Abstract
The title compound, {[ZnCl(C13H12F2N6O)2]Cl·2H2O}n, is a two-dimensional coordination polymer. The ZnII atom is six-coordinated by four N atoms from four 2-(2,4-difluorophenyl)-1,3-bis(1,2,4-triazol-1-yl)propan-2-ol (HFlu) ligands and by two Cl atoms in a distorted octahedral geometry. Two Cl atoms bridge two ZnII atoms, forming a centrosymmetric dinuclear unit. The HFlu ligands connect the dinuclear units into a 44 net parallel to (001) when the dinuclear unit is considered as a node. O—H⋯O and O—H⋯Cl hydrogen bonds link the cationic layer, free chloride anions and lattice water molecules. Intralayer π–π interactions between the triazole rings are observed [centroid–centroid distance = 3.716 (6) Å].
Related literature
For background to this class of compounds, see: Han et al. (2006a
▶,b
▶). For related structures, see: Gao et al. (2001 ▶); Zhang et al. (2007 ▶).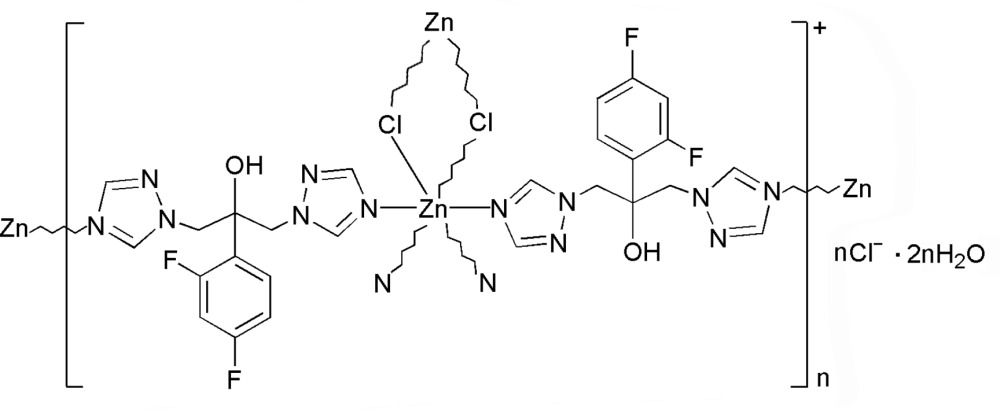
Experimental
Crystal data
[ZnCl(C13H12F2N6O)2]Cl·2H2O
M r = 784.89
Triclinic,
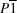
a = 10.2310 (6) Å
b = 11.8118 (6) Å
c = 14.3588 (9) Å
α = 91.191 (7)°
β = 107.481 (5)°
γ = 106.074 (6)°
V = 1580.11 (18) Å3
Z = 2
Mo Kα radiation
μ = 1.03 mm−1
T = 296 K
0.25 × 0.25 × 0.21 mm
Data collection
Bruker SMART 1000 CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.784, T max = 0.813
8374 measured reflections
5465 independent reflections
3137 reflections with I > 2σ(I)
R int = 0.064
Refinement
R[F 2 > 2σ(F 2)] = 0.080
wR(F 2) = 0.306
S = 1.07
5465 reflections
444 parameters
H-atom parameters constrained
Δρmax = 0.92 e Å−3
Δρmin = −1.09 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg, 1999 ▶); software used to prepare material for publication: SHELXTL (Sheldrick, 2008 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536813026524/hy2637sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813026524/hy2637Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯Cl2i | 0.82 | 2.29 | 3.103 (7) | 172 |
| O2—H2⋯O4i | 0.82 | 1.87 | 2.653 (9) | 160 |
| O3—H3A⋯Cl2ii | 0.85 | 2.32 | 3.163 (11) | 170 |
| O3—H3B⋯Cl1iii | 0.85 | 2.38 | 3.221 (10) | 170 |
| O4—H4A⋯O2iv | 0.85 | 2.24 | 2.784 (9) | 122 |
| O4—H4B⋯Cl2 | 0.85 | 2.29 | 3.101 (8) | 160 |
Symmetry codes: (i) 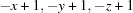 ; (ii)
; (ii) 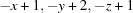 ; (iii)
; (iii) 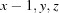 ; (iv)
; (iv) 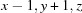 .
.
Acknowledgments
This work was supported by the Innovation Project of Guangxi University for Nationalities.
supplementary crystallographic information
1. Comment
Fluconazole 2-(2,4-difluorophenyl)-1,3-bis(1,2,4-triazol-1-yl) propan-2-ol, which is a 1,2,4-triazole derivative, is not only a widely used antifungal medicine but also a good flexible ligand to construct metal-organic polymers with optical properties and medical applications (Han et al., 2006a,b). Fluconazole can coordinate to metal ions in different configurations. We here report a new coordination polymer based on fluconazole.
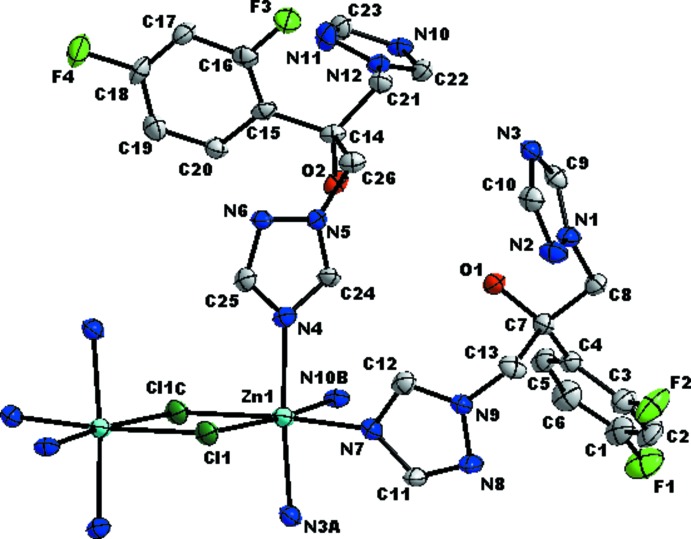
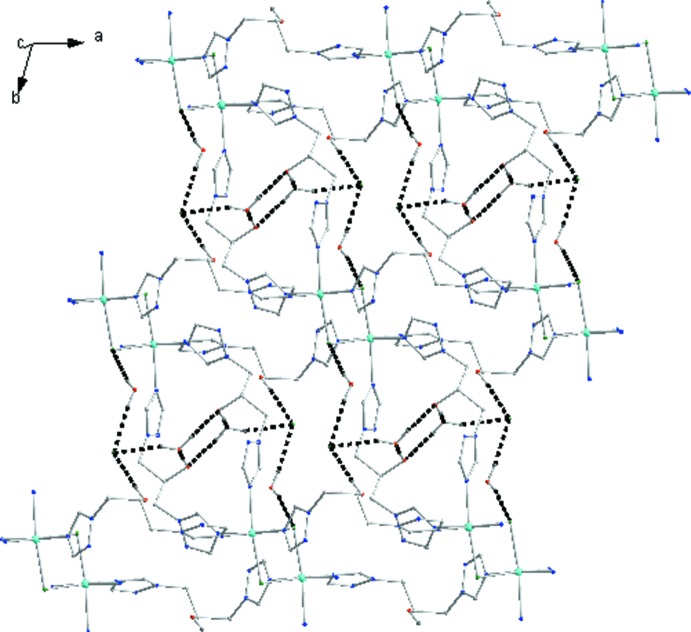
The asymmetric unit of the title compound contains one ZnII ion, two 2-(2,4-difluorophenyl)-1,3-bis(1,2,4-triazol-1-yl)propan-2-ol (HFlu) ligands, one coordinated Cl-1 anion, one free Cl-1 anion and two free water molecules. As shown in Fig. 1, the ZnII ion is six-coordinated by four N atoms from four HFlu ligands and two bridging Cl-1 anions. The Zn—N bond lengths range from 2.127 (8) to 2.197 (7) Å, and the Zn—Cl bond distances are 2.418 (3) and 2.732 (3) Å. The Zn—N bond lengths are in the normal range as observed in other Zn(II) complexes (Zhang et al., 2007). However, the Zn—Cl bond distances are longer than those as observed in other Zn(II) complexe (Gao et al., 2001). Two ZnII ions are connected by two HFlu ligands, forming a Zn2(HFlu)2 macrocycle, in which the Zn···Zn distance is 11.297 (2)Å. The other two HFlu ligands link the macrocycle along the a axis with a Zn···Zn distance of 10.231 (2) Å to form a grid unit with dimensions of 11.30 × 10.23 Å2 (Fig. 2a). These grid units are further connected by two Cl-1 anions with a Zn···Zn distance of 3.879 (1) Å into a two-dimensional structure (Fig. 2b), in which another type of grid with dimensions of 10.231 (2) × 3.879 (1) Å2 is formed.
Free water molecules and free Cl-1 anions are accommodated in the residual empties to shrink the void space and stabilize the structure. Moreover, there are intermolecular O—H···O and O—H···Cl hydrogen bonds involving the cationic layer, free Cl-1 anions and water molecules (Table 1, Fig. 3). These interactions further stabilize the structural framework.
2. Experimental
A mixture of fluconazole (153 mg, 0.5 mmol), ZnCl2 (136 mg, 1.0 mmol), 15 ml H2O, and 3 ml ethanol was placed in a Parr Teflon-lined stainless steel vessel (30 ml), and then the vessel was sealed and heated at 423 K for 3 days. After the mixture was slowly cooled to room temperature, colorless block-shaped crystals of the title compound was obtained. Analysis, calculated for C26H28Cl2F4N12O4Zn: C 39.79, H 3.60, N 21.42%; found: C 39.66, H 3.52, N 21.28%.
3. Refinement
H atoms were positioned geometrically and refined as riding atoms, with C—H = 0.93 (aromatic), 0.97(methylene) and O—H = 0.82 Å and with Uiso(H) = 1.2(1.5 for hydroxyl)Ueq(C,O). H atoms of the water molecules were located in a difference Fourier map and refined as riding, with O—H = 0.85 Å and Uiso(H) = 1.2Ueq(O). The maximum remaining electron density was found 1.00 Å from Zn1 and the minimum density 0.83 Å from Zn1.
Figures
Fig. 1.
The coordination environment around the ZnII atom in the title compound (uncoordinated Cl atom, water molecules and H atoms have been omitted for clarity). Displacement ellipsoids are drawn at the 30% probability level. [Symmetry codes: (A) 1+x, y, z; (B) 2-x, -y, 1-z; (C) 3-x, 1-y, 1-z.]
Fig. 2.
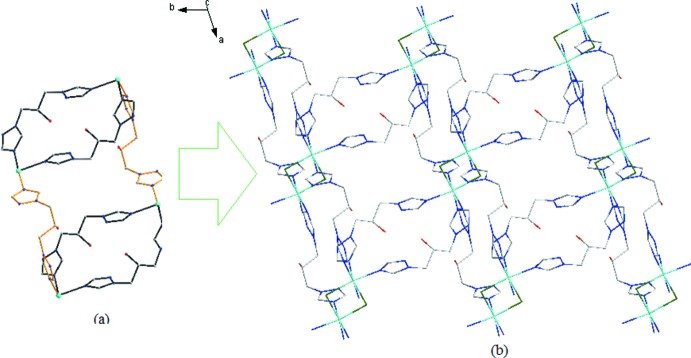
(a) A view of the grid unit in the title compound. (b) A view of the two-dimensional framework. H atoms and difluorophenyl groups of HFlu are omitted for clarity.
Fig. 3.
A view of hydrogen bonding interactions (dashed lines) in the title compound. H atoms and difluorophenyl groups of HFlu are omitted for clarity.
Crystal data
| [ZnCl(C13H12F2N6O)2]Cl·2H2O | Z = 2 |
| Mr = 784.89 | F(000) = 800 |
| Triclinic, P1 | Dx = 1.650 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.2310 (6) Å | Cell parameters from 859 reflections |
| b = 11.8118 (6) Å | θ = 2.2–22.1° |
| c = 14.3588 (9) Å | µ = 1.03 mm−1 |
| α = 91.191 (7)° | T = 296 K |
| β = 107.481 (5)° | Block, colorless |
| γ = 106.074 (6)° | 0.25 × 0.25 × 0.21 mm |
| V = 1580.11 (18) Å3 |
Data collection
| Bruker SMART 1000 CCD diffractometer | 5465 independent reflections |
| Radiation source: fine-focus sealed tube | 3137 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.064 |
| φ and ω scans | θmax = 25.0°, θmin = 1.5° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −12→12 |
| Tmin = 0.784, Tmax = 0.813 | k = −14→12 |
| 8374 measured reflections | l = −14→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.080 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.306 | H-atom parameters constrained |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.1513P)2 + 4.0805P] where P = (Fo2 + 2Fc2)/3 |
| 5465 reflections | (Δ/σ)max < 0.001 |
| 444 parameters | Δρmax = 0.92 e Å−3 |
| 0 restraints | Δρmin = −1.09 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Zn1 | 1.32995 (12) | 0.41377 (10) | 0.39336 (8) | 0.0404 (4) | |
| Cl1 | 1.4508 (3) | 0.6199 (2) | 0.45651 (17) | 0.0448 (6) | |
| Cl2 | 0.4395 (3) | 0.9322 (2) | 0.6822 (2) | 0.0584 (8) | |
| F1 | 0.7971 (10) | −0.0762 (7) | −0.0678 (5) | 0.086 (2) | |
| F2 | 0.6122 (9) | 0.2436 (7) | −0.0664 (4) | 0.083 (2) | |
| F3 | 0.8098 (7) | 0.2576 (6) | 0.7540 (4) | 0.0657 (18) | |
| F4 | 1.2544 (8) | 0.3386 (7) | 0.9953 (4) | 0.079 (2) | |
| N1 | 0.5148 (8) | 0.3554 (8) | 0.1793 (5) | 0.043 (2) | |
| N2 | 0.5348 (9) | 0.4724 (7) | 0.1718 (6) | 0.048 (2) | |
| N3 | 0.4536 (8) | 0.4152 (7) | 0.2976 (5) | 0.041 (2) | |
| N4 | 1.1903 (9) | 0.4025 (7) | 0.4804 (5) | 0.041 (2) | |
| N5 | 1.0061 (8) | 0.3403 (6) | 0.5307 (5) | 0.0335 (18) | |
| N6 | 1.0891 (8) | 0.4430 (6) | 0.5911 (5) | 0.0345 (18) | |
| N7 | 1.1616 (8) | 0.4340 (7) | 0.2632 (5) | 0.041 (2) | |
| N8 | 1.0182 (9) | 0.4016 (9) | 0.1051 (6) | 0.054 (2) | |
| N9 | 0.9478 (8) | 0.4220 (7) | 0.1671 (5) | 0.0388 (19) | |
| N10 | 0.7509 (8) | −0.2201 (7) | 0.6379 (5) | 0.0393 (19) | |
| N11 | 0.8367 (9) | −0.0419 (7) | 0.7239 (6) | 0.048 (2) | |
| N12 | 0.7677 (8) | −0.0339 (7) | 0.6293 (5) | 0.0381 (19) | |
| O1 | 0.7534 (7) | 0.2765 (6) | 0.2468 (4) | 0.0446 (17) | |
| H1 | 0.6956 | 0.2212 | 0.2602 | 0.067* | |
| O2 | 0.9802 (7) | 0.1089 (6) | 0.5651 (4) | 0.0417 (16) | |
| H2 | 0.9323 | 0.0797 | 0.5087 | 0.063* | |
| O3 | 0.5759 (12) | 0.8041 (10) | 0.3197 (8) | 0.106 (4) | |
| H3A | 0.5660 | 0.8728 | 0.3247 | 0.127* | |
| H3B | 0.5394 | 0.7628 | 0.3585 | 0.127* | |
| O4 | 0.1267 (8) | 0.9438 (6) | 0.6285 (5) | 0.055 (2) | |
| H4A | 0.0960 | 0.9921 | 0.6543 | 0.065* | |
| H4B | 0.2165 | 0.9582 | 0.6394 | 0.065* | |
| C1 | 0.7769 (15) | 0.0150 (10) | −0.0176 (9) | 0.061 (3) | |
| C2 | 0.7008 (14) | 0.0812 (11) | −0.0685 (8) | 0.061 (3) | |
| H2A | 0.6611 | 0.0669 | −0.1366 | 0.073* | |
| C3 | 0.6831 (12) | 0.1716 (11) | −0.0163 (8) | 0.055 (3) | |
| C4 | 0.7305 (10) | 0.1914 (8) | 0.0866 (6) | 0.037 (2) | |
| C5 | 0.8047 (12) | 0.1152 (9) | 0.1326 (7) | 0.048 (3) | |
| H5 | 0.8379 | 0.1228 | 0.2009 | 0.058* | |
| C6 | 0.8309 (14) | 0.0295 (9) | 0.0817 (8) | 0.059 (3) | |
| H6 | 0.8848 | −0.0178 | 0.1146 | 0.071* | |
| C7 | 0.7064 (10) | 0.2849 (8) | 0.1446 (6) | 0.038 (2) | |
| C8 | 0.5481 (11) | 0.2778 (9) | 0.1112 (7) | 0.044 (2) | |
| H8A | 0.5219 | 0.3018 | 0.0456 | 0.053* | |
| H8B | 0.4911 | 0.1963 | 0.1083 | 0.053* | |
| C9 | 0.4661 (10) | 0.3227 (9) | 0.2527 (7) | 0.043 (2) | |
| H9 | 0.4441 | 0.2460 | 0.2701 | 0.051* | |
| C10 | 0.4963 (11) | 0.5041 (9) | 0.2440 (7) | 0.045 (2) | |
| H10 | 0.4978 | 0.5816 | 0.2584 | 0.054* | |
| C11 | 1.1463 (11) | 0.4096 (10) | 0.1657 (7) | 0.050 (3) | |
| H11 | 1.2202 | 0.3997 | 0.1446 | 0.060* | |
| C12 | 1.0311 (11) | 0.4394 (8) | 0.2601 (7) | 0.042 (2) | |
| H12 | 1.0035 | 0.4532 | 0.3144 | 0.051* | |
| C13 | 0.7955 (10) | 0.4106 (9) | 0.1330 (7) | 0.045 (3) | |
| H13A | 0.7669 | 0.4244 | 0.0644 | 0.054* | |
| H13B | 0.7766 | 0.4699 | 0.1706 | 0.054* | |
| C14 | 0.9018 (10) | 0.1666 (8) | 0.6051 (6) | 0.035 (2) | |
| C15 | 0.9956 (10) | 0.2149 (8) | 0.7112 (6) | 0.036 (2) | |
| C16 | 0.9466 (11) | 0.2552 (8) | 0.7784 (7) | 0.043 (2) | |
| C17 | 1.0282 (12) | 0.2959 (9) | 0.8753 (6) | 0.047 (3) | |
| H17 | 0.9900 | 0.3211 | 0.9202 | 0.056* | |
| C18 | 1.1686 (12) | 0.2967 (10) | 0.9008 (6) | 0.050 (3) | |
| C19 | 1.2280 (12) | 0.2644 (9) | 0.8356 (7) | 0.052 (3) | |
| H19 | 1.3248 | 0.2691 | 0.8550 | 0.062* | |
| C20 | 1.1427 (10) | 0.2248 (8) | 0.7411 (7) | 0.040 (2) | |
| H20 | 1.1826 | 0.2039 | 0.6955 | 0.048* | |
| C21 | 0.7561 (10) | 0.0805 (8) | 0.5970 (7) | 0.043 (2) | |
| H21A | 0.7120 | 0.1149 | 0.6367 | 0.052* | |
| H21B | 0.6944 | 0.0683 | 0.5292 | 0.052* | |
| C22 | 0.7170 (11) | −0.1387 (9) | 0.5789 (7) | 0.045 (3) | |
| H22 | 0.6653 | −0.1546 | 0.5122 | 0.054* | |
| C23 | 0.8252 (11) | −0.1510 (9) | 0.7257 (7) | 0.046 (3) | |
| H23 | 0.8644 | −0.1822 | 0.7827 | 0.056* | |
| C24 | 1.0684 (10) | 0.3188 (8) | 0.4666 (6) | 0.038 (2) | |
| H24 | 1.0314 | 0.2539 | 0.4187 | 0.046* | |
| C25 | 1.1980 (11) | 0.4760 (8) | 0.5568 (6) | 0.038 (2) | |
| H25 | 1.2742 | 0.5445 | 0.5829 | 0.045* | |
| C26 | 0.8744 (10) | 0.2674 (8) | 0.5427 (7) | 0.038 (2) | |
| H26A | 0.8054 | 0.2338 | 0.4787 | 0.046* | |
| H26B | 0.8341 | 0.3160 | 0.5745 | 0.046* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Zn1 | 0.0408 (7) | 0.0385 (7) | 0.0419 (7) | 0.0027 (5) | 0.0216 (5) | −0.0007 (5) |
| Cl1 | 0.0473 (14) | 0.0335 (13) | 0.0487 (13) | 0.0027 (11) | 0.0168 (11) | 0.0009 (10) |
| Cl2 | 0.0528 (16) | 0.0554 (17) | 0.0658 (17) | 0.0127 (14) | 0.0199 (13) | 0.0137 (13) |
| F1 | 0.126 (7) | 0.067 (5) | 0.085 (5) | 0.039 (5) | 0.054 (5) | −0.006 (4) |
| F2 | 0.127 (7) | 0.099 (6) | 0.043 (4) | 0.071 (5) | 0.020 (4) | 0.013 (3) |
| F3 | 0.048 (4) | 0.095 (5) | 0.059 (4) | 0.027 (4) | 0.019 (3) | −0.005 (3) |
| F4 | 0.073 (5) | 0.105 (6) | 0.040 (3) | 0.023 (4) | −0.002 (3) | −0.014 (3) |
| N1 | 0.037 (4) | 0.057 (6) | 0.040 (4) | 0.018 (4) | 0.017 (3) | 0.006 (4) |
| N2 | 0.052 (5) | 0.038 (5) | 0.060 (5) | 0.008 (4) | 0.030 (4) | 0.008 (4) |
| N3 | 0.031 (4) | 0.048 (5) | 0.045 (4) | 0.010 (4) | 0.014 (3) | 0.009 (4) |
| N4 | 0.044 (5) | 0.045 (5) | 0.037 (4) | 0.012 (4) | 0.019 (3) | 0.009 (4) |
| N5 | 0.035 (4) | 0.034 (4) | 0.031 (4) | 0.007 (3) | 0.012 (3) | 0.002 (3) |
| N6 | 0.035 (4) | 0.037 (4) | 0.036 (4) | 0.009 (4) | 0.019 (3) | 0.003 (3) |
| N7 | 0.033 (4) | 0.053 (5) | 0.039 (4) | 0.012 (4) | 0.013 (3) | 0.005 (4) |
| N8 | 0.039 (5) | 0.091 (7) | 0.038 (4) | 0.022 (5) | 0.021 (4) | 0.004 (4) |
| N9 | 0.031 (4) | 0.051 (5) | 0.037 (4) | 0.010 (4) | 0.015 (3) | 0.007 (4) |
| N10 | 0.042 (5) | 0.039 (5) | 0.042 (4) | 0.009 (4) | 0.023 (4) | 0.011 (4) |
| N11 | 0.058 (5) | 0.025 (4) | 0.045 (4) | −0.005 (4) | 0.008 (4) | −0.001 (3) |
| N12 | 0.043 (5) | 0.036 (5) | 0.033 (4) | 0.005 (4) | 0.014 (3) | 0.009 (3) |
| O1 | 0.048 (4) | 0.046 (4) | 0.036 (3) | 0.002 (3) | 0.018 (3) | −0.002 (3) |
| O2 | 0.054 (4) | 0.043 (4) | 0.034 (3) | 0.023 (3) | 0.015 (3) | 0.005 (3) |
| O3 | 0.112 (9) | 0.096 (8) | 0.138 (9) | 0.039 (7) | 0.071 (7) | 0.047 (7) |
| O4 | 0.060 (5) | 0.052 (5) | 0.051 (4) | 0.025 (4) | 0.010 (3) | −0.002 (3) |
| C1 | 0.083 (9) | 0.036 (6) | 0.061 (7) | 0.006 (6) | 0.031 (6) | −0.010 (5) |
| C2 | 0.080 (9) | 0.064 (8) | 0.036 (6) | 0.018 (7) | 0.020 (5) | −0.011 (5) |
| C3 | 0.057 (7) | 0.067 (8) | 0.045 (6) | 0.023 (6) | 0.017 (5) | 0.011 (5) |
| C4 | 0.038 (5) | 0.040 (5) | 0.035 (5) | 0.011 (4) | 0.015 (4) | 0.007 (4) |
| C5 | 0.061 (7) | 0.040 (6) | 0.040 (5) | 0.007 (5) | 0.020 (5) | −0.005 (4) |
| C6 | 0.076 (8) | 0.035 (6) | 0.067 (7) | 0.019 (6) | 0.024 (6) | 0.002 (5) |
| C7 | 0.035 (5) | 0.039 (5) | 0.038 (5) | 0.009 (4) | 0.013 (4) | 0.002 (4) |
| C8 | 0.046 (6) | 0.047 (6) | 0.039 (5) | 0.007 (5) | 0.020 (4) | −0.009 (4) |
| C9 | 0.035 (5) | 0.043 (6) | 0.053 (6) | 0.014 (5) | 0.014 (4) | 0.005 (5) |
| C10 | 0.050 (6) | 0.031 (5) | 0.053 (6) | 0.007 (5) | 0.020 (5) | −0.002 (4) |
| C11 | 0.037 (6) | 0.083 (8) | 0.036 (5) | 0.020 (6) | 0.020 (4) | 0.010 (5) |
| C12 | 0.051 (6) | 0.036 (5) | 0.040 (5) | 0.009 (5) | 0.017 (4) | 0.001 (4) |
| C13 | 0.041 (6) | 0.049 (6) | 0.056 (6) | 0.023 (5) | 0.020 (5) | 0.015 (5) |
| C14 | 0.042 (5) | 0.036 (5) | 0.037 (5) | 0.015 (4) | 0.023 (4) | 0.008 (4) |
| C15 | 0.048 (6) | 0.031 (5) | 0.034 (5) | 0.010 (4) | 0.021 (4) | 0.011 (4) |
| C16 | 0.047 (6) | 0.032 (5) | 0.050 (6) | 0.006 (5) | 0.022 (5) | 0.008 (4) |
| C17 | 0.057 (7) | 0.057 (7) | 0.030 (5) | 0.019 (5) | 0.018 (4) | 0.001 (4) |
| C18 | 0.063 (7) | 0.058 (7) | 0.023 (5) | 0.019 (6) | 0.005 (5) | 0.006 (4) |
| C19 | 0.044 (6) | 0.049 (6) | 0.051 (6) | 0.003 (5) | 0.009 (5) | −0.001 (5) |
| C20 | 0.035 (5) | 0.044 (6) | 0.044 (5) | 0.011 (5) | 0.018 (4) | 0.004 (4) |
| C21 | 0.039 (5) | 0.047 (6) | 0.040 (5) | 0.006 (5) | 0.014 (4) | 0.010 (4) |
| C22 | 0.048 (6) | 0.046 (6) | 0.034 (5) | 0.001 (5) | 0.016 (4) | −0.004 (4) |
| C23 | 0.053 (6) | 0.038 (6) | 0.040 (5) | 0.001 (5) | 0.014 (5) | 0.001 (4) |
| C24 | 0.041 (5) | 0.037 (5) | 0.037 (5) | 0.002 (4) | 0.021 (4) | 0.006 (4) |
| C25 | 0.046 (6) | 0.025 (5) | 0.039 (5) | 0.005 (4) | 0.015 (4) | −0.002 (4) |
| C26 | 0.045 (6) | 0.035 (5) | 0.042 (5) | 0.016 (5) | 0.020 (4) | 0.010 (4) |
Geometric parameters (Å, º)
| Zn1—N3i | 2.127 (8) | O4—H4B | 0.8500 |
| Zn1—N4 | 2.144 (8) | C1—C2 | 1.331 (16) |
| Zn1—N10ii | 2.192 (8) | C1—C6 | 1.353 (15) |
| Zn1—N7 | 2.197 (7) | C2—C3 | 1.374 (15) |
| Zn1—Cl1 | 2.418 (3) | C2—H2A | 0.9300 |
| Zn1—Cl1iii | 2.732 (3) | C3—C4 | 1.402 (13) |
| F1—C1 | 1.380 (13) | C4—C5 | 1.387 (13) |
| F2—C3 | 1.349 (12) | C4—C7 | 1.487 (13) |
| F3—C16 | 1.345 (12) | C5—C6 | 1.367 (15) |
| F4—C18 | 1.368 (11) | C5—H5 | 0.9300 |
| N1—C9 | 1.317 (12) | C6—H6 | 0.9300 |
| N1—N2 | 1.351 (11) | C7—C8 | 1.521 (13) |
| N1—C8 | 1.500 (12) | C7—C13 | 1.554 (13) |
| N2—C10 | 1.297 (13) | C8—H8A | 0.9700 |
| N3—C9 | 1.312 (13) | C8—H8B | 0.9700 |
| N3—C10 | 1.366 (12) | C9—H9 | 0.9300 |
| N4—C24 | 1.317 (12) | C10—H10 | 0.9300 |
| N4—C25 | 1.354 (12) | C11—H11 | 0.9300 |
| N5—C24 | 1.323 (12) | C12—H12 | 0.9300 |
| N5—N6 | 1.375 (10) | C13—H13A | 0.9700 |
| N5—C26 | 1.446 (12) | C13—H13B | 0.9700 |
| N6—C25 | 1.317 (12) | C14—C21 | 1.524 (13) |
| N7—C12 | 1.342 (13) | C14—C15 | 1.533 (12) |
| N7—C11 | 1.376 (12) | C14—C26 | 1.542 (11) |
| N8—C11 | 1.314 (12) | C15—C16 | 1.346 (14) |
| N8—N9 | 1.356 (11) | C15—C20 | 1.406 (13) |
| N9—C12 | 1.327 (11) | C16—C17 | 1.384 (13) |
| N9—C13 | 1.452 (12) | C17—C18 | 1.369 (15) |
| N10—C22 | 1.343 (12) | C17—H17 | 0.9300 |
| N10—C23 | 1.367 (12) | C18—C19 | 1.359 (16) |
| N11—C23 | 1.262 (12) | C19—C20 | 1.363 (13) |
| N11—N12 | 1.350 (10) | C19—H19 | 0.9300 |
| N12—C22 | 1.306 (12) | C20—H20 | 0.9300 |
| N12—C21 | 1.462 (11) | C21—H21A | 0.9700 |
| O1—C7 | 1.415 (10) | C21—H21B | 0.9700 |
| O1—H1 | 0.8200 | C22—H22 | 0.9300 |
| O2—C14 | 1.415 (11) | C23—H23 | 0.9300 |
| O2—H2 | 0.8200 | C24—H24 | 0.9300 |
| O3—H3A | 0.8499 | C25—H25 | 0.9300 |
| O3—H3B | 0.8500 | C26—H26A | 0.9700 |
| O4—H4A | 0.8500 | C26—H26B | 0.9700 |
| N3i—Zn1—N4 | 175.2 (3) | N1—C8—C7 | 111.8 (8) |
| N3i—Zn1—N10ii | 88.7 (3) | N1—C8—H8A | 109.3 |
| N4—Zn1—N10ii | 88.0 (3) | C7—C8—H8A | 109.3 |
| N3i—Zn1—N7 | 86.0 (3) | N1—C8—H8B | 109.3 |
| N4—Zn1—N7 | 90.5 (3) | C7—C8—H8B | 109.3 |
| N10ii—Zn1—N7 | 91.2 (3) | H8A—C8—H8B | 107.9 |
| N3i—Zn1—Cl1 | 93.0 (2) | N3—C9—N1 | 109.5 (9) |
| N4—Zn1—Cl1 | 90.9 (2) | N3—C9—H9 | 125.3 |
| N10ii—Zn1—Cl1 | 168.89 (19) | N1—C9—H9 | 125.3 |
| N7—Zn1—Cl1 | 99.8 (2) | N2—C10—N3 | 115.1 (9) |
| N3i—Zn1—Cl1iii | 89.1 (2) | N2—C10—H10 | 122.5 |
| N4—Zn1—Cl1iii | 94.2 (2) | N3—C10—H10 | 122.5 |
| N10ii—Zn1—Cl1iii | 86.6 (2) | N8—C11—N7 | 114.2 (10) |
| N7—Zn1—Cl1iii | 174.7 (2) | N8—C11—H11 | 122.9 |
| Cl1—Zn1—Cl1iii | 82.44 (8) | N7—C11—H11 | 122.9 |
| Zn1—Cl1—Zn1iii | 97.56 (8) | N9—C12—N7 | 108.9 (9) |
| C9—N1—N2 | 111.4 (9) | N9—C12—H12 | 125.6 |
| C9—N1—C8 | 126.6 (9) | N7—C12—H12 | 125.6 |
| N2—N1—C8 | 122.0 (8) | N9—C13—C7 | 110.6 (7) |
| C10—N2—N1 | 101.5 (8) | N9—C13—H13A | 109.5 |
| C9—N3—C10 | 102.6 (8) | C7—C13—H13A | 109.5 |
| C9—N3—Zn1iv | 127.0 (7) | N9—C13—H13B | 109.5 |
| C10—N3—Zn1iv | 127.3 (7) | C7—C13—H13B | 109.5 |
| C24—N4—C25 | 103.4 (8) | H13A—C13—H13B | 108.1 |
| C24—N4—Zn1 | 125.4 (7) | O2—C14—C21 | 109.9 (8) |
| C25—N4—Zn1 | 131.1 (7) | O2—C14—C15 | 106.4 (7) |
| C24—N5—N6 | 110.1 (7) | C21—C14—C15 | 113.6 (7) |
| C24—N5—C26 | 127.4 (8) | O2—C14—C26 | 107.9 (7) |
| N6—N5—C26 | 122.4 (8) | C21—C14—C26 | 107.4 (7) |
| C25—N6—N5 | 101.7 (7) | C15—C14—C26 | 111.5 (8) |
| C12—N7—C11 | 102.9 (8) | C16—C15—C20 | 116.5 (9) |
| C12—N7—Zn1 | 126.3 (6) | C16—C15—C14 | 123.5 (9) |
| C11—N7—Zn1 | 128.3 (7) | C20—C15—C14 | 119.9 (9) |
| C11—N8—N9 | 102.4 (8) | C15—C16—F3 | 120.6 (9) |
| C12—N9—N8 | 111.5 (8) | C15—C16—C17 | 124.4 (10) |
| C12—N9—C13 | 126.0 (9) | F3—C16—C17 | 115.0 (10) |
| N8—N9—C13 | 122.1 (7) | C18—C17—C16 | 115.8 (10) |
| C22—N10—C23 | 101.2 (8) | C18—C17—H17 | 122.1 |
| C22—N10—Zn1ii | 128.2 (7) | C16—C17—H17 | 122.1 |
| C23—N10—Zn1ii | 129.6 (6) | C19—C18—C17 | 123.1 (9) |
| C23—N11—N12 | 103.6 (7) | C19—C18—F4 | 118.8 (10) |
| C22—N12—N11 | 110.1 (7) | C17—C18—F4 | 118.0 (10) |
| C22—N12—C21 | 129.4 (8) | C18—C19—C20 | 118.6 (10) |
| N11—N12—C21 | 120.5 (8) | C18—C19—H19 | 120.7 |
| C7—O1—H1 | 109.5 | C20—C19—H19 | 120.7 |
| C14—O2—H2 | 109.5 | C19—C20—C15 | 121.3 (10) |
| H3A—O3—H3B | 108.7 | C19—C20—H20 | 119.4 |
| H4A—O4—H4B | 119.4 | C15—C20—H20 | 119.4 |
| C2—C1—C6 | 123.2 (12) | N12—C21—C14 | 112.1 (8) |
| C2—C1—F1 | 118.7 (10) | N12—C21—H21A | 109.2 |
| C6—C1—F1 | 118.0 (11) | C14—C21—H21A | 109.2 |
| C1—C2—C3 | 117.2 (10) | N12—C21—H21B | 109.2 |
| C1—C2—H2A | 121.4 | C14—C21—H21B | 109.2 |
| C3—C2—H2A | 121.4 | H21A—C21—H21B | 107.9 |
| F2—C3—C2 | 118.5 (9) | N12—C22—N10 | 109.7 (8) |
| F2—C3—C4 | 117.7 (10) | N12—C22—H22 | 125.1 |
| C2—C3—C4 | 123.8 (9) | N10—C22—H22 | 125.1 |
| C5—C4—C3 | 114.2 (10) | N11—C23—N10 | 115.4 (8) |
| C5—C4—C7 | 121.1 (8) | N11—C23—H23 | 122.3 |
| C3—C4—C7 | 124.7 (8) | N10—C23—H23 | 122.3 |
| C6—C5—C4 | 122.7 (9) | N4—C24—N5 | 110.2 (9) |
| C6—C5—H5 | 118.6 | N4—C24—H24 | 124.9 |
| C4—C5—H5 | 118.6 | N5—C24—H24 | 124.9 |
| C1—C6—C5 | 118.6 (10) | N6—C25—N4 | 114.7 (8) |
| C1—C6—H6 | 120.7 | N6—C25—H25 | 122.6 |
| C5—C6—H6 | 120.7 | N4—C25—H25 | 122.6 |
| O1—C7—C4 | 112.2 (7) | N5—C26—C14 | 110.9 (7) |
| O1—C7—C8 | 109.6 (8) | N5—C26—H26A | 109.5 |
| C4—C7—C8 | 110.1 (8) | C14—C26—H26A | 109.5 |
| O1—C7—C13 | 104.5 (7) | N5—C26—H26B | 109.5 |
| C4—C7—C13 | 111.2 (8) | C14—C26—H26B | 109.5 |
| C8—C7—C13 | 109.0 (7) | H26A—C26—H26B | 108.1 |
Symmetry codes: (i) x+1, y, z; (ii) −x+2, −y, −z+1; (iii) −x+3, −y+1, −z+1; (iv) x−1, y, z.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···Cl2v | 0.82 | 2.29 | 3.103 (7) | 172 |
| O2—H2···O4v | 0.82 | 1.87 | 2.653 (9) | 160 |
| O3—H3A···Cl2vi | 0.85 | 2.32 | 3.163 (11) | 170 |
| O3—H3B···Cl1iv | 0.85 | 2.38 | 3.221 (10) | 170 |
| O4—H4A···O2vii | 0.85 | 2.24 | 2.784 (9) | 122 |
| O4—H4B···Cl2 | 0.85 | 2.29 | 3.101 (8) | 160 |
Symmetry codes: (iv) x−1, y, z; (v) −x+1, −y+1, −z+1; (vi) −x+1, −y+2, −z+1; (vii) x−1, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HY2637).
References
- Brandenburg, K. (1999). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2001). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Gao, J.-S., Ma, D.-S., Ma, Z.-G., Chen, G.-R., Hou, Y.-J. & Ye, L. (2001). Chin. J. Mol. Sci. 17, 17–22.
- Han, H., Song, Y., Hou, H., Fan, Y. & Zhu, Y. (2006a). Dalton Trans. pp. 1972–1980. [DOI] [PubMed]
- Han, H., Zhang, S., Hou, H., Fan, Y. & Zhu, Y. (2006b). Eur. J. Inorg. Chem. pp. 1594–1600.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Zhang, L., Ling, Y., Peng, F. & Du, M. (2007). J. Mol. Struct. 829, 161–167.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536813026524/hy2637sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813026524/hy2637Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report