldehyde and butanedione were toxic at 1% and 0.1% concentrations and were not further analyzed.
Acetoin.
These results therefore demonstrate that several E. coli volatile compounds influence a variety of bacterial phenotypes in aerially exposed bacteria. Considering its activity on a broad range of bacteria, we decided to focus the rest of our study on the TMA-dependent phenotype.
Biogenic volatile TMA produced from TMAO is active on distantly exposed bacteria.
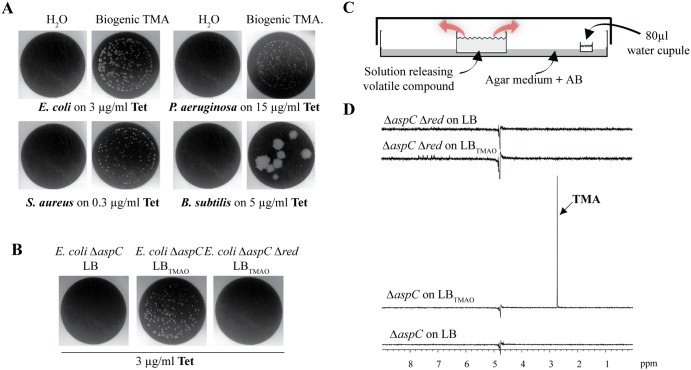
Trimethylamine (TMA; C3H9N) is a tertiary volatile amine that contributes to the odor of spoiling fish produced upon biogenic reduction of trimethylamine oxide (TMAO), a compound present in many animal and plant tissues (20, 27–29). While produced both under aerobic and anaerobic conditions, TMA is used as an alternative electron acceptor during respiration in the absence of oxygen (21). While our initial screen revealed increased resistance to tetracycline using TMA emitted from pure solutions, we next determined whether biogenic TMA produced and released upon spontaneous bacterial reduction in TMAO-containing lysogeny broth medium (LBTMAO) would have similar bioactivity. In order to avoid interference with ammonia naturally emitted from high-cell-density populations, we used an E. coli TG1 ∆aspC mutant impaired in ammonia production (17). We used a 2-petri-dish device to expose E. coli, P. aeruginosa, S. aureus, and B. subtilis to volatile compounds naturally emitted from filtered E. coli ∆aspC stationary-phase cultures grown in LBTMAO, and we demonstrated that all bacteria exposed to biogenic TMA displayed increased resistance to tetracycline (Fig. 2A). To confirm that this phenotype was due to emission of volatile TMA, we studied TMAO conversion into TMA in E. coli TG1 ∆aspC ∆red, containing mutations in all known TMAO reductases, namely torA, torYZ, dmsA, and ynfEFGH dmsD (21, 30, 31). E. coli aerially exposed to volatile compounds emitted from filtered ammonia and TMA-deficient TG1 ∆aspC ∆red stationary-phase cultures grown in LBTMAO did not display increased resistance to tetracycline (Fig. 2B). Consistently, nuclear magnetic resonance (NMR) analysis performed on TMA solubilized into water exposed for 24 h at the site of bacterial inoculation (see the experimental setup described in Fig. 2C) showed that, whereas E. coli TG1 ∆aspC grown in LBTMAO produced TMA, TG1 ∆aspC ∆red was unable to convert TMAO into TMA (Fig. 2D).
FIG 2 .
Biogenic production of TMA by E. coli from TMAO increases tetracycline resistance in representative bacteria. (A) Biogenic TMA produced by E. coli from TMAO induces increased tetracycline resistance of all tested Gram-positive and Gram-negative bacteria. Shown is growth of E. coli MG1655, P. aeruginosa PAO1, S. aureus HG001, and B. subtilis 168 on inhibitory concentrations of tetracycline when aerially exposed to H2O or to filtered E. coli TG1 ∆aspC cultures grown in LBTMAO. (B) E. coli MG1655 growth on 3 µg/ml tetracycline when aerially exposed to filtered E. coli TG1 ∆aspC cultures grown with or without TMAO substrate or exposed to a filtered culture of TMAO reductase multiple mutant grown in LBTMAO. (C) Principle of the experimental design used to compare the amounts of TMA emitted from various pure and biogenic TMA solutions when resolubilized in 80 µl of water exposed in the 2-petri-dish assay at the same distance from the source of tested volatile compounds as the exposed bacteria. (D) Biogenic production of TMA by E. coli TG1 ∆aspC when grown in the presence of TMAO substrate. One-dimensional proton spectra are shown. The ∆aspC ∆red strain (containing mutations in all known TMAO reductases) grown in LB was ammonia negative without TMAO in the culture. The ∆aspC ∆red strain grown in LBTMAO was ammonia negative with TMAO in the culture (pH 8.8). The ∆aspC strain grown in LBTMAO was ammonia negative with TMAO in the culture (reduction of TMAO in TMA). A single peak resonating at 2.73 ppm corresponding to TMA is indicated. The ∆aspC strain grown in LB was ammonia negative without TMAO in the culture.
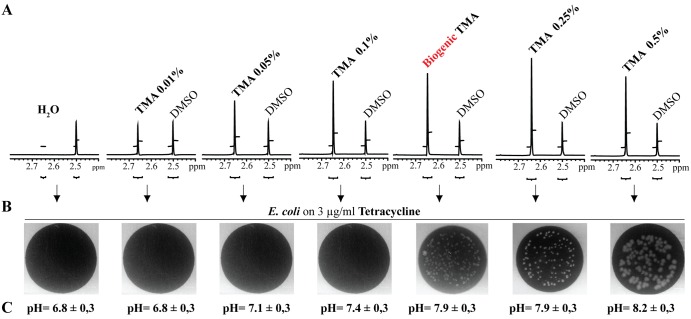
We then used a dimethyl sulfoxide (DMSO) standard and nuclear magnetic resonance (NMR) analysis to compare biogenic TMA with various emitting solutions prepared with increasing concentrations of pure TMA. This relative quantification analysis showed that increased tetracycline resistance of bacteria exposed to increasing concentrations of pure or biogenic TMA-emitting solutions also correlates with increased TMA solubilized into water exposed for 24 h at the site of bacterial inoculation (Fig. 3A and B). Moreover, although a saturation plateau is reached in our experimental condition after a 0.25% TMA-emitting solution, the quantity (Fig. 3A) and activity (Fig. 3B) of biogenic TMA produced by E. coli TG1 ∆aspC grown in LBTMAO are similar to those of 0.25% pure TMA solutions. These results therefore indicate that active TMA-emitting solutions used in our study (0.25% and 0.5%) are biologically relevant and close to those reached upon biogenic production of TMA by bacteria. Finally, we compared increased resistance to tetracycline in E. coli exposed to volatile compounds emitted either from TG1 grown in LB (only emitting ammonia) or from TG1 grown in LBTMAO (emitting ammonia and TMA), and we observed slightly increased resistance to tetracycline in the latter case, suggesting limited potential synergy between the two volatile compounds (see Fig. S1 in the supplemental material).
FIG 3 .
Phenotypic and quantitative comparison of pure and biogenic TMA solutions. Relative quantification of TMA produced from biogenic and various pure TMA-emitting solutions (0.01, 0.05, 0.1, 0.25, and 0.5%) was performed by proton NMR analyses using an internal standard of DMSO of a known concentration (1.08 mM). (A) The TMA concentration in the biogenic sample was evaluated by taking into account that two methyl groups contribute to the DMSO signal, whereas three methyl groups contribute to the TMA signal. A saturation plateau occurs around 0.25% TMA, giving only a lower limit of TMA concentration. (B) Increased tetracycline resistance of E. coli exposed to biogenic and various pure TMA-emitting solutions (0.01, 0.05, 0.1, 0.25, and 0.5%). (C) Determination of the pH in the 80-µl water recipient exposed in the 2-petri-dish assay to biogenic and various pure TMA-emitting solutions (0.01, 0.05, 0.1, 0.25, and 0.5%).
TMA and NH3 modes of action are distinct and lead to different phenotypes.
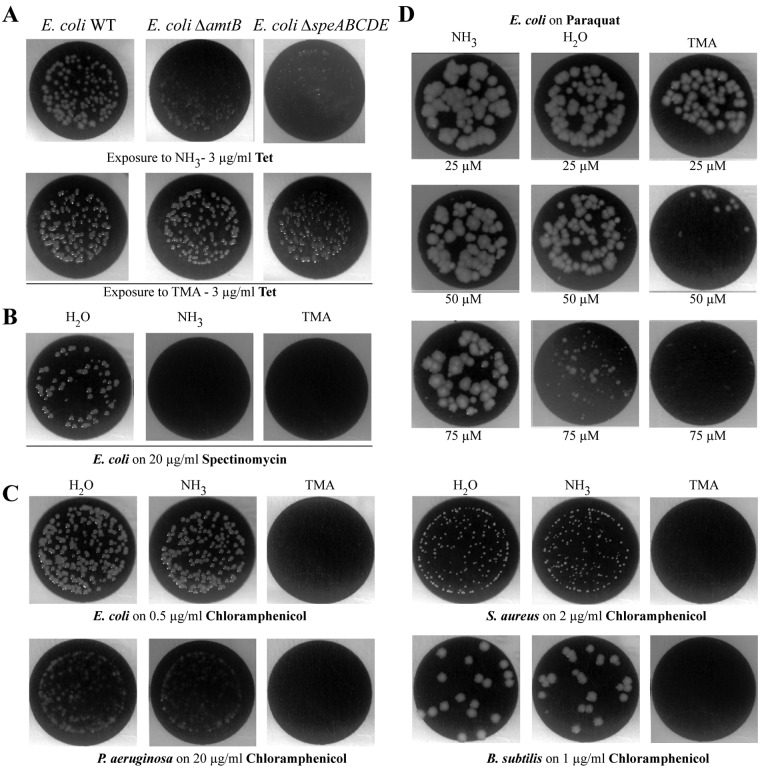
In E. coli, increased resistance to tetracycline upon exposure to ammonia was previously shown to require the presence of the Amt membrane channel and synthesis of the intracellular polyamines putrescine and spermidine (Fig. 4A) (17). However, exposure of corresponding amtB (Amt-negative) and speABCDE (polyamine-negative) E. coli mutants to TMA still conferred increased resistance to tetracycline (Fig. 4A). In addition, although both ammonia and TMA promoted increased resistance to tetracycline and decreased resistance to aminoglycosides, such as spectinomycin, aerial exposure to TMA also decreased resistance to chloramphenicol in all tested bacteria (Fig. 4B and C) (data not shown). Finally, while we previously showed that ammonia decreases oxidative stress induced by paraquat (17), exposure to volatile TMA instead led to increased paraquat toxicity in E. coli (Fig. 4D). These results therefore suggested that TMA perception and its mode of action are distinct from those of ammonia.
FIG 4 .
Differences between NH3- and TMA-induced phenotypes. (A) TMA mode of action does not depend on Amt channel or polyamines. Growth of E. coli MG1655 and corresponding amt and speABCDE mutants on 3 µg/ml tetracycline when exposed to NH3 (1%) or TMA (0.5%). (B) Exposure to TMA and NH3 decreased E. coli resistance to the aminoglycoside spectinomycin. E. coli MG1655 growth on 20 µg/ml spectinomycin when exposed to H2O, NH3 (1%), or TMA (0.5%). (C) Unlike NH3, TMA decreased resistance to chloramphenicol of aerially exposed bacteria. Shown is growth of E. coli MG1655, P. aeruginosa PAO1, S. aureus HG001, and B. subtilis 168 on subinhibitory concentrations of chloramphenicol upon exposure to H2O, NH3 (1%), or TMA (0.5%). (D) Exposure to TMA decreased paraquat toxicity. Shown is growth on increasing paraquat concentrations of E. coli MG1655 exposed to NH3 (1%), H2O, or TMA (0.5%).
TMA and chemically distinct volatile amines induce similar phenotypes.
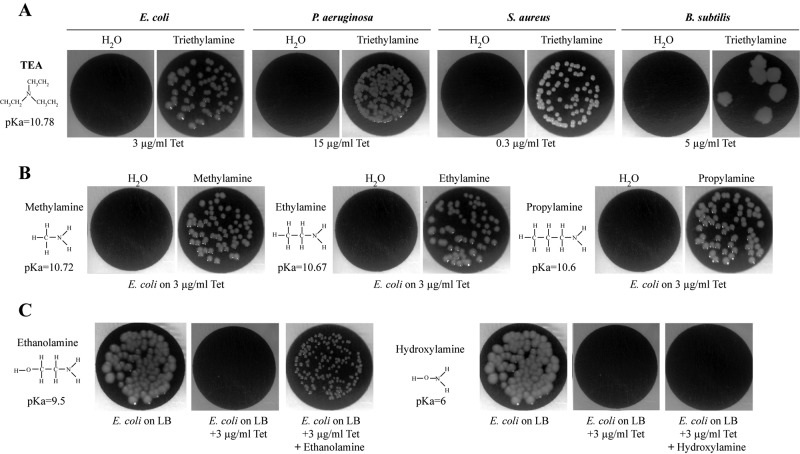
To further investigate genetic requirements for TMA perception, we tested whether transporters of structurally related nonvolatile trimethylammonium compounds such as betaine, choline, and carnithine could contribute to TMA perception in bacteria exposed to TMA (32). However, none of the tested mutants corresponding to the choline, betaine, or carnithine transporter (betT and caiT) and their identified paralogs in E. coli (proU, proP, yeaV, yehX, yhjE, yiaM, shiA, putP, afuC, metN, glnQ, kgtP, potA, potG, ycjV, and ydfJ) (33) were found to affect volatile TMA-dependent tetracycline resistance in mutant E. coli recipient bacteria (data not shown). Alternatively, we tested the activities of structurally related volatile amines, including triethylamine, methylamine, ethylamine, propylamine, ethanolamine, and hydroxylamine. We first verified that aerial exposure to these compounds did not affect the growth of E. coli, P. aeruginosa, S. aureus, or B. subtilis bacteria in tetracycline-free medium (data not shown). Interestingly, exposure to triethylamine, methylamine, ethylamine, and propylamine produced patterns of antibiotic and oxidative stress tolerance identical to those obtained with TMA (Fig. 5A and B) (data not shown). While ethanolamine and hydroxylamine lack activity, we hypothesized that this could be due to their low volatility. To test this, we provided them directly to recipient cells by supplementing LB-tetracycline (LB-tet) agar medium with nontoxic concentrations, along with TMA, NH3, and water as controls. We observed that ethanolamine, but not hydroxylamine, enabled E. coli to grow on tetracycline (Fig. 5C) (data not shown).
FIG 5 .
Structurally related volatile amines increase tetracycline resistance in representative bacteria. (A) Aerial exposure to triethylamine (TEA) increases resistance to tetracycline in all tested bacteria. Shown is growth of E. coli MG1655, P. aeruginosa PAO1, S. aureus HG001, and B. subtilis 168 on inhibitory concentrations of tetracycline upon exposure to H2O or 1% TEA. (B) Aerial exposure to 3 structurally related volatile amines increases E. coli resistance to tetracycline. Growth of E. coli MG1655 on 3 µg/ml tetracycline upon exposure to H2O, methylamine (1%), ethylamine (1%), or propylamine (1%). (C) Ethanolamine but not hydroxylamine added to agar medium increases tetracycline resistance in E. coli. Shown is growth of E. coli MG1655 on LB agar medium, LB agar medium with tetracyline (LB-tet), or LB-tet directly supplemented with ethanolamine (1%) or hydroxylamine (1%).
Volatile TMA increases pH of the growth medium of distantly exposed bacteria.
To further investigate the TMA mode of action, we compared active and inactive amines with respect to TMA-like phenotypes, and we noticed that all active amines (TMA, triethylamine, methylamine, ethylamine, propylamine, and ethanolamine) had a higher pKa (>9.5) than hydroxylamine (pKa of 6) (Fig. 4). As this suggested a role for pH in the effect of active amines on recipient bacteria, we used the experimental setup described in Fig. 2C and measured the pH of water in an 80-µl recipient exposed in the 2-petri dish assay at the same distance from the source of tested volatile compounds as the exposed bacteria. While aerial exposure to ammonia did not affect pH (6.6 ± 0.3 to 6.8 ± 0.3), exposure to purified or biogenic TMA led to a pH increase in aerially exposed water from 6.8 ± 0.3 up to 7.9 ± 0.3 for a 0.25% TMA-emitting solution and for biogenic TMA and to 8.2 ± 0.3 for a 0.5% TMA-emitting solution (Fig. 3C). A similar increase was also observed with the other active volatile amines (data not shown).
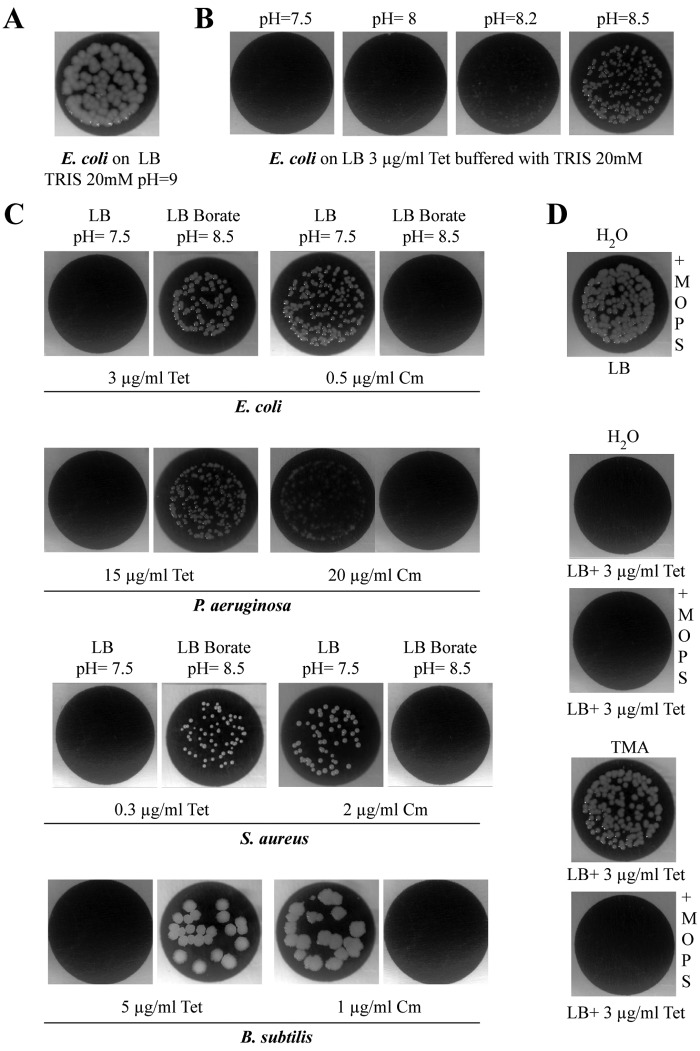
To determine whether the observed increase of pH in medium surrounding exposed bacteria could account for the TMA-associated phenotypes, we grew E. coli on medium buffered with 20 mM Tris-base buffer and adjusted the LB agar medium to pHs ranging from 7.5 to 8.5. While no growth inhibition was observed in this range of pHs (Fig. 6A), E. coli growth on medium adjusted to a pH above 8 led to phenotypes similar to those induced upon aerial exposure to TMA and other aminated volatile compounds, including increased resistance to tetracycline, decreased resistance to aminoglycosides and chloramphenicol, and increased sensitivity to paraquat (Fig. 6B) (data not shown). Since Tris is itself a primary amine [Tris-hydroxymethylaminomethane; NH2C(CH2OH)3; pKa of 8.3], we also used a nonaminated 50 mM borate buffer (KCl/H2BO3-NaOH) to adjust the pH of the LB agar medium to 8.5, and we obtained similar results with E. coli, P. aeruginosa, S. aureus, and B. subtilis (Fig. 6C). Finally, we showed that buffering the growth medium to pH 7 with 100 mM MOPS (morpholinepropanesulfonic acid) buffer prevented TMA and to other volatile amines from affecting bacteria aerially exposed in the 2-petri-dish assay (Fig. 6D) (data not shown). These results therefore demonstrated a pH-dependent mode of action for TMA.
FIG 6 .
Exposure to TMA increases pH of aerially exposed growth media. (A) High-pH medium does not affect E. coli growth. Shown is E. coli MG1655 growth on LB supplemented with Tris buffer and corresponding to pH 9. (B) Increasing pH with Tris buffer mimics the increased tetracycline resistance of E. coli observed upon exposure to TMA (0.5%). (C) pH increase of the agar growth medium leads to the same phenotype as those induced by TMA exposure, including increased resistance to tetracycline (Tet) and decreased resistance to chloramphenicol (Cm), in all tested bacteria. The pH was increased to 8.5 by adding borate buffer to LB agar medium containing inhibitory concentrations of tetracycline or chloramphenicol. Shown is growth of E. coli MG1655, P. aeruginosa PAO1, S. aureus HG001, and B. subtilis 168 on LB agar medium plus antibiotics supplemented or not with borate buffer. (D) Buffering of LB medium to pH 7 by adding MOPS buffer inhibits the increased tetracycline resistance induced by TMA.
A TMA-dependent increase in pH alters the tetracycline influx in exposed E. coli bacteria
Previous studies showed that medium alkalinization (pH range 7 to 11) produced no significant reduction in the tetracycline half-life (34), thereby suggesting that the TMA-dependent increase in tetracycline resistance is not due to antibiotic degradation on medium available to exposed bacteria. On the other hand, analysis of the effect of ionophores on tetracycline uptake by E. coli showed that tetracycline accumulation did not depend on membrane electrical potential but was a ΔpH-dependent process in which the higher the pH, the lower the tetracycline transport into recipient bacteria (35). To confirm that aerial exposure to TMA led to a pH-dependent reduction in the tetracycline entry into recipient bacteria, we used LB medium containing subinhibitory concentrations of tetracycline and a tetracycline-sensitive E. coli strain carrying a chromosomal tetR-ptetO lacZ transcriptional fusion, as a reporter of intracellular concentrations of tetracycline (36). In the presence of tetracycline, the tight TetR repressor was released from the tet operators (tetO) of the tetA promoter, leading to lacZ expression and induction of β-galactosidase production. Comparison of the β-galactosidase activities of bacteria exposed or not to TMA revealed a 2-fold reduction in activity compared to that in bacteria exposed to water or ammonia. Consistently, use of the TMA-like amines triethylamine, methylamine, ethylamine, and propylamine also reduced the activity of tetR-ptetO lacZ transcriptional fusion in exposed bacteria (Fig. 7). All together, these results showed a reduction in tetracycline uptake in E. coli cells aerially exposed to TMA, consistent with distant pH-dependent modification of tetracycline uptake.
FIG 7 .
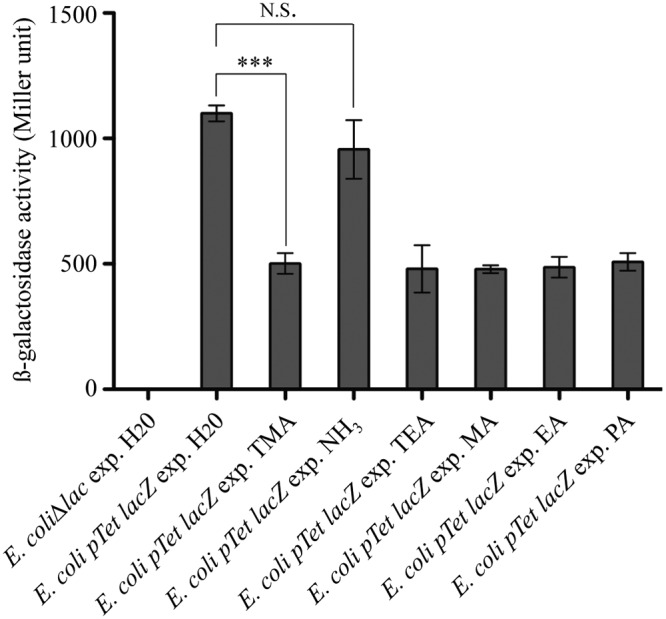
Exposure to TMA and to structurally related volatile amines reduces tetracycline entry into E. coli. β-Galactosidase activity measurements of tetR-ptetO lacZ transcriptional fusion after exposure to H2O, NH3, TMA, and other structurally related volatile amines. An E. coli strain carrying a chromosomal tetR-ptetO lacZ transcriptional fusion was grown on 1.5 µg/ml tetracycline and exposed to (exp.) H2O, TMA (0.5%), NH3 (1%), triethylamine (1%) (TEA), methylamine (1%) (MA), ethylamine (1%) (EA), and propylamine (1%) (PA) for 24 h; β-galactosidase activities were measured as described in Materials and Methods. An E. coli strain with the lac operon deleted was used as the negative control. The data represent means ± standard deviations (SD) of 3 independent experiments. For statistical analysis, P < 0.05 (*), P < 0.01 (**), or P < 0.0001 (***) in comparison with H2O by one-tailed unpaired Student’s t test.
DISCUSSION
Interactions between microorganisms are key determinants of their distribution and activity in most ecosystems (1). Soluble bacterial secondary metabolites contribute to microbial competition and cooperation or trigger bacterial adaptive behavior to changing environments (37). However, bacteria also release complex growth condition-dependent blends of volatile molecules, whose impact on bacterial biological processes remains poorly characterized (4, 8, 38). Here we investigated the role of a subset of volatile compounds produced by E. coli, and we show that several of them might influence motility, adhesion, and antibiotic resistance.
For instance, exposure to indole, a molecule previously demonstrated to be involved in interbacterial communication (39–41), inhibits biofilm formation in aerially exposed E. coli and P. aeruginosa, whereas it stimulates S. aureus biofilm formation. In the case of gaseous ammonia, which was shown to stimulate pigment production and biofilm formation of Bacillus licheniformis (15), it only slightly stimulates biofilm formation of B. subtilis and S. aureus. These results therefore suggest that, depending on their concentrations, the physiological state of recipient bacteria, and the structure of the environment, volatile molecules might have opposing, possibly contradictory roles. Interestingly, a very recent study investigating the impact of B. subtilis volatile compounds on E. coli gene expression showed that 2,3-butanedione inhibits bacterial motility (19). Although the authors identified changes in motility-related gene expression, our observation that exposure to 2,3-butanedione reduces bacterial growth raises the possibility that inhibition of E. coli motility upon exposure to 2,3-butanedione could be partly due to growth inhibition. The biological relevance of the tested concentration, at which some of the tested volatile compounds could affect bacterial motility and biofilm formation, remains to be determined in natural environments, including soil, gut, and biofilm interstitial spaces. However, our study amply illustrates the potential of bacterial volatile compounds to act as chemical cues influencing the development of bacterial communities.
We recently demonstrated that gaseous ammonia modulates tolerance of neighboring bacterial cells to several antibiotics (17). In this study, we showed that exposure to TMA, a volatile compound produced by E. coli and many other Gram-negative bacteria, also modulates antibiotic tolerance in all tested bacteria, although by a radically distinct mode of action. While none of the bacterial factors required for ammonia sensing (Amt transporter and polyamine synthesis) were involved in TMA-dependent phenotypes, we established that volatile TMA acts by changing the pH of the medium surrounding exposed bacteria and that the TMA effect could be mimicked by nonspecific medium alkalinization.
Increasing the pH of extracellular medium was previously shown to increase uptake of aminoglycosides and chloramphenicol and to reduce uptake of tetracycline due to modification of transmembrane ∆pH and proton motive force (35, 42–45). These results are in agreement with our observation of increased sensitivity of bacteria to aminoglycosides and chloramphenicol and reduced sensitivity to tetracycline upon exposure to TMA and pH alkalinization. Moreover, the observed increased sensitivity to paraquat in bacteria exposed to TMA could also be a consequence of alkalinization of the environment, since alkaline conditions enhance paraquat uptake in E. coli (46).
Under anaerobic conditions, E. coli and other Gram-negative bacteria can use TMAO as an alternate electron acceptor to produce energy (47). However, TMAO conversion into TMA by bacterial TMAO reductases can occur under both aerobic and anaerobic conditions, and the physiological role of TMAO reduction during aerobiosis is not well understood. Several studies suggested that TMA production and the associated increase in pH could contribute to pH homeostasis in response to sudden acidification of bacterial environments due to, for instance, the presence of large amounts of acetate in the intestine (22, 48). The TMA-dependent increase in tetracycline resistance of aerially exposed bacteria can also be observed under both anaerobic (data not shown) and aerobic conditions. This suggests that TMA and pH-dependent modifications in antibiotic resistance could have profound ecological consequences in a variety of TMAO-rich environments, such as animal gut and tissues rich in phosphatidylcholine, choline, betaine, and carnitine (6, 20, 27, 28, 49). Production of TMA in these environments could alter concentration-dependent bacterial responses to antibiotic stresses and cues (50). Moreover, modification of sensitivity to antibiotic gradients in the presence of TMA could alter biofilm formation in response to sublethal concentrations of aminoglycosides and therefore influence community structure and function (51, 52).
Several volatile compounds and gases have now been shown to impact bacterial antibiotic resistance and tolerance, including indole, hydrogen sulfide (H2S), nitric oxide (NO), and ammonia (17, 53–55). As is the case for indole, which induces efflux pumps and oxidative stress protective mechanisms that benefit the entire population (54), H2S, NO, and ammonia display various protective activities against oxidative stress and the lethal effect of antibiotics, which could contribute to community-based antibiotic tolerance and the spread of such tolerance (50). Similarly, the transient increase in resistance to antibiotic stress associated with medium alkalinization upon TMA production could also contribute to the evolution of antibiotic tolerance in bacterial populations. Moreover, modification of the pH of physically separated niches by biogenic TMA could constitute a biological cue picked up at a distance by competing microorganisms. For instance, increased pH was shown to influence S. aureus biofilm formation (56), hyphal formation, and virulence in pathogenic yeasts and other fungi (57–59).
Although we investigated only a limited subset of volatile molecules under restrained test conditions, our results further illustrate the biological relevance of bacterial gases and volatile compounds as chemical cues mediating bacterial interactions in a variety of colonized environments. In addition to cross-kingdom interactions with plants, nematodes, and fungi, distant airborne chemical interactions trigger functional responses in exposed bacteria and could therefore play an important role in the development of bacterial communities.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 2. The following antibiotics were used as indicated: kanamycin (50 µg/ml), chloramphenicol (25 µg/ml), and tetracycline (15 µg/ml). All experiments were performed in lysogeny broth (LB) medium and incubated at 37°C. All chemicals were purchased from Sigma-Aldrich and dissolved in water, and aqueous dilutions of 1% vol/vol or wt/vol (or 0.5% for TMA and 0.15% wt/vol for indole) were used as sources of volatile compounds. For biogenic production of TMA, 0.4% TMAO (wt/vol) was added to the LB culture.
TABLE 2 .
Strains used in the study
| Strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Gram negative | ||
| Escherichia coli | ||
| MG1655 | E. coli K-12 derivative (poorly motile) | 65 |
| MG1655-motility+ | Motile MG1655 | Gift from D. Murat |
| MG1655 F′ tet | F+, biofilm-forming strain, Tetr | 66 |
| MG1655 ΔamtB | ΔamtB | 17 |
| MG1655 ΔspeABCDE | ΔspeABCDE | 17 |
| TG1 ∆KCP | Source of the chloramphenicol resistance cassette | Laboratory collection |
| TG1 ΔaspC | ΔaspC | 17 |
| TG1 Δred | ΔtorA ΔdmsA ΔtorYZ ΔynEFGH dmsD Kmr Cmr Specr | This study |
| TG1 ∆aspC ∆red | TG1 Δred ΔaspC Kmr Cmr Specr Tetr | This study |
| BW25113 | E. coli K-12 derivative | 60 |
| BW25113 ΔbetT | ΔbetT Kmr | 60 |
| BW25113 ΔcaiT | ΔcaiT Kmr | 60 |
| BW25113 ΔproU | ΔproU Kmr | 60 |
| BW25113 ΔproP | ΔproP Kmr | 60 |
| BW25113 ΔyeaV | ΔyeaV Kmr | 60 |
| BW25113 ΔyehX | ΔyehX Kmr | 60 |
| BW25113 ΔyhjE | ΔyhjE Kmr | 60 |
| BW25113 ΔyiaM | ΔyiaM Kmr | 60 |
| BW25113 ΔshiA | ΔshiA Kmr | 60 |
| BW25113 ΔputP | ΔputP Kmr | 60 |
| BW25113 ΔafuC | ΔafuC Kmr | 60 |
| BW25113 ΔmetN | ΔmetN Kmr | 60 |
| BW25113 ΔglnQ | ΔglnQ Kmr | 60 |
| BW25113 ΔkgtP | ΔkgtP Kmr | 60 |
| BW25113 ΔpotA | ΔpotA Kmr | 60 |
| W25113 ΔpotG | ΔpotG Kmr | 60 |
| BW25113 ΔycjV | ΔycjV Kmr | 60 |
| BW25113 ΔydfJ | ΔydfJ Kmr | 60 |
| MG1655 tetR-ptetO lacZ | tetR-ptetO lacZ transcriptional fusion (Tets) | 36 |
| P. aeruginosa PAO1 | Laboratory collection | |
| Gram positive | ||
| S. aureus HG001 | 67 | |
| B. subtilis | ||
| 168 | 68 | |
| 3610 | Laboratory collection |
Mutant construction.
The E. coli deletion mutants used in this study originated either from the Keio Collection (60) or were carried out using λ-Red linear DNA gene inactivation with pKOBEG plasmid derivatives (61, 62). When required, kanamycin resistance markers flanked by two FLP recombination target (FRT) sites were removed using Flp recombinase (63). Strain TG1 Δred, corresponding to the genotype ∆torA ∆dmsA ∆torYZ ∆ynfEFGH dmsD, was built via multiple steps as follows. The ∆torA::Km FRT mutation from strain JW0982 (Keio Collection) was introduced into the strain TG1 genetic background by P1vir phage transduction (TG1 ∆torA::Km FRT), in which the kanamycin marker was then removed by the Flp recombinase (TG1 ∆torA::∆FRT). Subsequently, the ∆dmsA::Km FRT mutation was transferred by P1vir from strain JW5118 (Keio Collection) into TG1 ∆torA::∆FRT, resulting in the intermediate strain TG1 ∆torA::∆FRT ∆dmsA::Km FRT. Deletion of torYZ genes was performed by the λ-Red linear DNA gene inactivation method with the following primers: torYZ.Cat.L5 (ACAATGATTTCGGTGTCCAGTAATTTAATTAGAGGAATCTCCTGTGACGGAAGATCACTTCGCA) and torYZ.Cat.L3 (TGCATAACCTCAGCGCCCGTTTCCGGGCGCTATTCACGTCTTACGCCCCGCCCTGCCACTCATC). Strain TG1 ∆IKCP was used for the PCR as the source of the chloramphenicol resistance marker. The deletion mutation ∆torYZ::Cat was first introduced into TG1, giving TG1 ∆torYZ::C, and then was transferred by P1vir into TG1 ∆torA::∆FRT ∆dmsA::Km FRT to create TG1 ∆torA::∆FRT ∆dmsA::Km FRT ∆torYZ::Cat. Deletion of genes ynfEFGH and dmsD was performed using primers ynfEFGHdmsD.Spec.L5 (TATAAACTTTTATATAACGATAAAGAACAGGGAGTGAGTTGCGTCAATTGCGCCGAATAAATAC) and ynfEFGHdmsD.Spec.L3 (TTGCGCGTAATCGTCACCATCCGGCAATATTACGGTGATCTCATTGGCTGGCACCAAGCAGTTT). Strain DH5α pir (pSW25oriT::ccdB) was used for the PCR as the source of the spectinomycin resistance marker (Spec), and the mutation was introduced into MG1655, creating MG1655 ∆ynfEFGHdmsD::Spec. The ∆ynfEFGHdmsD::Spec mutation was then transferred by P1vir into TG1 ∆torA::∆FRT ∆dmsA::Km FRT ∆torYZ::Cat, resulting in TG1 ∆torA::∆FRT ∆dmsA::Km FRT ∆torYZ::Cat ∆ynfEFGHdmsD::Spec (TG1Δred). The primers used to construct TG1 ∆aspC ∆red, corresponding to introduction of the ∆aspC mutation into TG1 Δred are aspC.FrtTET.L-3 (GTGAACACAGCGATAGACGGCCTCCATGACGAGGTTCCATTATGGTTAC) and aspC.FrtTET.L-5 (CAAGATGTATCCGGTTACCGACTGGCATTAAAAACAATGAAGCCCGCTG). All mutations were confirmed by PCR and sequencing analysis.
Screening for volatile-mediated phenotypes.
To evaluate the activity of a volatile compound on various bacterial phenotypes, we used variations of a previously described 2-petri-dish assay (Fig. 1A) (17). Three milliliters of a 1% (0.5% for TMA and 0.15% for indole) aqueous dilution of all tested volatile compounds was introduced in the smaller middle petri dish as the sources of volatile molecules. The large petri dish was then closed and incubated for 24 h at 37°C, and volatile activity was estimated as indicated below.
(i) Test of antibiotic resistance.
Recipient test bacteria were spotted on the external agar ring as 20-µl drops of a 10−5 dilution of an O/N culture adjusted to an optical density at 600 nm (OD600) of 1, which corresponded to approximately 100 CFU for E. coli, P. aeruginosa, and S. aureus. Due to rapid spread and coalescence of B. subtilis colonies on plate, we used only 20 CFU for the B. subtilis inoculum. Volatile activity was visually estimated as a function of the growth of recipient bacteria on the external ring containing an inhibitory concentration of an antibiotic previously determined in the absence of exposure to a bacterial volatile source.
(ii) Test of bacterial growth and morphotype.
Recipient test bacteria were spotted on the external agar ring (without antibiotics) as 20-µl drops of a 10−5 dilution of an O/N culture adjusted to an OD600 of 1, which corresponded to approximately 100 CFU for E. coli, P. aeruginosa, and S. aureus but only 20 CFU for B. subtilis. Volatile activity was qualitatively estimated as a function of bacterial growth and colony morphotype, compared to bacteria exposed only to water placed in the middle petri dish.
(iii) Test of motility.
The external ring was filled with 20 ml of motility 0.3% agar (10 g/liter Bacto tryptone, 5 g/liter NaCl, 3 g/liter agar). Recipient test bacteria were spotted on the external 0.3% agar ring as a 2-µl drop of a 10−6 dilution of an O/N culture for all strains. Volatile activity was visually estimated by comparing bacterial motility after exposure to volatile compounds with that after exposure to water.
(iv) Test of biofilm capacity.
Overnight cultures were adjusted to an OD600 of 0.01 before inoculation of 100 µl into part (6 wells) of a 96-well polyvinyl chloride (PVC) plate placed in the external ring of the 2-petri-dish assay (Fig. 1A). After 24 h, part of the PVC plate was removed from the large petri dish and rinsed, and 125 µl of a 1% solution of crystal violet was added to each well. These were incubated 15 min at room temperature and rinsed 3 times. Biofilm formation was estimated by solubilization of crystal violet by adding 150 µl of ethanol-acetone (80:20) and determining the OD595 after 30 min. Volatile activity was estimated by comparing biofilm formation from bacteria exposed to volatile compounds to that of bacteria exposed to water.
Measurement of pH.
The pH of water in an 80-µl recipient exposed and positioned in the 2-petri dish assay (Fig. 2C) was determined using Duotest pH paper (Macherey-Nagel).
NMR analyses.
The experimental setup used to analyze TMA produced from different cultures is described in Fig. 2C. NMR analyses were performed on water exposed to E. coli TG1 ∆aspC stationary-phase cultures grown in LB or LBTMAO and to E. coli TG1 ∆aspC ∆red stationary-phase culture grown in LBTMAO. Proton NMR spectra were acquired on a Varian NMR system 500 spectrometer (Agilent Technologies, Santa Clara, CA) equipped with a triple resonance 1H[13C/15N] pulsed-field gradient (PFG) probe with a proton resonating frequency of 499.4 MHz. Spectra were recorded at 25°C and referenced to sodium 4,4-dimethyl-4-silapentane-1-sulfonate following IUPAC recommendations. Data were collected using VnmrJ 2.3 A (Agilent Technologies). 1H NMR spectroscopy included double pulsed-field gradient spin echo (DPFGSE) to suppress the water signal. For each sample, 10% D2O was added to the 600 microliters of sample for internal lock. Quantification of the TMA produced was performed using an internal standard of DMSO of known concentration (1.08 mM) for samples exposed to variable quantities of TMA (0.01, 0.05, 0.1, 0.25, and 0.5%).
β-Galactosidase assay.
β-Galactosidase enzymatic activity was determined by spotting 20-µl drops of a 10−5 dilution of an O/N E. coli MG1655 tetR-Km-ptetO::lacZ culture adjusted to an OD600 of 1. Bacteria were spotted on LB agar containing a subinhibitory concentration of tetracycline (1.5 µg/ml) and grown at 37°C upon exposure to 3 ml of 1% (0.5% for TMA) aqueous dilution of ammonia, TMA, triethylamine, methylamine, ethylamine, or propylamine. β-Galactosidase enzymatic assays were performed as previously described (64).
Statistical analysis.
One-tailed unpaired Student’s t test analyses were performed using Prism 5.0 for Mac Os X (GraphPad Software). Each experiment was performed at least three times.
SUPPLEMENTAL MATERIAL
Synergy between ammonia and TMA. Shown is a comparison of the increased tolerance to tetracycline in E. coli exposed to volatile compounds emitted either from TG1 grown in LBTMAO (emitting ammonia and TMA) or from TG1 grown in LB (only emitting ammonia). (Upper row) E. coli recipient cells grown on 3 µg/ml tetracycline. (Lower row) E. coli recipient cells grown on 4 µg/ml tetracycline. Download
ACKNOWLEDGMENTS
We thank David Lebeaux, Christophe Beloin, and Olaya Rendueles for helpful discussions and critical reading of the manuscript. We thank Catherine Simenel for technical help.
This work was supported by grants from the Institut Pasteur and from the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (grant no. ANR-10-LABX-62-IBEID). S.P.B. was the recipient of a postdoctoral fellowship from the Canadian Louis Pasteur Foundation.
Footnotes
Citation Létoffé S, Audrain B, Bernier SP, Delepierre M, Ghigo J-M. 2014. Aerial exposure to the bacterial volatile compound trimethylamine modifies antibiotic resistance of physically separated bacteria by raising culture medium pH. mBio 5(1):e00944-13. doi:10.1128/mBio.00944-13.
REFERENCES
- 1.Keller L, Surette MG. 2006. Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4:249–258 [DOI] [PubMed] [Google Scholar]
- 2.Monds RD, O’Toole GA. 2008. Metabolites as intracellular signals for regulation of community level traits, p 105–129 In Winans SC, Bassler BL. (ed), Chemical communication among bacteria. ASM Press, Washington, DC [Google Scholar]
- 3.Straight PD, Kolter R. 2009. Interspecies chemical communication in bacterial development. Annu. Rev. Microbiol. 63:99–118 [DOI] [PubMed] [Google Scholar]
- 4.Schulz S, Dickschat JS. 2007. Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 24:814–842 [DOI] [PubMed] [Google Scholar]
- 5.Dunkel M, Schmidt U, Struck S, Berger L, Gruening B, Hossbach J, Jaeger IS, Effmert U, Piechulla B, Eriksson R, Knudsen J, Preissner R. 2009. Superscent—a database of flavors and scents. Nucleic Acids Res. 37:D291–D294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos LDJ, Sterk PJ, Schultz MJ. 2013. Volatile metabolites of pathogens: a systematic review. PLoS Pathog. 9:e1003311. 10.1371/journal.ppat.1003311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams DH, Stone MJ, Hauck PR, Rahman SK. 1989. Why are secondary metabolites (natural products) biosynthesized? J. Nat. Prod. 52:1189–1208 [DOI] [PubMed] [Google Scholar]
- 8.Kai M, Haustein M, Molina F, Petri A, Scholz B, Piechulla B. 2009. Bacterial volatiles and their action potential. Appl. Microbiol. Biotechnol. 81:1001–1012 [DOI] [PubMed] [Google Scholar]
- 9.Niu Q, Huang X, Zhang L, Xu J, Yang D, Wei K, Niu X, An Z, Bennett JW, Zou C, Yang J, Zhang KQ. 2010. A Trojan horse mechanism of bacterial pathogenesis against nematodes. Proc. Natl. Acad. Sci. U. S. A. 107:16631–16636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blom D, Fabbri C, Eberl L, Weisskopf L. 2011. Volatile-mediated killing of Arabidopsis thaliana by bacteria is mainly due to hydrogen cyanide. Appl. Environ. Microbiol. 77:1000–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Effmert U, Kalderas J, Warnke R, Piechulla B. 2012. Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol. 38:665–703 [DOI] [PubMed] [Google Scholar]
- 12.Ma Q, Fonseca A, Liu W, Fields AT, Pimsler ML, Spindola AF, Tarone AM, Crippen TL, Tomberlin JK, Wood TK. 2012. Proteus mirabilis interkingdom swarming signals attract blow flies. ISME J. 6:1356–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heal RD, Parsons AT. 2002. Novel intercellular communication system in Escherichia coli that confers antibiotic resistance between physically separated populations. J. Appl. Microbiol. 92:1116–1122 [DOI] [PubMed] [Google Scholar]
- 14.Cepl JJ, Patkova I, Blahuskova A, Cvrckova F, Markos A. 2010. Patterning of mutually interacting bacterial bodies: close contacts and airborne signals. BMC Microbiol. 10:139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nijland R, Burgess JG. 2010. Bacterial olfaction. Biotechnol. J. 5:974–977 [DOI] [PubMed] [Google Scholar]
- 16.Lee JH, Lee J. 2010. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 34:426–444. 10.1111/j.1574-6976.2009.00204.x [DOI] [PubMed] [Google Scholar]
- 17.Bernier SP, Letoffe S, Delepierre M, Ghigo JM. 2011. Biogenic ammonia modifies antibiotic resistance at a distance in physically separated bacteria. Mol. Microbiol. 81:705–716 [DOI] [PubMed] [Google Scholar]
- 18.Kesarwani M, Hazan R, He J, Que YA, Apidianakis Y, Lesic B, Xiao G, Dekimpe V, Milot S, Deziel E, Lepine F, Rahme LG. 2011. A quorum sensing regulated small volatile molecule reduces acute virulence and promotes chronic infection phenotypes. PLoS Pathog. 7:e1002192. 10.1371/journal.ppat.1002192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim KS, Lee S, Ryu CM. 2013. Interspecific bacterial sensing through airborne signals modulates locomotion and drug resistance. Nat. Commun 4:1809. 10.1038/ncomms2789 [DOI] [PubMed] [Google Scholar]
- 20.Barrett EL, Kwan HS. 1985. Bacterial reduction of trimethylamine oxide. Annu. Rev. Microbiol. 39:131–149 [DOI] [PubMed] [Google Scholar]
- 21.McCrindle SL, Kappler U, McEwan AG. 2005. Microbial dimethylsulfoxide and trimethylamine-N-oxide respiration. Adv. Microb. Physiol. 50:147–198 [DOI] [PubMed] [Google Scholar]
- 22.Ansaldi M, Theraulaz L, Baraquet C, Panis G, Mejean V. 2007. Aerobic TMAO respiration in Escherichia coli. Mol. Microbiol. 66:484–494 [DOI] [PubMed] [Google Scholar]
- 23.Joffraud JJ, Leroi F, Roy C, Berdague JL. 2001. Characterisation of volatile compounds produced by bacteria isolated from the spoilage flora of cold-smoked salmon. Int. J. Food Microbiol. 66:175–184 [DOI] [PubMed] [Google Scholar]
- 24.Farag MA, Ryu CM, Sumner LW, Pare PW. 2006. GC-MS SPME profiling of rhizobacterial volatiles reveals prospective inducers of growth promotion and induced systemic resistance in plants. Phytochemistry 67:2262–2268 [DOI] [PubMed] [Google Scholar]
- 25.Bunge M, Araghipour N, Mikoviny T, Dunkl J, Schnitzhofer R, Hansel A, Schinner F, Wisthaler A, Margesin R, Mark TD. 2008. On-line monitoring of microbial volatile metabolites by proton transfer reaction-mass spectrometry. Appl. Environ. Microbiol. 74:2179–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorn RM, Reynolds DM, Greenman J. 2011. Multivariate analysis of bacterial volatile compound profiles for discrimination between selected species and strains in vitro. J. Microbiol. Methods 84:258–264 [DOI] [PubMed] [Google Scholar]
- 27.Rebouche CJ, Seim H. 1998. Carnitine metabolism and its regulation in microorganisms and mammals. Annu. Rev. Nutr. 18:39–61 [DOI] [PubMed] [Google Scholar]
- 28.al-Waiz M, Mikov M, Mitchell SC, Smith RL. 1992. The exogenous origin of trimethylamine in the mouse. Metab. Clin. Exp. 41:135–136 [DOI] [PubMed] [Google Scholar]
- 29.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, Didonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. 2013. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19:576–585. 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sargent F, Berks BC, Palmer T. 2002. Assembly of membrane-bound respiratory complexes by the Tat protein-transport system. Arch. Microbiol. 178:77–84 [DOI] [PubMed] [Google Scholar]
- 31.Lubitz SP, Weiner JH. 2003. The Escherichia coli ynfEFGHI operon encodes polypeptides which are paralogues of dimethyl sulfoxide reductase (DmsABC). Arch. Biochem. Biophys. 418:205–216 [DOI] [PubMed] [Google Scholar]
- 32.Kappes RM, Kempf B, Bremer E. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziegler C, Bremer E, Kramer R. 2010. The BCCT family of carriers: from physiology to crystal structure. Mol. Microbiol. 78:13–34. 10.1111/j.1365-2958.2010.07332.x [DOI] [PubMed] [Google Scholar]
- 34.Loftin KA, Adams CD, Meyer MT, Surampalli R. 2008. Effects of ionic strength, temperature, and pH on degradation of selected antibiotics. J. Environ. Qual. 37:378–386 [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi A, Ohmori H, Kaneko-Ohdera M, Nomura T, Sawai T. 1991. Delta pH-dependent accumulation of tetracycline in Escherichia coli. Antimicrob. Agents Chemother. 35:53–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Da Re S, Le Quere B, Ghigo JM, Beloin C. 2007. Tight modulation of Escherichia coli bacterial biofilm formation through controlled expression of adhesion factors. Appl. Environ. Microbiol. 73:3391–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korpi A, Pasanen AL, Pasanen P. 1998. Volatile compounds originating from mixed microbial cultures on building materials under various humidity conditions. Appl. Environ. Microbiol. 64:2914–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheatley RE. 2002. The consequences of volatile organic compound mediated bacterial and fungal interactions. Antonie Van Leeuwenhoek 81:357–364 [DOI] [PubMed] [Google Scholar]
- 39.Di Martino P, Fursy R, Bret L, Sundararaju B, Phillips RS. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49:443–449 [DOI] [PubMed] [Google Scholar]
- 40.Domka J, Lee J, Wood TK. 2006. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 72:2449–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu M, Zhang C, Mu Y, Shen Q, Feng Y. 2010. Indole affects biofilm formation in bacteria. Indian J. Microbiol. 50:362–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burns JL, Smith AL. 1987. Chloramphenicol accumulation by Haemophilus influenzae. Antimicrob. Agents Chemother. 31:686–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taber HW, Mueller JP, Miller PF, Arrow AS. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51:439–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Speer BS, Shoemaker NB, Salyers AA. 1992. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 5:387–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikaido H. 1993. Transport across the bacterial outer membrane. J. Bioenerget. Biomembr. 25:581–589 [DOI] [PubMed] [Google Scholar]
- 46.Minakami H, Kitzler JW, Fridovich I. 1990. Effects of pH, glucose, and chelating agents on lethality of paraquat to Escherichia coli. J. Bacteriol. 172:691–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gennis RB, Stewart V. 2005. Chapter 17. Respiration. In Curtiss R, III, Böck A, Ingraham JL, Kaper JB, Neidhardt FC, Riley M, Squires CL. (ed), EcoSal—Escherichia coli and Salmonella. Cellular and molecular biology, ASM Press, Washington, DC. 10.1126/ecosal.17 [DOI] [Google Scholar]
- 48.Bordi C, Theraulaz L, Mejean V, Jourlin-Castelli C. 2003. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol. Microbiol. 48:211–223 [DOI] [PubMed] [Google Scholar]
- 49.Seibel BA, Walsh PJ. 2002. Trimethylamine oxide accumulation in marine animals: relationship to acylglycerol storage. J. Exp. Biol. 205:297–306 [DOI] [PubMed] [Google Scholar]
- 50.Bernier SP, Surette MG. 2013. Concentration-dependent activity of antibiotics in natural environments. Front. Microbiol. 4:20. 10.3389/fmicb.2013.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. U. S. A. 99:17025–17030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175 [DOI] [PubMed] [Google Scholar]
- 53.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. 2009. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325:1380–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee HH, Molla MN, Cantor CR, Collins JJ. 2010. Bacterial charity work leads to population-wide resistance. Nature 467:82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shatalin K, Shatalina E, Mironov A, Nudler E. 2011. H2S: a universal defense against antibiotics in bacteria. Science 334:986–990 [DOI] [PubMed] [Google Scholar]
- 56.Nostro A, Cellini L, Di Giulio M, D’Arrigo M, Marino A, Blanco AR, Favaloro A, Cutroneo G, Bisignano G. 2012. Effect of alkaline pH on staphylococcal biofilm formation. APMIS 120:733–742 [DOI] [PubMed] [Google Scholar]
- 57.Piccirillo S, White MG, Murphy JC, Law DJ, Honigberg SM. 2010. The Rim101p/PacC pathway and alkaline pH regulate pattern formation in yeast colonies. Genetics 184:707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. 2011. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio 2(3):e00055-11. 10.1128/mBio.00055-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9:737–748 [DOI] [PubMed] [Google Scholar]
- 60.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:0008. 10.1038/msb41000500008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaveroche MK, Ghigo JM, d’Enfert C. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. 10.1093/nar/28.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 64.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 65.Guyer MS, Reed RR, Steitz JA, Low KB. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb. Symp. Quant. Biol. 45:135–140 [DOI] [PubMed] [Google Scholar]
- 66.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445 [DOI] [PubMed] [Google Scholar]
- 67.Herbert S, Ziebandt AK, Ohlsen K, Schafer T, Hecker M, Albrecht D, Novick R, Gotz F. 2010. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 78:2877–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burkholder PR, Giles NH., Jr. 1947. Induced biochemical mutations in Bacillus subtilis. Am. J. Bot. 34:345–348 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Synergy between ammonia and TMA. Shown is a comparison of the increased tolerance to tetracycline in E. coli exposed to volatile compounds emitted either from TG1 grown in LBTMAO (emitting ammonia and TMA) or from TG1 grown in LB (only emitting ammonia). (Upper row) E. coli recipient cells grown on 3 µg/ml tetracycline. (Lower row) E. coli recipient cells grown on 4 µg/ml tetracycline. Download