Abstract
AIM
To assess a novel method of three-dimensional (3D) co-registration of prostate cancer digital histology and in-vivo multiparametric magnetic resonance imaging (mpMRI) image sets for clinical usefulness.
MATERIAL AND METHODS
A software platform was developed to achieve 3D co- registration. This software was prospectively applied to three patients who underwent radical prostatectomy. Data comprised in-vivo mpMRI [T2-weighted, dynamic contrast-enhanced weighted images (DCE); apparent diffusion coefficient (ADC)], ex-vivo T2-weighted imaging, 3D-rebuilt pathological specimen, and digital histology. Internal landmarks from zonal anatomy served as reference points for assessing co-registration accuracy and precision.
RESULTS
Applying a method of deformable transformation based on 22 internal landmarks, a 1.6 mm accuracy was reached to align T2-weighted images and the 3D-rebuilt pathological specimen, an improvement over rigid transformation of 32% (p = 0.003). The 22 zonal anatomy landmarks were more accurately mapped using deformable transformation than rigid transformation (p = 0.0008). An automatic method based on mutual information, enabled automation of the process and to include perfusion and diffusion MRI images. Evaluation of co-registration accuracy using the volume overlap index (Dice index) met clinically relevant requirements, ranging from 0.81–0.96 for sequences tested. Ex-vivo images of the specimen did not significantly improve co-registration accuracy.
CONCLUSION
This preliminary analysis suggests that deformable transformation based on zonal anatomy landmarks is accurate in the co-registration of mpMRI and histology. Including diffusion and perfusion sequences in the same 3D space as histology is essential further clinical information. The ability to localize cancer in 3D space may improve targeting for image-guided biopsy, focal therapy, and disease quantification in surveillance protocols.
Introduction
Contemporary methods of multiparametric magnetic resonance imaging (mpMRI) of the prostate have greatly improved the ability of urologists to localize prostate cancer for detection and targeting.1,2 In evaluating the accuracy of mpMRI in disease localization, simple, reproducible methods for correlating imaging findings with histology are lacking. Such co-registration methods aim to map different image sets within the same “space.” Therefore, a geometrical transformation must be computed for optimal image alignment. These computational methods may be based on geometrical features of the image, incorporating various anatomical landmarks internal or external to the prostate, or be based on image intensity values that are generally assessed via an automated process. In addition, the transformation used to achieve image alignment can be attained using a rigid or deformable approach. The more commonly applied rigid transformation performs alignment of objects without their modification, whereas the more advanced deformable approach allows and compensates for changes in object size and/or shape. In view of these various, and possibly complex, methods available for achieving co-registration, the present pilot study was undertaken to assess a novel image-based method of automatically co-registering mpMRI with three-dimensional (3D) reconstructed prostate histology data sets.
Materials and methods
Three patients (57–73 years old) with a diagnosis of localized prostate cancer were prospectively enrolled according to an ethics committee-approved protocol. The patients’ prostate-specific antigen (PSA) values ranged from 5.1–7.7 ng/ml. Informed consent was obtained for each study subject.
In-vivo and ex-vivo mpMRI acquisition
The patients underwent a previously described protocol of mpMRI3 using a 3 T system (Magnetom Trio, Siemens Healthcare, Erlangen, Germany) equipped with a pelvic phased-array coil. MRI sequences included T2-weighted imaging (T2WI; 3 mm section thickness), dynamic contrast-enhanced (DCE) imaging, and diffusion-weighted images (DWI) acquired at 3 mm section thickness (b-values 0 and 1000 s/mm2), with in-line generation of apparent diffusion coefficient (ADC) maps.
All three patients underwent robotic-assisted prostatectomy at NYU Medical Center. Within 12 h of radical prostatectomy and prior to sectioning, ex-vivo MRI of the fresh specimen was performed, before any fixation or histopathology process.
Pathology processing
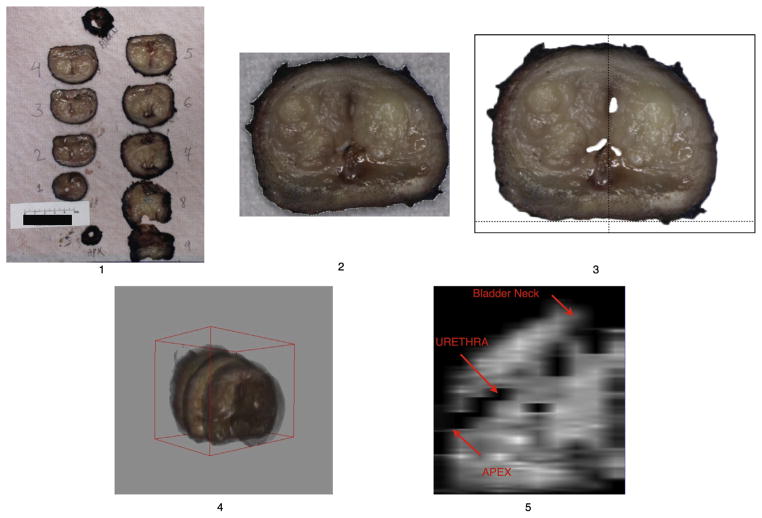
Pathology processing follows a standard institutional protocol. Fresh gross specimens were sliced perpendicular to rectal wall at regular intervals. All slices were then digitally photographed with a linear scale indication. A digital 3D surgical specimen was then rebuilt (Fig 1) using assembled digital photographs of the gross specimen slices, leading to a 3D dataset compatible for registration to MRI. The rebuilding process was fully independent of the MRI findings, with the aim of reproducing the slicing process of the specimen, which is perpendicular to the posterior capsule and distal urethral axis. The images of the gross specimen slices were assembled in a digital 3D model (Fig 1) using five steps: (1) photographs of fresh slices of the specimen were taken with a ruler included in the field of view; (2) each slice was saved as a separate digital image and loaded in Adobe Photoshop in a prefixed numeric matrix. The pixel size was based on ruler marks; (3) the slices were segmented from the background using global thresholding. Images were then rotated in two-dimensions so that the posterior capsule becomes the horizontal axis, and the urethra matches the prefixed vertical axis. Images were rotated and translated using Adobe Photoshop; (4) the aligned slices were saved in TIFF format and then imported in Image J software (V 1.44e, NIH). The slices were assigned original thickness and the entire 3D volume was exported from image J in Analyze 7.5 format; (5) to verify the continuity of the urethra, images were reconstructed in midsagittal view after importation in the dedicated platform to enable co-registration.
Figure 1.
Rebuilding of 3D fresh gross specimen after cutting in contiguous parallel slices, perpendicular to the posterior capsule. See text for detailed description of the five steps illustrated in the figure.
The 3D surgical specimen offers access to volumetry with detailed zonal anatomy of the gland. Following fixing and staining, all histopathology slides were digitized using a whole-slide scanner (Leica SCN 400, Leica Microsystems, Germany), allowing virtual whole-mount sections (Fig 2). Digital histopathology was performed using high-resolution square pixels of 25 × 25 μm, sufficient for diagnosis. The reassembled histology cross-sections [see Fig 2] were saved in Analyze 7.5 format.
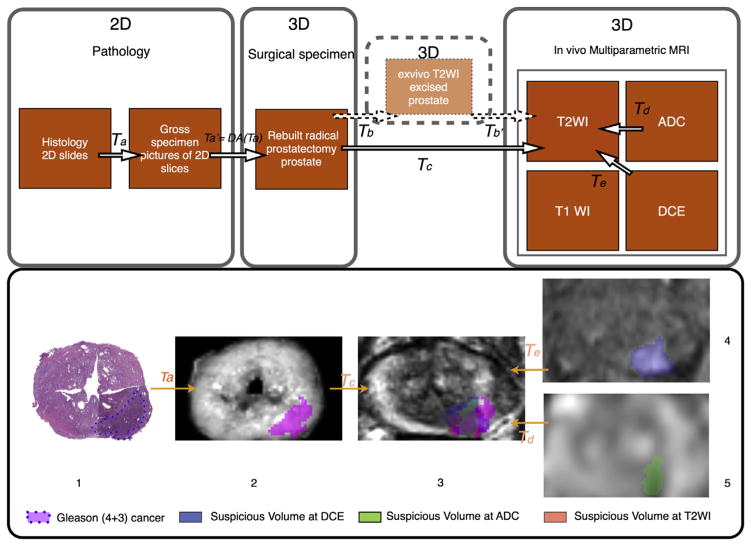
Figure 2.
Co-registration workflow applied to cancer focus. 1st row. Workflow for pathology–mpMRI co-registration in a surgical 3D space. 2nd row. 3D deformable co-registration of virtual whole-mount histology (1), fresh specimen (2), T2WI (3), perfusion (4), and diffusion (5) sequences (ADC) applied to prostate cancer Gleason score 7 (4 + 3). The T2WI is the common space. Data extracted from the 3D volume (2,3,4,5). mpMRI, multiparametric magnetic resonance imaging; T2WI, T2-weighted imaging; ADC, apparent diffusion coefficient; T1WI, T1-weighted imaging; DCE, dynamic contrast-enhanced weighted images; 2D, two-dimensional 3D, three-dimensional; T(x), transformation x.
Co-registration method
Co-registration of the MRI and histology images was carried out using a software platform with in-house code. Two methods of co-registration were tested: (1) a manually directed landmark-based method,4 and (2) an automatic, voxel-based, mutual information5,6 (MI)-based method. Each method was tested by performing rigid transformation, which aligns without volume modification, and affine transformation (computed with 12 degrees of freedom), which compensates for changes in volume and shape.
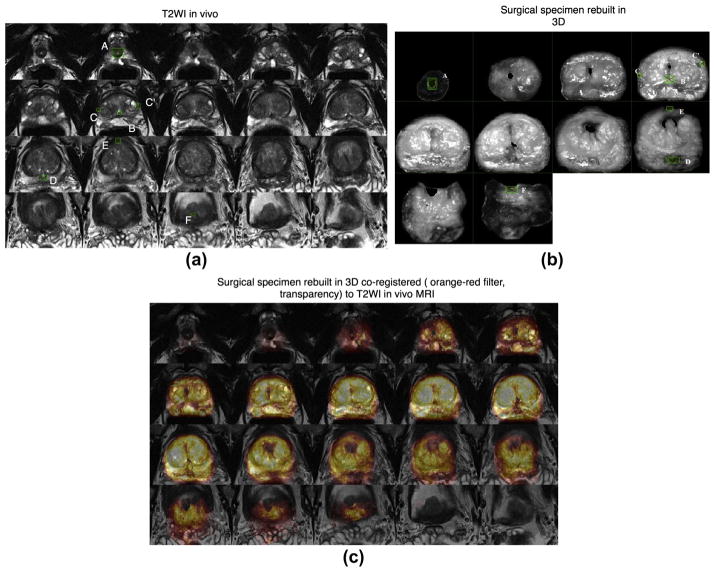
For the landmark-based method, the operator indicated a set of identical internal points of interest within each technique to be co-registered (e.g., Fig 3). Observation of prostate zonal anatomy enables detection of identical landmarks in mpMRI and the 3D rebuilt specimen. The co-registration is carried out to achieve alignment of the defined landmarks within the same space, thus realizing co-registration of the whole image. The MI-based automated method enabled co-registration in 3D between the in-vivo T2WI and the 3D rebuilt specimen based upon individual voxels of each image. The MI-based automated method performed iterative adjustment of the transformation parameters to maximize the similarity between the mpMRI and rebuilt pathology datasets in 3D. Initially, the in-vivo T2W MRI was co-registered with the 3D rebuilt specimen. Subsequently, the functional MRI sequences (ADC maps and DCE-MRI) were co-registered with the T2WI. The automatic MI-based method was employed for the functional sequences; landmark-based co-registration was not implemented owing to less precise definition of anatomical landmarks on these images. Thus, T2WI provided the final 3D space for analysis of all techniques and sequences.
Figure 3.
Zonal anatomy landmarks and examples of landmark-based co-registration in 3D. 3D landmarks are in green boxes plotted by an operator and used for transformation computation. The landmarks were A: distal point of urethra at apex; B: veru montanum; C & C′: anterior-most point of the right and left peripheral zone horn; D: ejaculatory duct; E: anterior-most point of the gland overall; F: proximal end of urethra at the prostate base, adjacent to the bladder. MRI, magnetic resonance imaging; T2WI, T2-weighted images; 3D, three dimensional. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The software enabled the operator to analyse the consistency of each transformation by overlaying the source and the target data and modifying each in real-time. The processes required to reach optimal alignment and deformation compensation are presented in Fig 2. Anatomical techniques provided the link between ex-vivo and in-vivo prostate images.
Data analysis
For the landmark-matching method, the residual error was computed as the 3D distance between the transformed source and the target zonal anatomy landmarks. The residual error was computed for each zonal anatomy landmark, which was recorded for both rigid and affine transformation. This produced a set of 22 paired data points for comparison. The overall residual root mean square (RMS) distance was computed and recorded by the software. RMS was considered to represent a good measure of utility of the technique because the zonal anatomy landmarks were clinically relevant, for example, in biopsy targeting,7 and they were derived from the whole gland, thus avoiding sampling error.
The automatic method was evaluated by the overlap between the prostate volume, delineated within the software, in each technique after alignment using rigid or affine transformation. The overlap across techniques for each method was computed as a volume overlap index (Dice index).8 Dice index is the ratio between twice the volume of gland overlap across two methods and the sum of the gland volume for those two methods. It is a similarity index that reflects volume overlap. This provides an additional measure for evaluating the success of the co-registration method, in addition to linear displacement as measured by RMS.
The two co-registration methods (MI-based and landmark-based) were evaluated to transform the 3D rebuilt specimen to T2WI with Dice index. This enabled comparison of the automatic and manual methods. Internal consistency of the automatic framework and evaluation process was tested using a phantom test. T2WI was co-registered to itself rotated by 5° in the y-axis to test rigid registration. Similarly, the ability of affine transformation to co-register T2WI to itself rotated by 5° in the y-axis was tested after a 20% size reduction in the x, y, and z directions. Paired t-tests were used to compare residual error for rigid and affine 3D transformation for each of the 22 zonal anatomy landmarks; the overall RMS for landmark-based transformation (either rigid or affine) as a measure of linear displacement error; and Dice index calculated for manual and automatic methods of volume overlap. The method was also tested both with and without the use of the fresh ex-vivo specimen MRI images, in order to evaluate the value of incorporating the ex-vivo imaging in the workflow as a method of improving co-registration accuracy. All p-values are two-sided and considered statistically significant at p < 0.05. Shapiro–Wilk’s test was used to check normality of the variable if required. Statistical analysis was performed using R software (R, version 2.14.0, CRAN, Vienne, Austria).
Results
Final histopathology revealed pT3aR0 stage in two cases and pT2cR0 stage in the remaining case. Specimen Gleason’s scores were 7 (3 + 4) in two cases and 7 (4 + 3) in one case. Prostate volumes at T2WI were 13.89, 41.39, and 73.17 cm3. In all three cases, MRI-detected cancer was observed on all sequences. For each case, all techniques were transformed to a common 3D space, as illustrated in Fig 2.
Twenty-two landmarks extracted from the zonal anatomy were used to co-register the rebuilt surgical specimen with the in-vivo T2WI. Anatomical landmarks were more precisely mapped using affine transformation than rigid transformation. In mapping the surgical specimen to T2WI, the mean residual error for zonal anatomy landmarks was significantly lower for affine than for rigid transformation [1.47 mm (range 0.59–2.88) versus 2.18 mm (range 0.55–3.64), respectively p = 0.0008, mean difference (MD) between methods of 0.71 mm, 95% confidence interval: −1.0877 to −0.329], improving precision by a median of 38%.
An average RMS error of 1.6 mm (range 1.51–1.85 mm) was observed when mapping in-vivo MRI to the surgical specimen using affine landmark-based transformation, which represented an improvement in RMS error compared with the rigid method of 2.35 mm (range 2.17–2.58 mm; p = 0.003, MD −0.73, 95% CI: −0.9039 to −0.5561), and corresponded to a 32% improvement.
Table 1 displays the results for each transformation (Ta, Tb’, Tc; referring to Fig 2), modelled by incorporating volume changes. For these transformations, the affine method performed better than the rigid method in the prostate workflow, with respective p-values of 0.01 (MD −1.2, 95% CI: −1.6303 to −0.7697), 0.03 (MD −0.56, 95% CI: −1.0362 to −0.0905) and 0.003 (MD −0.73, 95% CI: −0.9039 to −0.5561). The automatic framework (MI-based method) achieved the same performance for overlap (Dice index) as the landmark-based method (Table 1). Dice index D measurements were D = 0.89 versus D = 0.86 (p = 0.48).
Table 1.
Results for each transformation.
| Landmark-based registration
|
Mutual information-based registration
|
|||||
|---|---|---|---|---|---|---|
| Ta
|
Tc
|
Td
|
Te
|
Test
|
||
| Specimen–pathology | 3D specimen–T2WI in vivo | ADC–T2WI in vivo | DCE–T2WI in vivo | T2WI–phantom T2WI | ||
| Best transformation type | Affine | Affine | Affine | Affine/rigid | Rigid | Rigid/affine |
| Mean RMS (range) | 0.9mm (0.7–1.2) | 1.61mm (1.51–1.85) | ||||
| Mean similarity Dice index (range) | 0.86 (0.83–0.88) | 0.89 (0.84–0.97) | 0.81 (0.71–0.91) | 0.86 (0.74–0.94) | 0.96 (0.95–0.98) | |
T2WI, T2-weighted imaging; DCE, dynamic contrast enhanced weighted images; ADC, apparent diffusion coefficient; 3D, three-dimensional; RMS, root mean square distance.
The inter-sequence MRI registration showed good co-registration accuracy, with a Dice index of 0.81 for the ADC map and 0.89 for DCE (Table 1). This indicates that the automatic framework achieved alignment in the same 3D space as the functional and anatomical sequences with enough similarity to be useful.
The internal consistency of the automatic framework and evaluation process was excellent and able to reach D = 0.96 and D = 0.95, respectively, for the rigid and affine phantom studies.
The use of ex-vivo acquired MRI images for co-registration to histology was found to yield a 3.5 mm RMS error, suggesting inferior accuracy as compared to co-registration using the in-vivo mpMRI. Moreover, adding the ex-vivo step did not further decrease the error in the observed alignment between the ex-vivo and in-vivo MRI.
Discussion
In-vivo mpMRI has evolved as the most promising non-invasive method of visualizing prostate anatomy and for detecting tumours.1 Modern mpMRI, incorporating ADC and DCE, has been shown to have a significant role in localizing dominant3 tumours and potentially excluding clinically significant disease.1 In addition, ADC values obtained from DWI correlate with the Gleason score.1,9
Regarding the validation of MRI accuracy in predicting the location of cancer, no simple method for correlating MRI to the histological sections has been devised, and localization in 3D space remains a problem. This also limits the ability to implement focal therapy strategies in which it has been shown repeatedly that localization of suspicious lesions in 3D surgical space is essential.10 Adequate therapeutic targeting requires the ability to delineate the target volume in 3D, which is difficult with two-dimensional (2D) assessment alone. Moreover, working in two-dimensions assumes that each slice of the prostate will exactly match an axial MRI section. Good accuracy was demonstrated for an automated system of 3D co-registration using transformation of internal landmarks seen within each image. Although the method was applied between mpMRI sequences and histology, the technique could be easily applied to any image sequences used in diagnosis or treatment of prostate cancer.
The prostate undergoes shape changes after surgical removal and during histology processing. Specimen formalin fixation and paraffin embedding induces variable tissue shrinkage.11 Various authors have used either 2D image transformation8 or prostate moulds12 created before sectioning the specimen to overcome deformation. To overcome discordance between the MRI acquisition plane13 and the plane used in cutting the surgical specimen, ex-vivo MRI-based cutting devices13 and software for recognition of injected landmarks14 have been developed. Although these methods result in accurate transformation as measured by Dice index, prostate moulds and customized cutting devices are labour intensive, costly, and do not fully compensate for tissue deformation.
Chappelow et al.15 describe a method based on mutual information that co-registers T2WI, DCE, and ADC. However, this method is based on 2D histology and requires considerable expertise to determine the correspondence between histology and 3D MRI volume. As such, wide implementation of this method may have variable outcomes.
To the authors’ knowledge, the present co-registration method is the first to create a 3D counterpart within the same reference space between histology and both anatomical and functional sequences provided by prostate mpMRI. The method enables a true, deformable transformation and provides 1–2 mm accuracy. This is less than twice the thickness of an 18 G prostate biopsy needle and matches the diameter of laser fibres already used in focal therapy trials.16 The observed 3D Dice index (0.89) enables adequate accuracy for targeting cancers >0.18 cm3, suggesting it would enable detection of those cancers typically defined as clinically significant.1 Moreover, Ven et al.17 concluded that co-registration accuracy between another technique and MRI had to reach 1.9 mm in linear displacement to correctly co-register the smallest volume of interest detected by the ADC map, in order to achieve clinical utility. The present method fulfils this objective.
Both manual landmark-based and automatic methods for alignment of T2WI and the 3D rebuilt specimen were tested, using Dice index to measure similarity. The automatic method achieved the same performance as the manual method and is independent of the user experience in either pathology or image interpretation. This is critical to achieve objective correlation between pathology and MRI.
The proposed method is not subject to limitations relating to slicing location and cut angle variation, because the rigid transformation corrects for the orientation of the prostate specimen in 3D. Affine transformation furthermore compensates for any change in volume, slice thickness, and shape between MRI and histology. By adding only one step in the clinical workflow—taking pictures of the sliced specimen—this image-based method is widely transferable.
Internal consistency was excellent with a mean Dice index of 0.95–0.96 for the phantom tests. This indicates that variation in Dice index relies on differences in the image sequence, for instance, between T2WI and the ADC map. However, mpMRI with ADC and DCE still requires further standardization across institutions. This may affect the reliability of the presented technique in other centres. The presented workflow is nonetheless attractive because there is no need for an intermediate image representing an ex-vivo MRI, an elaborate and time-consuming process. The anatomical landmarks, extracted from McNeal zonal anatomy, achieved a bridge between in-vivo and ex-vivo techniques. These were highly discernible at T2WI. Accuracy for the distinguishing parts of the gland are essential owing to their central role in procedural guidance.7 Although the method was tested with very different prostate volumes and for numerous anatomical landmarks, more cases are needed for clinical validation.
The inter-sequence registration of MRI is decisive for quantitative assessment of functional aspects in 3D. The aim of this part of the protocol is to allow the conjunction of all relevant sequences in the same 3D space. Therefore, it is essential to be able to co-register both ADC and DCE, as these each represent different tissue properties.18 The co-registration method is critical considering prostate motion,19 especially related to rectal peristalsis. Good accuracy was achieved in this domain, with a Dice index of 0.81 for ADC and 0.89 for DCE. Rigid registration was sufficient to correct motion and, therefore, affine transformation was not required (Table 1). Functional sequences were then correctly mapped to anatomical sequences in the same 3D space, allowing objective comparison between these. As shown previously,20 co-registration of different sequences can improve cancer detection. As some tumours are primarily detected on functional sequences without clear anatomical correlation at T2WI, greater accuracy in targeting can be achieved through simultaneous registration of sequences in 3D.
An immediate clinical use of the present method could be to facilitate achieving efficient, yet accurate, correlation between MRI and pathological findings, which is critical for quality assurance in any prostate MRI program and may assist in the education of radiologists and clinicians involved in MRI interpretation. For instance, the present technique may be used to more reliably investigate discrepancies, in terms of both false-positive and false-negative interpretations, between MRI and pathology. In addition, the present preliminary work suggests that any anatomical changes to the prostate induced by the prostatectomy procedure itself may be compensated and fit to the MRI images by deformable transformation. Also, further studies may explore a potential role of the described algorithm in performing co-registration of prostate images obtained before and after ablative therapies, which can cause gross anatomical deformation of the prostate; the present co-registration method may impact the reliability of follow-up imaging. Finally, a natural extrapolation of the work will be the co-registration of diagnostic sequences in 3D for procedure guidance. The technique requires limited preprocessing, making it widely transferable to a number of therapeutic strategies.
In conclusion, the present workflow for 3D co-registration between preoperative prostate MRI and pathology produced clinically acceptable accuracy in terms of zonal anatomy. The system is able to account for changes in the ex-vivo specimen using non-rigid (affine) transformation. The use of an accurate 3D automatic method could offer greater accuracy and reproducibility than existing 2D methods, suggesting the potential for wide implementation. The present findings will be validated in larger datasets and implemented into clinical diagnostic and therapeutic research workflows.
Acknowledgments
The authors acknowledge the support of The Joseph and Diane Steinberg Charitable Trust and the Grant 1UL1RR029893 from the National Center for Research Resources, National Institutes of Health. C.O., H.R., A.M., F.M.D., J.M., and A.B.R. have nothing to disclose. S.S.T. is a consultant for Eigen, consultant and scientific investigator for GTX, scientific investigator for Steba Biotech, speaker for Janssen, and receives royalties from Elsevier.
Footnotes
No direct financial conflict of interest is identified.
References
- 1.Dickinson L, Ahmed HU, Allen C, et al. Magnetic resonance imaging for the detection, localisation, and characterisation of prostate cancer: recommendations from a European consensus meeting. Eur Urol. 2011;59:477–94. doi: 10.1016/j.eururo.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Hoeks CMA, Barentsz JO, Hambrock T, et al. Prostate cancer: multi-parametric MR imaging for detection, localization, and staging. Radiology. 2011;261:46–66. doi: 10.1148/radiol.11091822. [DOI] [PubMed] [Google Scholar]
- 3.Rosenkrantz AB, Deng F-M, Kim S, et al. Prostate cancer: multi-parametric MRI for index lesion localization–a multiple-reader study. AJR Am J Roentgenol. 2012;199:830–7. doi: 10.2214/AJR.11.8446. [DOI] [PubMed] [Google Scholar]
- 4.Fitzpatrick JM, West JB, Maurer CR., Jr Predicting error in rigid-body point-based registration. IEEE Trans Med Imaging. 1998;17:694–702. doi: 10.1109/42.736021. [DOI] [PubMed] [Google Scholar]
- 5.Wells WM, 3rd, Viola P, Atsumi H, et al. Multi-modal volume registration by maximization of mutual information. Med Image Anal. 1996;1:35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- 6.Maes F, Collignon A, Vandermeulen D, et al. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16:187–98. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 7.Ouzzane A, Puech P, Lemaitre L, et al. Combined multiparametric MRI and targeted biopsies improve anterior prostate cancer detection, staging, and grading. Urology. 2011;78:1356–62. doi: 10.1016/j.urology.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 8.Mazaheri Y, Bokacheva L, Kroon D-J, et al. Semi-automatic deformable registration of prostate MR images to pathological slices. J Magn Reson Imaging. 2010;32:1149–57. doi: 10.1002/jmri.22347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hambrock T, Somford DM, Huisman HJ, et al. Relationship between apparent diffusion coefficients at 3. 0-T MR imaging and Gleason grade in peripheral zone prostate cancer. Radiology. 2011;259:453–61. doi: 10.1148/radiol.11091409. [DOI] [PubMed] [Google Scholar]
- 10.Ukimura O, Gill IS. Reply from authors re: Mark Emberton. Tissue preservation may offer a harm-reduction strategy for men with early prostate cancer. Eur Urol. 2012;62:66–67. doi: 10.1016/j.eururo.2012.03.043. [DOI] [PubMed] [Google Scholar]
- 11.Schned AR, Wheeler KJ, Hodorowski CA, et al. Tissue-shrinkage correction factor in the calculation of prostate cancer volume. Am J Surg Pathol. 1996;20:1501–6. doi: 10.1097/00000478-199612000-00009. [DOI] [PubMed] [Google Scholar]
- 12.Trivedi H, Turkbey B, Rastinehad AR, et al. Use of patient-specific MRI-based prostate mold for validation of multiparametric MRI in localization of prostate cancer. Urology. 2012;79:233–9. doi: 10.1016/j.urology.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimm SY, Tarin TV, Lee JH, et al. Methods for registration of magnetic resonance images of ex vivo prostate specimens with histology. J Magn Reson Imaging. 2012;36:206–12. doi: 10.1002/jmri.23614. [DOI] [PubMed] [Google Scholar]
- 14.Ward AD, Crukley C, McKenzie CA, et al. Prostate: registration of digital histopathologic images to in vivo MR images acquired by using endorectal receive coil. Radiology. 2012;263:856–64. doi: 10.1148/radiol.12102294. [DOI] [PubMed] [Google Scholar]
- 15.Chappelow J, Bloch BN, Rofsky N, et al. Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information. Med Phys. 2011;38:2005. doi: 10.1118/1.3560879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenkrantz AB, Scionti SM, Mendrinos S, et al. Role of MRI in minimally invasive focal ablative therapy for prostate cancer. AJR Am J Roentgenol. 2011;197:W90–6. doi: 10.2214/AJR.10.5946. [DOI] [PubMed] [Google Scholar]
- 17.Ven WJM, Hulsbergen-van de Kaa CA, Hambrock T, et al. Simulated required accuracy of image registration tools for targeting high-grade cancer components with prostate biopsies. Eur Radiol. 2013;23:1401–7. doi: 10.1007/s00330-012-2701-1. [DOI] [PubMed] [Google Scholar]
- 18.Rosenkrantz AB, Mendrinos S, Babb JS, et al. Prostate cancer foci detected on multiparametric magnetic resonance imaging are histologically distinct from those not detected. J Urol. 2012;187:2032–8. doi: 10.1016/j.juro.2012.01.074. [DOI] [PubMed] [Google Scholar]
- 19.Van Herk M, Bruce A, Kroes AP, et al. Quantification of organ motion during conformal radiotherapy of the prostate by three dimensional image registration. Int J Radiat Oncol Biol Phys. 1995;33:1311–20. doi: 10.1016/0360-3016(95)00116-6. [DOI] [PubMed] [Google Scholar]
- 20.Rosenkrantz AB, Mannelli L, Kong X, et al. Prostate cancer: utility of fusion of T2-weighted and high b-value diffusion-weighted images for peripheral zone tumor detection and localization. J Magn Reson Imaging. 2011;34:95–100. doi: 10.1002/jmri.22598. [DOI] [PubMed] [Google Scholar]