Abstract
Asthma is characterized by lung inflammation caused by complex interaction between the immune system and environmental factors such as allergens and inorganic pollutants. Recent research in this field is focused on discovering new biomarkers associated with asthma pathogenesis. This review illustrates updated research associating biomarkers of allergic asthma and their potential use in systems biology of the disease. We focus on biomolecules with altered expression, which may serve as inflammatory, diagnostic and therapeutic biomarkers of asthma discovered in human or experimental asthma model using genomic, proteomic and epigenomic approaches for gene and protein expression profiling. These include high-throughput technologies such as state of the art microarray and proteomics Mass Spectrometry (MS) platforms. Emerging concepts of molecular interactions and pathways may provide new insights in searching potential clinical biomarkers. We summarized certain pathways with significant linkage to asthma pathophysiology by analyzing the compiled biomarkers. Systems approaches with this data can identify the regulating networks, which will eventually identify the key biomarkers to be used for diagnostics and drug discovery.
Keywords: allergic asthma, biomarker, DAAB, TH-2 cytokines and ROS pathway
Introduction
Asthma is a chronic immunological disorder of lung characterized by reversible airway obstruction, airway inflammation and increased airway hyperresponsiveness in response to provocative challenge. Physiological changes of the disease include the accumulation of inflammatory cells, especially the eosinophils, goblet cell metaplasia of lung epithelium with a mucus secreting phenotype (Laitinen et al., 1985). The worldwide incidence rate of asthma has been estimated to be 2.65 to 4/1000 per year and is more common among children with age less than 5 years where it ranges from 8.1 to 14/1000 per year (Gergen and Weiss, 1995). This case study also says that according to report presented by National Health and Nutrition Examination Survey (NHANES-2), this prevalence is higher in African-Americans (12.27) than in Caucasian (10.47) respectively. In Asia, adult asthma prevalence rate ranges from 3.6% in Hongkong, 2.4% in India, 0.1% in Singapore, 2.4% in Taiwan, and 2.91% in Thailand (Subbarao et al., 2009). Thus, asthma is arguably a major health problem worldwide deteriorating the quality of life of individuals affected and places a burden on their family and even the society. Indirect losses are due to disability, absenteeism and health care management. Focus of the present asthma biomarkers has been in the risk assessment before diagnosis, to determine the stage, grade of the disease during diagnosis and monitoring therapy or recurrent disease in the later stage of treatment. The biomolecules that undergo cellular, biochemical or molecular alterations in asthma patients vs. healthy subjects that are measurable in biological samples such as Broncho alveolar lavage Fluid (BALF), Nasal lavage fluid (NLF), blood or lung tissues may be considered as asthma biomarkers. These biomarkers are used for disease diagnosis and prognosis. A few native proteins that are targets for “hit” by a drug to achieve desirable therapeutic effects are another class of biomolecules which are known as “drug targets.” There has been a continuous quest for developing diagnostic biomarker to differentiate “allergic asthma” from other pulmonary inflammations and also to develop more biologic drugs by targeting biomolecules playing a key role in regulating asthma pathogenesis which may be more effective than traditional chemical drugs such as steroids (Murugan et al., 2009). Current research is focused on identifying key regulators and molecular pathways, which are associated in asthma pathogenesis. Systems approaches including genomics, proteomics, epigenomics and further integrating these attempts provide deeper understanding of the disease prognosis (Strimbu and Tavel, 2010). In this mini review, we gave a brief overview of different systems level approaches studied related to asthma biomarkers and we further focused on pathways, biological processes and molecular functions of these classes of biomarkers.
Molecular biomarkers in allergic asthma
Genomic approach
Genomic studies have reported large number of candidate biomarkers through both high and low throughput techniques. Experiments were done on human, mouse, monkey and rat model systems by comparing the expression of genes through challenging them with inhalant allergens and monitoring at different time intervals or by using resistant and susceptible strains of animals. Microarray based experiments reported hundreds of differentially expressed genes and hence plethora of information. Different samples like bronchial epithelial cells, eosinophils, CD4+ T-cells, mouse lung tissues have been employed in the experimental designs. The genes which showed significant differential expression were found to be linked with airway remodeling, production of mucus, macrophages and shifting the immune response toward Th2 phenotype thus enhancing asthma exacerbation (Laprise et al., 2004; Woodruff et al., 2007; Siddiqui et al., 2013). In most microarray experiments the differentially expressed genes were further validated either by RT-PCR or western blot. Genome Wide Association Study and Candidate gene approach have identified several regions on human chromosome which are linked to asthma phenotype. Nucleotide substitution in promoter region and ORF of IL4 receptor, IL13, HLA-II alleles, RANTES and CC-chemokine ligands were found to be strongly associated with asthma (Toda and Ono, 2002). We have compiled fifteen biomarkers from Database of Allergy and Asthma Biomarkers (DAAB)1 having more than two citations and listed in Table 1, out of which 11 were obtained from genomics.
Table 1.
List of asthma biomarkers cited in two or more times in Database of Allergy and Asthma Biomarkers (DAAB).
| Gene Symbol | Name of the genes/proteins | Sample | Organism | Approach | References |
|---|---|---|---|---|---|
| ARG1 | Arginase 1 | BAL macrophages, BAL Fluid | Mouse, human | GHL, PHL | Siddiqui et al., 2013 [GH] |
| Wu et al., 2005 [PH] | |||||
| Torrone et al., 2012 [PH | |||||
| Cloots et al., 2013 [GL] | |||||
| North et al., 2009 [PL] | |||||
| BPIFA1 | Palate lung nasal epithelial clone | BALFluid and nasal lavage fluid | Human | PHL | Wu et al., 2005 [PH] |
| Ghafouri et al., 2006 [PH] | |||||
| Chu et al., 2007 [PL] | |||||
| CPA3 | Carboxypeptidase A3 | Airway epithelial cells, bronchoscopy tissue sample | Human, mouse | GHL | Woodruff et al., 2007 [GH] |
| Laprise et al., 2004 [GH] | |||||
| Balzar et al., 2011 [GL] | |||||
| CCL8 | Chemokine (C-C motif) ligand 8 | Left lung tissue, BAL macrophages | Mouse | GHL | Park et al., 2008 [GH] |
| Siddiqui et al., 2013 [GH] | |||||
| Fu et al., 2013 [GL] | |||||
| Chi3l3 | Chitinase 3-like3 | BALFluid | Mouse | PHL | Greenlee et al., 2006 [PH] |
| Zhao et al., 2005 [PH] | |||||
| Louten et al., 2012 [PL] | |||||
| Chi3l4 | Chitinase 3-like 4 | BAL macrophages, BAL Fluid, | Human, mouse | GHL, PHL | Siddiqui et al., 2013 [GH] |
| Webb et al., 2001 [GL] | |||||
| Greenlee et al., 2006 [PH] | |||||
| Zhao et al., 2005 [PH] | |||||
| Louten et al., 2012 [PL] | |||||
| CLCA3 | Calcium activated chloride channel -3 | Airway epithelial cells, left lung tissue | Mouse | GHL | Woodruff et al., 2007 [GH] |
| Park et al., 2008 [GH] | |||||
| Zhou et al., 2001 [GL] | |||||
| Cxcl15 | Chemokine (C-X-C motif) ligand 15 | BAL Fluid | Mouse | PH | Greenlee et al., 2006 [PH] |
| Zhao et al., 2005 [PH] | |||||
| IL10 | Interleukin 10 | Lung tissue, CD4+T Cell | Mouse, human | rGHL | López et al., 2011 [GH] |
| Hansel et al., 2008 [GH] | |||||
| Lyon et al., 2004 [GL] | |||||
| IL13 | Interleukin 13 | CD4+T Cell, | Human | GHL, EL | Hansel et al., 2008 [GH] |
| Durham et al., 2011 [GH] | |||||
| Kanoh et al., 2011 [GL] | |||||
| MUC5AC | Mucin 5AC | Bronchoscopy tissue sample, Left lung tissue | Mouse | GHL | Laprise et al., 2004 [GH] |
| Park et al., 2008 [GH] | |||||
| Ordonez et al., 2001 [GL] | |||||
| NOS2A | Nitric oxide synthase | Bronchoscopy tissue sample | Mouse | GHL, EL | Laprise et al., 2004 [GH] |
| Torrone et al., 2012 [E] | |||||
| Pascual et al., 2008 [GL] | |||||
| Retnla | Resistin like alpha | Lung eosinophil, BAL macrophage | Mouse | GHL | Siddiqui et al., 2013 [GH] |
| Tumes et al., 2009 [GH] | |||||
| Doherty et al., 2012 [GL] | |||||
| SERPINB | Serpin peptidase inhibitor, clade B | Bronchoscopy tissue sample, airway epithelial cells | Human, mouse | GHL | Woodruff et al., 2007 [GH] |
| Laprise et al., 2004 [GH] | |||||
| Karaaslan et al., 2012 [GL] | |||||
| S100A9 | Calcium binding protein A9 | CD3+T cell | Human | PHL | Wu et al., 2005 [PH] |
| Jeong et al., 2007 [PH] | |||||
| Lee et al., 2013 [PL] |
G, Genomics; P, Proteomics; E, Epigenetics; BAL, Broncho alveolar lavage; H, High-throughput; L, Low-throughput.
In genomic studies of asthma several genes have been found to be significantly induced, of which some significant biomarkers are Chemokine ligands (CCL8, CCL5, CCL11, and CCL24), SERPINs (SERPINB2, SERPINB4, and SERPINA1) and CarboxypeptidaseA3. These three genes have not been studied earlier in detail however they have the potential of being used as asthma biomarkers. Chemokine ligands are potent attractants of Th2 lymphocytes at the site of lung inflammation in atopic asthma (Lukacs, 2001). SERPINS are members serine protease inhibitors family which inhibit neutrophil protease cathepsin G and mast cell chymase and protects the lower respiratory tract from damage caused by proteolytic enzymes. Thus, it can be used as potent diagnostic marker of asthma attack (Zou et al., 2002). Carboxypeptidase A3 is an asthma associated protease identified in lung epithelium and is a significant mast cell marker and was found to be upregulated in 42 non-smoking asthma patients (Woodruff et al., 2007). Retnla, also known as Fizz (found in inflammatory zone) protein is an inducible product of bronchial epithelial cell. This is considered as a marker of alternatively activated macrophages and highly polarized Th2 responses. In Retnla deficient mice the severity of atopic response is increased dramatically, whereas the IL13 response is suppressed by Retnla in airway hyper-responsiveness (Pesce et al., 2009). NOS2A is a gene that encodes inducible nitric oxide synthase, iNOS which produce nitric oxide (NO) from T lymphocytes in response to proinflammatory cytokines in an asthma model (Ricciardolo et al., 2004). This NO assists in the development of reactive nitrogen species such as peroxynitrites leading to cellular injury in the airways (Gabazza et al., 2003). NOS2A was found to be upregulated in bronchial biopsies in a microarray study (Laprise et al., 2004) and a (CCTTT)n polymorphism in the promoter region was associated with asthma phenotype studied in White population (Pascual et al., 2008) and some SNP's were found on asthmatic children having Latino and Caucasian ancestry (Islam et al., 2010). These genes together with other mediators contribute to epithelial cell activation and dysfunction (Dougherty et al., 2010).
Proteomic approach
Proteomic approaches are widely used to identify the expression level and modification of proteins to understand the pathophysiology of asthma. Proteomic signatures of lung parenchyma, BAL fluid, Immune cells (CD3+T cells or CD4+ T cells) from human or animal model have been used in different studies after experimental allergen challenge or after natural exposure to inhalant allergens. The advancement of proteomic techniques from earlier 2D gel based approach to recently more advanced LC-MS/MS based analysis resulted in precise identification of candidate proteins involved in asthma inflammation. In Table 1 we have listed six proteins identified in asthma proteomics studies, which can be analyzed in more detail to use them as clinical biomarkers. Similarly in asthma proteomics, a number of protein biomarkers have been identified, three of these potential biomarkers include AMcase (Chia, Chi3l3, Chi3l4, Chi3l1, and ChiT1), Calcium binding protein (S100A8 and S100A9), and Arginase (Arg1 and Arg2). These three proteins and their corresponding genes need further investigation at system level to reveal their use as potential diagnostic biomarkers.
AMcases are human chitinases induced via Th2 specific IL13 mediated pathway in aeroallergen challenged lung epithelium and macrophages as means of host defense. Th2 inflammation in asthma can be improved by targeted neutralization of these human chitinases (Zhu et al., 2004). A K(Lys)17R(Arg) polymorphism was identified in AMcase gene by genotyping study conducted on 322 pediatric asthma patients at University of Berlin and Freiburg (Bierbaum et al., 2005). Chi3l3 (Ym1) and Chi3l4 (Ym2) are other non-chitinolytic chitin binding proteins, have close linkage with asthma. Certain corticosteroids and leukotrienes receptor antagonist were shown to suppress the elevated pulmonary level of this protein (Zhu et al., 2004).
Calcium binding protein (S100A9/A8) form complex and inhibits macrophage activation and immunoglobulin synthesis by lymphocytes. Its homodimer also acts as a chemotactic agent for leukocytes and has pro-inflammatory activity on endothelial cell and inflammatory cells (Zhou et al., 2001). It is found in neutrophil cytoplasm and released upon cell activation (Cookson, 2002). This protein was found to be highly upregulated in endotoxin mediated response in non-smoking population challenged with endotoxin (Michel et al., 2013).
In asthmatic lung, Arginase expression is increased via Th2-induced, STAT6-dependent mechanism (Zimmermann et al., 2003). This affects arginine metabolism, and contribute to asthma pathogenesis through inhibition of NO generation and alterations of cell growth and collagen deposition (Shi et al., 2001). Association between four SNP's in this gene and atopic asthma were identified by genotyping 433 asthmatic case-parent triads in a public hospital of Mexican city (Huiling et al., 2006).
BPIFA1 (also known as SPLUNC), is highly expressed in the upper airways and nasopharyngeal regions and thought to be involved in inflammatory responses to irritants in the upper airways (Barnes et al., 2008). A Sialylated form of BPIFA1 was observed as post translational modification and was identified as being predominant in nasal lavage fluid (NLF) of allergy rhinitis patients (Ghafouri et al., 2006).
Epigenomic approach
Epigenomics has emerged as a promising field, and have addressed the gaps in our current understanding of the interaction between nature and nurture in the development of asthma. Epigenetic modification can alter the DNA structure (by methylation, acetylation), the chromatin structure (by altering the Scaffolding protein) and by small non-coding RNAs. It was found that reduced Histone Deacetylase (HDAC) activity and increased Histone acetyl transferase (HAT) activity jointly promotes the expression of multiple inflammatory genes associated with asthma, however inhaled steroids reduce HAT activity to the normal level (Ito et al., 2002). External stimuli such as allergen exposure, cigarette smoke, traffic exhaust and folate rich diet cause methylation mediated silencing of genes like IFNγ, Fox-P3, IL2, iNOS and hypomethylation mediated activation of genes like IL6, IL4, IL8, and Acyl CoA thus increasing the Th2 phenotype assisting in the development of asthma (Durham et al., 2011). Usually IFN-γ and FOX-P3 undergo H4 acetylation and demethylation mediated activation to prevent post natal asthma and in-utero atopicity, respectively (Lovinsky-Desir and Miller, 2012). In the promoter region and other cis-acting element of two important Th2 cytokines like IL4 and IL13 demethylation causes recruitment of STAT6 and GATA3 thereby enhancing their expression (Miller and Ho, 2008). In addition to that small non-coding RNA plays a crucial role in fine epigenetic tuning of genes which are key factors in asthma pathophysiology (Durham et al., 2011). These include let-7, miR-9, miR-21, miR-125, miR-146a, miR-147, and miR-155. For example let-7 families of micro RNA and mi R-155 are found to inhibit expression of IL 13. This miRNA was found to block the IL13 R alpha 1 and ultimately lower the expression of STAT 6 thus controlling the Th2/Th1 balance in macrophages (Kumar et al., 2011 and Martinez-Nunez et al., 2011). An overexpression of miR21 and an underexpression of miR1 were demonstrated in IL-13 induced transgenic mice. This miR-21 was also found to control expression of IL12, a molecule responsible for Th2 mediated cellular response (Lu et al., 2009). A G/C polymorphism in miRNA146a gene locus resulted in a functional variant that in turn can significantly modulate expression of genes such as TNF-α, IL-6, Cox-2, iNOS, and RANTES that are closely linked with asthma pathophysiology (Jiménez-Morales et al., 2012). This polymorphism was found to have statistically significant association with a pediatric Mexican cohort.
Integrated approaches
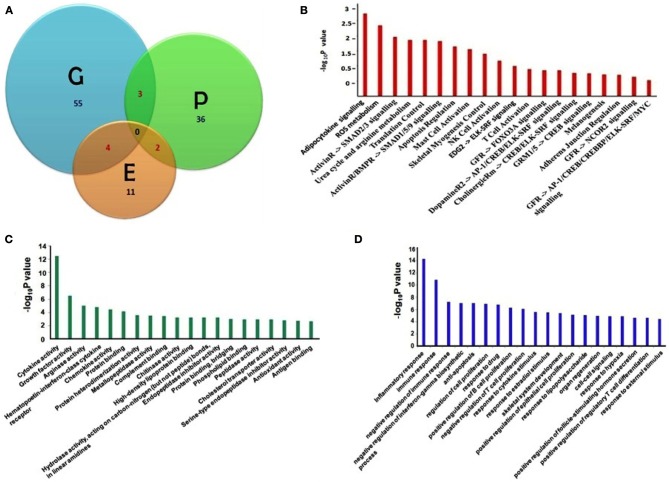
We have compiled the asthma biomarkers from different approaches including genomics, proteomics and epigenetics and have found little overlap amongst them as shown in Figure 1A. Detailed molecular information of all asthma related biomarkers are stored in DAAB. All the genes compiled from the high-throughput experiments have significant value (p = 0.05) of fold change, validated further by low-throughput techniques such as PCR, blotting and hold significantly close association with asthma pathophysiology.
Figure 1.
(A) Venn diagram showing asthma biomarkers identified in three different approaches of Genomics [G], Proteomics [P] and Epigenetic [E] studies with overlaps among the intersects. (B) Significant pathways; (C) enriched gene ontology molecular functions, and (D) biological function terms are listed which are linked with asthma biomarkers (Pathway and Gene Ontology analyses were done using Pathway studio 7.12, Ariadane Genomics, Rockville, MD, USA). Pathways are significant where (−log10P) ≥ 1.3 (0.05% significance).
Furthermore, we have listed fifteen genes in Table 1, which have been cited for two or more times in DAAB database. Asthma is dependent on many factors and thus it develops as a consequence of crosstalk among different pathways. Thus, we analyzed all the genes in our dataset compiled from several literatures in order to identify the pathways containing these biomarkers. Figure 1B shows cytokine pathways, ROS metabolism, NO metabolism and certain other metabolic pathways were significantly enriched (Detailed information of Figure 1B is shown in Table A1). In addition, Gene ontology of the biomarkers is shown in Figures 1C,D (Detailed information of GO terms are shown in Tables A2, A3). Cytokine activity, growth factor activity and Arginase activity were found to be significantly enriched in molecular function analysis. With respect to biological process inflammatory response, immune response and cell proliferation were found to be considerably predominating.
The most significant pathway triggering asthma has been the adipocyte signaling pathway. A few significant genes such as ACSL3, IL13, IL9, IL4, IL2, IL10, IFNA1, SOCS1, PON1, APOB, SOCS3, SCD, and NR1D1 were found to be the component of this pathway and associated with asthma pathogenesis (Tilg and Moschen, 2006; Diego et al., 2012). Adipokine or adipocytokine are cytokines secreted by the adipose tissues. These include Th2 cytokines and chemokines such as MCP1, RANTES, which are potent attractants of mast cells. There are also several clinical observations suggesting the role of obesity with asthma and one of the major conclusions so far has been the action of adipocytes derived cytokines which inhibit the activity of T-regs thus decreasing the tolerance (Theoharides et al., 2008). Cytokines such as TNFα, IL6 secreted by the adipocytes are important mediators of asthma. These molecules also affect vascular function by modulating nitric oxide and superoxide release. Some molecules such as leptin, adiponectin are the most abundantly expressed adipocytokines and are involved in classical cytokine pathway thus showing an asthmatic phenotype (Guzik et al., 2006).
Another significant pathway has been the ROS signaling pathway which is characterized by production of free radicals from molecular oxygen due to recruitment of activated inflammatory cells and associated with mitochondrial dysfunction that result in variety of physiological changes including increased airway reactivity, tissue injury and mucus production (Zuo and Clanton, 2005). Presently certain metabolites such as malondialdehyde, 8-isoprostane, exhaled NO, thiobarbituric acid are used as markers to measure the disease severity in sputum or exhaled air (Zuo et al., 2013). Several genes including MPO, PRDX6, SOD1, and CYBB as molecules involved in asthmatic responses and linked to ROS generation and hold the potential of using as biomarkers.
An additional significant pathway uncovered has been the Urea cycle and arginine metabolism. iNOS, ARG1, and ARG2 belong to this pathway and have also been found to be induced significantly in several genomic, proteomic, and epigenetic studies (North et al., 2009; Breton et al., 2011; Cloots et al., 2013). In asthmatic airway inducible NOS in inflammatory cells catalyses the production of NO from L-arginine, which results in the formation of reactive nitrogen species (RNS) that alters protein function by nitration of tyrosine residues thereby mediating inflammation and injury. In asthmatics upregulation of Arginase limits the availability of L-arg to iNOS thus generating peroxynitrite and concomitant nitration of proteins. It also enhances the level of L-ornithine which promotes airway remodeling by collagen deposition and excess cell proliferation (Ghosh and Erzurum, 2011).
Conclusion
In the last few decades efforts to understand the pathophysiogy of allergic asthma has been intensified to a great extent because of increased mortality and morbidity. The aim of the present review is to focus on genes or their products which can be used as biomarker for allergic asthma. Occurrence of allergic asthma involves multiple genes, environmental factors and epigenetic mechanisms. Presently the potential difficulties to diagnose this disease are due to (i) remarkable overlap in symptoms of other pulmonary diseases, (ii) high interindividual and interpopulation variation at genetic level leads to changes in the uniformity of molecular marker, and (iii) absence of discriminative molecular markers, specific to atopic asthma, since most of the biomarkers currently used or in clinical trial are indicative of asthmatic inflammation irrespective of atopic background. Some of the common features of asthma exacerbation are eosinophilic inflammation, collagenitis, mucus deposition and extracellular matrix formation. However, these are common characteristics of other lung inflammations such as Chronic Obstructive Pulmonary Disease (COPD) or, non-allergic asthma. Therefore, the genes involved in these phenotypes may also be induced in all kinds of lung inflammations. To develop diagnostic markers exclusively for “allergic asthma” it is necessary to identify upstream components of the molecular pathways initiated immediately after allergen sensitization. Researchers can use these biomarkers for screening and risk assessment before the disease assumes severity by (i) identifying polymorphisms in wide population and (ii) correlating them with the alteration of signaling pathways that ultimately lead to allergic asthma. Since application of single biomarker approach to asthma research may not be realistic, newly identified biomarkers can be integrated in a multidimensional way to strengthen the treatment. Our mini review is focused on biomarker discovery by systemic approach using high-throughput “OMICS” platforms including genomics, proteomics and epigenetics and further some of them are well-studied in low-throughput experiments. Application of systems biology as a discipline provides a way to investigate the pathophysiology of asthma by giving a closer look to the system components, its dynamics and response to any kind of perturbation in the population level. Systemic approaches may emerge as a promising strategy to zoom into the global mechanism and identify features specific to asthma for developing better diagnostics and therapeutics.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tanmoy Jana for retrieving data from DAAB. Gaurab Sircar and Bodhisattwa Saha acknowledge the financial support from Council of Scientific and Industrial Research (CSIR) and University Grant Commission (UGC) respectively. Sudipto Saha acknowledges the financial support of Department of Biotechnology (DBT)- Ramalingaswami Re-entry Fellowship (No. BT/RLF/Re-entry/11/2011). We also thank the support of Bioinformatics Centre at Bose Institute.
Appendix
Table A1.
The list of pathways that play key role in asthma pathogenesis, as evident from the biomarkers identified by genomics, proteomics and epigenomics approaches.
| Name of the pathway | Total entities | Expanded of entities | Overlap | Percent overlap | Overlapping entities | p-value | (−logP) |
|---|---|---|---|---|---|---|---|
| Adipocytokine signaling | 52 | 780 | 13 | 1 | ACSL3, IL13, IL9, IL4, IL2, IL10, IFNA1, SOCS1, PON1, APOB, SOCS3, SCD, NR1D1 | 0.001809 | 2.742605 |
| ROS metabolism | 43 | 74 | 3 | 4 | PRDX6, SOD1, CYBB | 0.004493 | 2.347436 |
| ActivinR -> SMAD2/3 signaling | 23 | 23 | 2 | 8 | INHBA, INHA | 0.010913 | 1.962064 |
| Urea cycle and arginine metabolism | 86 | 110 | 3 | 2 | NOS2A, ARG1, ARG2 | 0.013536 | 1.868503 |
| Translation control | 86 | 984 | 13 | 1 | CCL11, EGFR, IL13, IL9, IL4, IL2, IL10, CCL21, IFNA1, SOCS1, VEGFC, SOCS3, GNB2 | 0.013549 | 1.86809 |
| ActivinR/BMPR -> SMAD1/5/9 signaling | 27 | 27 | 2 | 7 | INHBA, INHA | 0.014876 | 1.827505 |
| Apoptosis regulation | 69 | 613 | 9 | 1 | IL13, IL9, IL4, TGFB1, IL2, IL10, IFNA1, INHBA, INHA | 0.022869 | 1.640747 |
| Mast cell activation | 64 | 529 | 8 | 1 | PTGS2, IL13, IL9, IL4, IL2, IL10, IFNA1, ALOX15 | 0.027347 | 1.563092 |
| Skeletal myogenesis control | 70 | 569 | 8 | 1 | EGFR, TGFB1, SOCS1, INHBA, CYBB, VEGFC, INHA, SOCS3 | 0.039923 | 1.39878 |
| NK Cell Activation | 59 | 523 | 7 | 1 | IL13, IL9, IL4, IL2, IL10, IFNA1, TYROBP | 0.067085 | 1.173375 |
| EDG2 -> ELK-SRF signaling | 33 | 78 | 2 | 2 | EGFR, GNB2 | 0.102033 | 0.991259 |
| T cell activation | 81 | 1100 | 11 | 1 | SGPP1, PTGS2, IL13, IL9, IL4, IL2, IL10, CTLA4, IFNA1, ALOX15, CNN1 | 0.129288 | 0.888442 |
| GFR -> FOXO3A signaling | 7 | 94 | 2 | 2 | EGFR, VEGFC | 0.138723 | 0.857852 |
| DopamineR2 -> AP-1/CREB/ELK-SRF signaling | 47 | 95 | 2 | 2 | EGFR, GNB2 | 0.141106 | 0.850455 |
| CholinergicRm -> CREB/ELK-SRF signaling | 41 | 107 | 2 | 1 | EGFR, GNB2 | 0.170351 | 0.768655 |
| GRM1/5 -> CREB signaling | 39 | 110 | 2 | 1 | EGFR, GNB2 | 0.177823 | 0.750012 |
| Melanogenesis | 51 | 682 | 7 | 1 | INMT, CCL11, EGFR, CCL21, PRDX6, VEGFC, GNB2 | 0.190612 | 0.71985 |
| Adherens junction regulation | 41 | 692 | 7 | 1 | EGFR, TGFB1, INHBA, VEGFC, CDH11, INHA, DSP | 0.20054 | 0.697799 |
| GFR -> NCOR2 signaling | 27 | 130 | 2 | 1 | EGFR, VEGFC | 0.228743 | 0.640652 |
| GFR -> AP-1/CREB/CREBBP/ELK-SRF/MYC signaling | 50 | 156 | 2 | 1 | EGFR, VEGFC | 0.296227 | 0.528375 |
The data was generated using Pathway studio 7.1, Ariadane Genomics, Rockville, MD, USA. The column names are: Name of the pathway; Total entities; expanded entities; overlap; percent overlap; overlapping entities; p-value and −log10 p-value.
Table A2.
The list of Gene Ontology Molecular Function (GOMF) terms that are significant in asthma pathogenesis, as evident from the biomarkers identified by genomics, proteomics and epigenomics approaches.
| Name of the GOMF terms | Total entities | Expanded number of entities | Overlap | Percent overlap | Overlapping entities | p-value | (−logP) |
|---|---|---|---|---|---|---|---|
| Cytokine activity | 217 | 217 | 13 | 5 | Ccl8, Cxcl15, CCL11, IL13, IL9, IL4, IL2, IL10, CCL21, IFNA1, INHBA, INHA, SCGB3A1 | 3.30E–13 | 12.48204 |
| Growth factor activity | 198 | 198 | 8 | 4 | IL9, IL4, TGFB1, IL2, INHBA, VEGFC, INHA, TFF2 | 3.10E–07 | 6.509308 |
| Arginase activity | 2 | 2 | 2 | 100 | ARG1, ARG2 | 1.08E–05 | 4.967132 |
| Hematopoietin-interferon-class (D200-domain) cytokine receptor binding | 47 | 47 | 4 | 8 | IL13, IL9, IL4, IFNA1 | 1.78E–05 | 4.750718 |
| Chemokine activity | 56 | 56 | 4 | 7 | Ccl8, Cxcl15, CCL11, CCL21 | 3.57E–05 | 4.446842 |
| Protein binding | 7274 | 7274 | 41 | 0 | Muc5ac, Serpinb3c, Apoa1, ACSL3, Igh-6, IGHG1, PTGS2, EGFR, ORM1, IL13, IL4, TGFB1, IL2, IL10, CTLA4, NOS2A, SERPINA1, SOCS1, SOD1, ARG2, APOB, CYBB, TIMP4, FCGR2B, POSTN, FOXP3, CDH11, INHA, S100A9, SOCS3, TYROBP, VIM, TCF21, FBN1, C4BPA, AATF, SCNN1G, HSPA1B, ITIH1, LCN1, GNB2 | 7.37E–05 | 4.132694 |
| Protein heterodimerization activity | 268 | 268 | 6 | 2 | EGFR, TGFB1, INHBA, APOB, CYBB, INHA | 0.000265 | 3.576101 |
| Metallopeptidase activity | 178 | 178 | 5 | 2 | CPA4, MMP12, ADAM33, ACE, ADAM8 | 0.000314 | 3.502791 |
| Complement binding | 9 | 9 | 2 | 22 | CFB, C4BPA | 0.000383 | 3.417369 |
| Chitinase activity | 11 | 11 | 2 | 18 | Chi3l3, CHIA | 0.000582 | 3.235176 |
| High-density lipoprotein binding | 11 | 11 | 2 | 18 | Apoa1, PON1 | 0.000582 | 3.235176 |
| Hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines | 11 | 11 | 2 | 18 | ARG1,ARG2 | 0.000582 | 3.235176 |
| Endopeptidase inhibitor activity | 118 | 118 | 4 | 3 | SERPINA1, SERPINB2, CSTA, ITIH1 | 0.000639 | 3.194323 |
| Protein binding, bridging | 60 | 60 | 3 | 5 | DSP, CSTA, COL11A1 | 0.001039 | 2.983192 |
| Peptidase activity | 633 | 633 | 8 | 1 | Slpi, CPA4, MMP12, SERPINA1, ADAM33, ACE, CFB, ADAM8 | 0.001171 | 2.93128 |
| Phospholipid binding | 64 | 64 | 3 | 4 | Apoa1, PON1, APOB | 0.001254 | 2.901838 |
| Cholesterol transporter activity | 16 | 16 | 2 | 12 | Apoa1, APOB | 0.001256 | 2.901021 |
| Serine-type endopeptidase inhibitor activity | 150 | 150 | 4 | 2 | Serpinb3c, SERPINA1, SERPINB2, ITIH1 | 0.001558 | 2.807388 |
| Antioxidant activity | 20 | 20 | 2 | 10 | PRDX6, SOD1 | 0.001972 | 2.705175 |
| Antigen binding | 79 | 79 | 3 | 3 | Igh-2, Igh-6, IGHG1 | 0.002296 | 2.63899 |
The data was generated using Pathway studio 7.1, Ariadane Genomics, Rockville, MD, USA. The column names are: Name of the GOMF terms; Total entities; expanded entities; overlap; percent overlap; overlapping entities; p-value and −log10 p-value.
Table A3.
The list of Gene Ontology Biological Process (GOBP) terms that are significant in asthma pathogenesis, as evident from the biomarkers identified by genomics, proteomics and epigenomics approaches.
| Name of GOBP terms | Total entities | Expanded number of entities | Overlap | Percent overlap | Overlapping entities | p-value | (−logP) |
|---|---|---|---|---|---|---|---|
| Inflammatory response | 293 | 15 | 5 | 5 | Ccl8, Cxcl15, CCL11, PTGS2, ORM1, IL13, IL9, IL4, TGFB1, IL10, NOS2A, CCL21, CYBB, ALOX15, S100A9 | 4.54E–15 | 14.34325 |
| Immune response | 604 | 16 | 2 | 2 | LILRA6, Ccl8, Cxcl15, IGHG1, CCL11, IL13, IL9, IL4, IL2, IL10, CTLA4, CCL21, FCGR2B, CFB, C4BPA, CHIA | 1.24E–11 | 10.90786 |
| Negative regulation of immune response | 14 | 4 | 28 | 28 | TGFB1, CTLA4, FCGR2B, FOXP3 | 5.90E–08 | 7.229314 |
| Negative regulation of interferon-gamma biosynthetic process | 4 | 3 | 75 | 75 | INHBA, FOXP3, INHA | 8.79E–08 | 7.055796 |
| Anti-apoptosis | 198 | 8 | 4 | 4 | IL2, IL10, SOD1, ALOX15, SOCS3, SERPINB2, AATF, HSPA1B | 9.69E–08 | 7.013485 |
| Regulation of cell proliferation | 135 | 7 | 5 | 5 | PTGS2, EGFR, TGFB1, NOS2A, ADAM33, INHA, SCGB3A1 | 1.18E–07 | 6.929752 |
| Response to drug | 295 | 9 | 3 | 3 | Apoa1, PTGS2, MMP12, TGFB1, SOCS1, SOD1, CYBB, TIMP4, SOCS3 | 1.62E–07 | 6.791126 |
| Positive regulation of B cell proliferation | 23 | 4 | 17 | 17 | Igh-6, IL13, IL4, IL2 | 5.12E–07 | 6.290996 |
| Negative regulation of T cell proliferation | 26 | 4 | 15 | 15 | TGFB1, IL10, CTLA4, FOXP3 | 8.58E–07 | 6.066354 |
| Response to cytokine stimulus | 77 | 5 | 6 | 6 | PTGS2, SERPINA1, SOCS1, TIMP4, SOCS3 | 2.73E–06 | 5.563052 |
| Response to estradiol stimulus | 79 | 5 | 6 | 6 | PTGS2, TGFB1, ERPINA1, SOCS1, SOCS3 | 3.11E–06 | 5.50781 |
| Skeletal system development | 147 | 6 | 4 | 4 | TGFB1, INHBA, POSTN, CDH11, INHA, FBN1 | 3.98E–06 | 5.400395 |
| Positive regulation of epithelial cell proliferation | 44 | 4 | 9 | 9 | EGFR, MMP12, TGFB1, VEGFC | 7.50E–06 | 5.125108 |
| Response to lipopolysaccharide | 99 | 5 | 5 | 5 | PTGS2, SERPINA1, SOCS1, TIMP4, SOCS3 | 9.43E–06 | 5.025503 |
| Organ regeneration | 49 | 4 | 8 | 8 | Apoa1, TGFB1, SOCS1, SOCS3 | 1.16E–05 | 4.936449 |
| Cell–cell signaling | 275 | 7 | 2 | 2 | IL13, IL2, IL10, CCL21, INHBA, INHA, S100A9 | 1.35E–05 | 4.869351 |
| Response to hypoxia | 184 | 6 | 3 | 3 | TGFB1, NOS2A, SERPINA1, ACE, SOCS3, SCNN1G | 1.44E–05 | 4.842389 |
| Positive regulation of folliclE-stimulating hormone secretion | 3 | 2 | 66 | 66 | INHBA, INHA | 2.38E–05 | 4.62283 |
| Positive regulation of regulatory T cell differentiation | 3 | 2 | 66 | 66 | IL2, FOXP3 | 2.38E–05 | 4.62283 |
| Response to external stimulus | 23 | 3 | 13 | 13 | INHBA, PON1, INHA | 3.75E–05 | 4.426548 |
The data was generated using Pathway studio 7.1, Ariadane Genomics, Rockville, MD, USA. The column names are: Name of the GOBP terms; Total entities; expanded entities; overlap; percent overlap; overlapping entities; p-value and −log10 p-value.
Footnotes
References
- Balzar S., Fajt M. L., Comhair S. A., Erzurum S. C., Bleecker E., Busse W. W., et al. (2011). Mast cell phenotype, location, and activation in severe asthma: data from the severe asthma research program. Am. J. Respir. Crit. Care Med. 183, 299–309 10.1164/rccm.201002-0295OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes F. A., Bingle L., Bingle C. D. (2008). Pulmonary genomics, proteomics, and PLUNCs. Am. J. Respir. Cell Mol. Biol. 38, 377–379 10.1165/rcmb.2007-0388TR [DOI] [PubMed] [Google Scholar]
- Bierbaum S., Nickel R., Koch A., Lau S., Deichmann K. A., Wahn U., et al. (2005). Polymorphisms and haplotypes of acid mammalian chitinase are associated with bronchial asthma. Am. J. Respir. Crit. Care Med. 172, 1505 10.1164/rccm.200506-890OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C. V., Byun H.-M., Wang X., Salam M. T., Siegmund K., Gilliland F. D. (2011). DNA methylation in the arginase–nitric oxide synthase pathway is associated with exhaled nitric oxide in children with asthma. Am. J. Respir. Crit. Care Med. 184, 191 10.1164/rccm.201012-2029OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H. W., Thaikoottathil J., Rino J. G., Zhang G., Wu Q., Moss T., et al. (2007). Function and regulation of SPLUNC1 protein in Mycoplasma infection and allergic inflammation. J. Immunol. 179, 3995–4002 [DOI] [PubMed] [Google Scholar]
- Cloots R. H., Sankaranarayanan S., De Theije C. C., Poynter M. E., Terwindt E., Van Dijk P., et al. (2013). Ablation of Arg1 in hematopoietic cells improves respiratory function of lung parenchyma, but not that of larger airways or inflammation in asthmatic mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 305, L364–L376 10.1152/ajplung.00341 [DOI] [PubMed] [Google Scholar]
- Cookson W. (2002). Genetics and genomics of asthma and allergic diseases. Immunol. Rev. 1, 195–206 10.1034/j.1600-065X.2002.19015.x [DOI] [PubMed] [Google Scholar]
- Diego D., Goto S., Fahy J. V., Erle D. J., Woodruff P. G., Wheelock Å. M., et al. (2012). Network analysis identifies a putative role for the PPAR and type 1 interferon pathways in glucocorticoid actions in asthmatics. BMC Med. Genomics 5:27 10.1186/1755-8794-5-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty T. A., Khorram N., Sugimoto K., Sheppard D., Rosenthal P., Cho J. Y., et al. (2012). Alternaria induces STAT6-dependent acute airway eosinophilia and epithelial FIZZ1 expression that promotes airway fibrosis and epithelial thickness. J. Immunol. 188, 2622–2629 10.4049/jimmunol.1101632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty R. H., Sidhu S. S., Raman K., Solon M., Solberg O. D., Caughey G. H., et al. (2010). Accumulation of intraepithelial mast cells with a unique protease phenotype in Th-2 high asthma. J. Allergy Clin. Immunol. 5, 1046–1053 10.1016/j.jaci.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham A. L., Wiegman C., Adcock I. M. (2011). Epigenetics of asthma. Biochim. Biophys. Acta 11, 1103–1109 10.1016/j.bbagen.2011.03.006 [DOI] [PubMed] [Google Scholar]
- Fu J. J., Baines K. J., Wood L. G., Gibson P. G. (2013). Systemic inflammation is associated with differential gene expression and airway neutrophilia in asthma. OMICS 17, 187–199 10.1089/omi.2012.0104 [DOI] [PubMed] [Google Scholar]
- Gabazza E. C., Taguchi O., Tamaki S., Murashima S., Kobayashi H., Yasui H., et al. (2003). Role of nitric oxide in airway remodelling. Clin. Sci. 98, 291–294 10.1042/CS19990176 [DOI] [PubMed] [Google Scholar]
- Gergen P. J., Weiss K. B. (1995). Epidemiology of asthma in Asthma and Rhinitis, Chap. 3, eds Busse W. W., Holgate S. T. (Boston, MA: Blackwell Scientific Publications; ), 17–32 [Google Scholar]
- Ghafouri B., Kristina I., John L., Christer T., Mats L. (2006). Comparative proteomics of nasal fluid in seasonal allergic rhinitis. J. Proteome Res. 5, 330–338 10.1021/pr050341h [DOI] [PubMed] [Google Scholar]
- Ghosh S., Erzurum S. C. (2011). Nitric oxide metabolism in asthma pathophysiology. Biochim. Biophys. Acta 11, 1008–1016 10.1016/j.bbagen.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee K. J., Corry D. B., Engler D. A., Matsunami R. K., Tessier P., Cook R. G., et al. (2006). Proteomic identification of in vivo substrates for matrix metalloproteinases 2 and 9 reveals a mechanism for resolution of inflammation. J. Immunol. 10, 7312–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik T. J., Mangalat D., Korbut R. (2006). Adipocytokines novel link between inflammation. J. Physiol. Pharmacol. 4, 505–528 [PubMed] [Google Scholar]
- Hansel N. N., Cheadle C., Diette G. B., Wright J., Thompson K. M., Barnes K. C., et al. (2008). Analysis of CD4+ T-cell gene expression in allergic subjects using two different microarray platforms. Allergy 3, 366–369 10.1111/j.1398-9995.2007.01540.x [DOI] [PubMed] [Google Scholar]
- Huiling L., Romieu I., Sienra-Monge J.-J., Ramirez-Aguilar M., Estela del Rio-Navarro B., Kistner E. O., et al. (2006). Genetic polymorphisms in Arginase I and II and childhood asthma and atopy. J. Allergy Clin. Immunol. 117, 119–126 10.1016/j.jaci.2005.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam T., Breton C., Salam M. T., McConnell R., Wenten M., Gauderman W. J., et al. (2010). Role of inducible nitric oxide synthase in asthma risk and lung function growth during adolescence. Thorax 65, 139–145 10.1136/thx.2009.114355 [DOI] [PubMed] [Google Scholar]
- Ito K., Caramori G., Lim S., Oates T., Chung K. F., Barnes P. J., et al. (2002). Expression and activity of histone deacetylases in human asthmatic airways. Am. J. Respir. Crit. Care Med. 3, 392–396 10.1164/rccm.2110060 [DOI] [PubMed] [Google Scholar]
- Jeong H. C., Lee S. Y., Lee E. J., Jung K. H., Kang E. H., Lee S. Y., et al. (2007). Proteomic analysis of peripheral T-lymphocytes in patients with asthma. Chest 132, 489–496 10.1378/chest.06-2980 [DOI] [PubMed] [Google Scholar]
- Jiménez-Morales S., Gamboa-Becerra R., Baca V., Río-Navarro D., López-Ley D. Y., Velázquez-Cruz R., et al. (2012). MiR-146a polymorphism is associated with asthma but not with systemic lupus erythematosus and juvenile rheumatoid arthritis in Mexican patients. Tissue Antigens 4, 317–321 10.1111/j.1399-0039.2012.01929.x [DOI] [PubMed] [Google Scholar]
- Kanoh S., Tanabe T., Rubin B. K. (2011). IL-13-induced MUC5AC production and goblet cell differentiation is steroid resistant in human airway cells. Clin. Exp. Allergy 41, 1747–1756 10.1111/j.1365-2222.2011.03852.x [DOI] [PubMed] [Google Scholar]
- Karaaslan C., Birben E., Keskin O., Sahiner U., Sackesen C., Kalayci O. (2012). The role of SCCA1 in asthma related physiological events in the airway epithelium and the effect of promoter variants on asthma and gene function. Respir. Med. 107, 368–379 10.1016/j.rmed.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Kumar M., Ahmad T., Sharma A., Mabalirajan U., Kulshreshtha A., Agrawal A., et al. (2011). Let-7 microRNA–mediated regulation of IL-13 and allergic airway inflammation. J. Allergy Clin. Immunol. 5, 1077–1085 10.1016/j.jaci.2011.04.034 [DOI] [PubMed] [Google Scholar]
- Laitinen L. A., Heino M., Laitinen A., Kava T., Haahtela T. (1985). Damage of the airway epithelium and bronchial reactivity in patients with asthma. Am. Rev. Respir. Dis. 4, 599–606 [DOI] [PubMed] [Google Scholar]
- Laprise C., Sladek R., Ponton A., Bernier M.-C., Hudson T. J., Laviolette M. (2004). Functional classes of bronchial mucosa genes that are differentially expressed in asthma. BMC Genomics 1:21 10.1186/1471-2164-5-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Jang A. S., Park J. S., Kim T. H., Choi Y. S., Shin H. R., et al. (2013). Elevation of S100 calcium binding protein A9 in sputum of neutrophilic inflammation in severe uncontrolled asthma. Ann. Allergy Asthma Immunol. 111, 268–275 10.1016/j.anai.2013.06.028 [DOI] [PubMed] [Google Scholar]
- López E., Zafra M. P., Sastre B., Gámez C., Lahoz C., del Pozo V. (2011). Gene expression profiling in lungs of chronic asthmatic mice treated with galectin-3: downregulation of inflammatory and regulatory genes. Mediat. Inflamm. 2011, 823279 10.1155/2011/823279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louten J., Mattson J. D., Malinao M. C., Li Y., Emson C., Vega F., et al. (2012). Biomarkers of disease and treatment in murine and cynomolgus models of chronic asthma. Biomark. Insights 7, 87 10.4137/BMI.S9776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovinsky-Desir S., Miller R. L. (2012). Epigenetics, asthma, and allergic diseases: a review of the latest advancements. Curr. Allergy Asthma Rep. 12, 211–220 10.1007/s11882-012-0257-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T. X., Munitz A., Rothenberg M. E. (2009). MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 8, 4994–5002 10.4049/jimmunol.0803560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs N. W. (2001). Role of chemokines in the pathogenesis of asthma. Nat. Rev. Immunol. 2, 108–116 10.1038/35100503 [DOI] [PubMed] [Google Scholar]
- Lyon H., Lange C., Lake S., Silverman E. K., Randolph A. G., Kwiatkowski D., et al. (2004). IL10 gene polymorphisms are associated with asthma phenotypes in children. Genet. Epidemiol. 26, 155–165 10.1002/gepi.10298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Nunez R. T., Louafi F., Sanchez-Elsner T. (2011). The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor α1 (IL13Rα1). J. Biol. Chem. 3, 1786–1794 10.1074/jbc.M110.169367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel O., Doyen V., Leroy B., Bopp B., Dinh D. H., Corazza F., et al. (2013). Expression of calgranulin A/B heterodimer after acute inhalation of endotoxin: proteomic approach and validation. BMC Pulm. Med. 13:65 10.1186/1471-2466-13-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. L., Ho S.-M. (2008). Environmental epigenetics and asthma: current concepts and call for studies. Am. J. Respir. Crit. Care Med. 177, 567 10.1164/rccm.200710-1511PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan A., Prys-Picard C., Calhoun W. J. (2009). Biomarkers in asthma. Curr. Opin. Pulm. Med. 1, 12–18 10.1097/MCP.0b013e32831de235 [DOI] [PubMed] [Google Scholar]
- North M. L., Khanna N., Marsden P. A., Grasemann H., Scott J. A. (2009). Functionally important role for arginase 1 in the airway hyperresponsiveness of asthma. Am. J. Physiol. Lung Cell. Mol. Physiol. 296, L911–L920 10.1152/ajplung.00025.2009 [DOI] [PubMed] [Google Scholar]
- Ordonez C. L., Khashayar R., Wong H. H., Ferrando R., Wu R., Hyde D. M., et al. (2001). Mild and moderate asthma is associated with airway goblet cell hyperplasia and abnormalities in mucin gene expression. Am. J. Respir. Crit. Care Med. 163, 517–523 10.1164/ajrccm.163.2.2004039 [DOI] [PubMed] [Google Scholar]
- Park S. G., Choi J.-W., Kim H. J., Roh G. S., Jeong B., Min J. G., et al. (2008). Genome-wide profiling of antigen-induced time course expression using murine models for acute and chronic asthma. Int. Arch. Allergy Immunol. 146, 44–56 10.1159/000112502 [DOI] [PubMed] [Google Scholar]
- Pascual M., Sanz C., Isidoro-Garcia M., Davila I., Moreno E., Laffond E., et al. (2008). (CCTTT) n polymorphism of NOS2A in nasal polyposis and asthma: a case-control study. J. Investig. Allergol. Clin. Immunol. 18, 239 [PubMed] [Google Scholar]
- Pesce J. T., Ramalingam T. R., Wilson M. S., Mentink-Kane M. M., Thompson R. W., Cheever A. W., et al. (2009). Retnla (relmα/fizz1) suppresses helminth-induced Th2-type immunity. PLoS Pathog. 5:e1000393 10.1371/journal.ppat.1000393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardolo F. L., Sterk P. J., Gaston B., Folkerts G. (2004). Nitric oxide in health and disease of the respiratory system. Physiol. Rev. 84, 731–765 10.1152/physrev.00034.2003 [DOI] [PubMed] [Google Scholar]
- Shi O., Morris S. M., Zoghbi H., Porter C. W., O'Brien W. E. (2001). Generation of a mouse model for arginase II deficiency by targeted disruption of the arginase II gene. Mol. Cell. Biol. 21, 811–813 10.1128/MCB.21.3.811-813.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui S., Eric R. S., Jr., Lawrence K. S. (2013). Broncho-alveolar macrophages express chemokines associated with leukocyte migration in a mouse model of asthma. Cell. Immunol. 2, 159–169 10.1016/j.cellimm.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Strimbu K., Tavel J. A. (2010). What are biomarkers? Curr. Opin. HIV AIDS 6, 463–466 10.1097/COH.0b013e32833ed177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao P., Mandhane P. J., Sears M. R. (2009). Asthma: epidemiology, etiology and risk factors. Can. Med. Assoc. J. 181, E181–E190 10.1503/cmaj.080612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theoharides T. C., Makris M., Kalogeromitros D. (2008). Allergic inflammation and adipocytokines. Int. J. Immunopathol. Pharmacol. 1, 1–4 [DOI] [PubMed] [Google Scholar]
- Tilg H., Moschen A. R. (2006). Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat. Rev. Immunol. 6, 772–783 10.1038/nri1937 [DOI] [PubMed] [Google Scholar]
- Toda M., Ono S. J. (2002). Genomics and proteomics of allergic disease. Immunology 106, 1–10 10.1046/j.1365-2567.2002.01407.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrone D. Z., Kuriakose J. S., Moors K., Jiang H., Niedzwiecki M. M., Perera F. F., et al. (2012). Reproducibility and intraindividual variation over days in buccal cell DNA methylation of two asthma genes, interferon g (IFNg) and inducible nitric oxide synthase (iNOS). Clin. Epigenet. 4:3 10.1186/1868-7083-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumes D. J., Connolly A., Dent L. A. (2009). Expression of survivin in lung eosinophils is associated with pathology in a mouse model of allergic asthma. Int. Immunol. 6, 633–644 10.1093/intimm/dxp032 [DOI] [PubMed] [Google Scholar]
- Webb D. C., McKenzie A. N., Foster P. S. (2001). Expression of the ym2 lectin-binding protein is dependent on interleukin (IL)-4 AND IL-13 signal transduction identification of a novel allergy-associated protein. J. Biol. Chem. 276, 41969–41976 10.1074/jbc.M106223200 [DOI] [PubMed] [Google Scholar]
- Woodruff P. G., Boushey H. A., Dolganov G. M., Barker C. S., Yang Y. H., Donnelly S., et al. (2007). Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc. Natl. Acad. Sci. U.S.A. 40, 15858–15863 10.1073/pnas.0707413104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Kobayashi M., Sousa E. A., Liu W., Cai J., Goldman S. J., et al. (2005). Differential proteomic analysis of bronchoalveolar lavage fluid in asthmatics following segmental antigen challenge. Mol. Cell. Proteomics 4, 1251–1264 10.1074/mcp.M500041-MCP200 [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhu H., Wong C. H., Leung K. Y., Wong W. S. (2005). Increased lungkine and chitinase levels in allergic airway inflammation: a proteomics approach. Proteomics 11, 2799–2807 10.1002/pmic.200401169 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Dong Q., Louahed J., Dragwa C., Savio D., Huang M., et al. (2001). Characterization of a calcium-activated chloride channel as a shared target of Th2 cytokine pathways and its potential involvement in asthma. Am. J. Respir. Cell Mol. Biol. 25, 486–491 10.1165/ajrcmb.25.4.4578 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Zheng T., Homer R. J., Kim Y. K., Chen N. Y., Cohn L., et al. (2004). Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science 5677, 1678–1682 10.1126/science.1095336 [DOI] [PubMed] [Google Scholar]
- Zimmermann N., King N. E., Laporte J., Yang M., Mishra A., Pope S. M., et al. (2003). Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J. Clin. Investig. 12, 1863–1874 10.1172/JCI200317912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Young S., Zhu F., Gheyas F., Skeans S., Wan Y., et al. (2002). Microarray profile of differentially expressed genes in a monkey model of allergic asthma. Genome Biol. 5, 20 10.1186/gb-2002-3-5-research0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L., Clanton T. L. (2005). Reactive oxygen species formation in the transition to hypoxia in skeletal muscle. Am. J. Physiol. Cell Physiol. 1, C207–C216 10.1152/ajpcell.00449.2004 [DOI] [PubMed] [Google Scholar]
- Zuo L., Otenbaker N. P., Rose B. A., Salisbury K. S. (2013). Molecular mechanisms of reactive oxygen species-related pulmonary inflammation and asthma. Mol. Immunol. 1, 57–63 10.1016/j.molimm.2013.04.002 [DOI] [PubMed] [Google Scholar]