Abstract
In the asymmetric unit of the title salt, C14H18N2O2+·2CF3O3S−, the components are linked by two N—H⋯O and one C—H⋯O hydrogen bonds. The dipyridinium salt demonstrates a skew conformation based upon C—O—C—C torsion angles of 61.5 (3) and 15.1 (4)°. A C—O—C angle of 119.3 (2)° and C—O bond distances of 1.364 (3) and 1.389 (3) Å are consistent with other dipyridyl ethers. The planes of the pyridyl rings exhibit a twist angle of 67.89 (8)°. One of the trifluoromethanesulfonate ions shows disorder of the F atoms [in a 0.52 (7):0.48 (7) occupancy ratio] and an O atom [0.64 (8):0.36 (8) occupancy ratio]. In the crystal, the components are linked by C—H⋯O interactions, which form chains along [101].
Related literature
For the structure of the unsubstituted 4,4′-oxybisdipyridine, see: Dunne et al. (1996 ▶). For the structure of bis[4′-(2,2′:6′,2′′-terpyridinyl)]ether, see: Constable et al. (1995 ▶). For the stuctures of the neutral ether 9,9′-oxybisacridine and its dication, see: Maas (1985 ▶). For a description of conformations in bridged diphenyls, see: van der Heijden et al. (1975 ▶).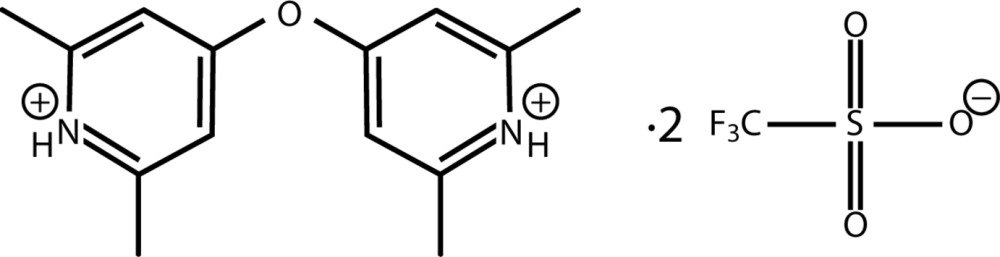
Experimental
Crystal data
C14H18N2O2+·2CF3O3S−
M r = 528.44
Monoclinic,
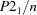
a = 12.7397 (18) Å
b = 11.3610 (16) Å
c = 15.611 (2) Å
β = 101.405 (4)°
V = 2214.8 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.33 mm−1
T = 100 K
0.24 × 0.18 × 0.10 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.925, T max = 0.968
15390 measured reflections
4360 independent reflections
3546 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.049
wR(F 2) = 0.125
S = 1.09
4360 reflections
316 parameters
53 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.94 e Å−3
Δρmin = −1.04 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2005 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S1600536813027505/ff2121sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813027505/ff2121Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813027505/ff2121Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O4 | 0.86 (2) | 1.93 (2) | 2.783 (3) | 171 (3) |
| N2—H2N⋯O7 | 0.87 (2) | 1.97 (2) | 2.826 (3) | 169 (3) |
| C2—H2A⋯O6i | 0.95 | 2.36 | 3.170 (4) | 142 |
| C6—H6B⋯O6i | 0.98 | 2.50 | 3.383 (4) | 149 |
| C7—H7B⋯O3ii | 0.98 | 2.47 | 3.421 (4) | 164 |
| C9—H9A⋯O3iii | 0.95 | 2.44 | 3.293 (4) | 149 |
| C12—H12A⋯O5iv | 0.95 | 2.26 | 3.168 (4) | 160 |
| C14—H14A⋯O6 | 0.98 | 2.52 | 3.436 (4) | 155 |
Symmetry codes: (i) 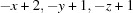 ; (ii)
; (ii) 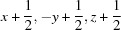 ; (iii)
; (iii) 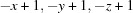 ; (iv)
; (iv) 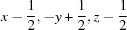 .
.
Acknowledgments
AWS thanks the Jean Dreyfus Boissevain Lectureship for Undergraduate Institutions, the UMass Dartmouth Office of Undergraduate Research Award, the Urban Massachusetts Louis Stokes Alliance for Minority Participation (UMLSAMP), the UMass Dartmouth Honors Program and the Northeast Section of the American Chemical Society Norris/Richards Summer Research Scholarship for funding. DRM gratefully acknowledges support from the UMass Dartmouth Chancellor’s Research Fund, the Joseph P. Healey Endowment and the National Science Foundation (CHE-1229339).
supplementary crystallographic information
1. Comment
The structures of bridged diaryls have been examined for many years and here we submit another structure into this data set. Based upon dissimilar C–O–C–C torsion angles of 61.5 (3)° and 15.1 (4)°, this structure exhibits a skew conformation (van der Heijden et al. 1975). The previously reported structures of 4,4'-oxybisdipyridyls and their cations (Dunne et al. 1996, Maas, 1985, Constable et al., 1995) have shown a twist structure, with torsion angles that are closer in size. Otherwise, the C–O–C angle of 119.3 (2)° and C–O bond distances of 1.364 (3) Å and 1.389 (3) Å are consistent with reported dipyridyl ethers
The structure of the title salt is shown in Figure 1. N–H···O hydrogen bonds between the dication and the two anions are seen between N1–H1N···O4 and N2–H2N···O7. There are no π-π interactions between pyridinium rings of the dications observed. One of the trifluoromethanesulfonate ions shows a disorder at the fluorines with a 52.0:48.0 percentage distribution and at one oxygen with a 64:36 percentage distribution.
2. Experimental
Colorless crystals of the title compound formed from the slow decomposition of neat 2,6-dimethyl-4-triflatopyridine.
3. Refinement
All non-hydrogen atoms were refined anisotropically by full matrix least squares on F2. Fluorine atoms F1, F2, and F3 were disordered over two positions (52.0/48.0) and were refined anisotropically with similar distances and amplitudes using SADI restraints and EADP constraints. Oxygen atom O2 was found to be disordered over two sites (63.5/36.5) and was refined with DFIX restraints for S–O bond length of 1.44(0.01) Å and O–O distances of 2.41(0.02) Å and ISOR restraint for O2 and O2'. Hydrogen atoms H1N and H2N were found from a Fourier difference map and were refined isotropically with N—H distance of 0.87 (2) Å and 1.20 Ueq of parent N atom. All other hydrogen atoms were placed in calculated positions with appropriate carbon hydrogen bond lengths; C—H(Ar) 0.950 Å and CH3 0.980 Å and 1.20 and 1.50 Ueq of parent C atom.
Figures
Fig. 1.
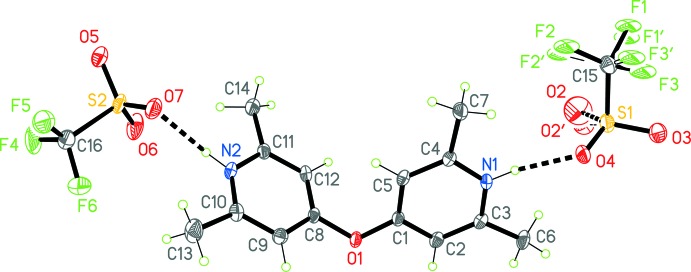
The molecular structure of the title compound, showing the atom labeling scheme, with displacement ellipsoids drawn at the 50% probability level. H atoms are presented as spheres of arbitrary radius. Hydrogen bonding is shown with dashed lines.
Crystal data
| C14H18N2O2+·2CF3O3S− | F(000) = 1080 |
| Mr = 528.44 | Dx = 1.585 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 5851 reflections |
| a = 12.7397 (18) Å | θ = 2.4–26.2° |
| b = 11.3610 (16) Å | µ = 0.33 mm−1 |
| c = 15.611 (2) Å | T = 100 K |
| β = 101.405 (4)° | Block, colourless |
| V = 2214.8 (6) Å3 | 0.24 × 0.18 × 0.10 mm |
| Z = 4 |
Data collection
| Bruker APEXII CCD diffractometer | 4360 independent reflections |
| Radiation source: fine-focus sealed tube | 3546 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.027 |
| φ and ω scans | θmax = 26.0°, θmin = 2.9° |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | h = −15→15 |
| Tmin = 0.925, Tmax = 0.968 | k = −14→10 |
| 15390 measured reflections | l = −19→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.125 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.0486P)2 + 3.6577P] where P = (Fo2 + 2Fc2)/3 |
| 4360 reflections | (Δ/σ)max = 0.023 |
| 316 parameters | Δρmax = 0.94 e Å−3 |
| 53 restraints | Δρmin = −1.04 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| S1 | 0.30074 (6) | 0.21804 (7) | 0.30451 (5) | 0.0244 (2) | |
| S2 | 1.28559 (6) | 0.28663 (8) | 0.84123 (5) | 0.0261 (2) | |
| F1 | 0.2582 (5) | −0.0159 (3) | 0.3316 (4) | 0.0595 (7) | 0.520 (7) |
| F2 | 0.3592 (5) | 0.0635 (5) | 0.4423 (4) | 0.0595 (7) | 0.520 (7) |
| F3 | 0.1942 (4) | 0.1172 (5) | 0.4014 (4) | 0.0595 (7) | 0.520 (7) |
| F1' | 0.2686 (5) | 0.0044 (4) | 0.3063 (4) | 0.0595 (7) | 0.480 (7) |
| F2' | 0.3863 (4) | 0.0710 (5) | 0.4133 (4) | 0.0595 (7) | 0.480 (7) |
| F3' | 0.2151 (5) | 0.0891 (5) | 0.4199 (3) | 0.0595 (7) | 0.480 (7) |
| F4 | 1.45831 (17) | 0.3944 (2) | 0.92267 (16) | 0.0540 (7) | |
| F5 | 1.33893 (18) | 0.36761 (19) | 1.00127 (13) | 0.0429 (6) | |
| F6 | 1.3158 (2) | 0.5016 (2) | 0.90176 (19) | 0.0642 (8) | |
| O1 | 0.81802 (15) | 0.55937 (18) | 0.51520 (13) | 0.0198 (5) | |
| O2 | 0.3883 (10) | 0.178 (3) | 0.268 (2) | 0.065 (4) | 0.64 (8) |
| O2' | 0.3796 (13) | 0.206 (3) | 0.2489 (14) | 0.039 (5) | 0.36 (8) |
| O3 | 0.19673 (19) | 0.2307 (2) | 0.24973 (15) | 0.0349 (6) | |
| O4 | 0.32838 (16) | 0.30914 (19) | 0.36998 (14) | 0.0247 (5) | |
| O5 | 1.33183 (18) | 0.1762 (2) | 0.87294 (14) | 0.0283 (5) | |
| O6 | 1.3101 (2) | 0.3260 (3) | 0.75980 (16) | 0.0445 (7) | |
| O7 | 1.17400 (17) | 0.3012 (2) | 0.84615 (16) | 0.0322 (6) | |
| N1 | 0.53677 (19) | 0.3908 (2) | 0.42520 (15) | 0.0166 (5) | |
| H1N | 0.4755 (19) | 0.358 (3) | 0.408 (2) | 0.020* | |
| N2 | 1.05589 (19) | 0.4233 (2) | 0.70080 (15) | 0.0180 (5) | |
| H2N | 1.099 (2) | 0.386 (3) | 0.7421 (18) | 0.022* | |
| C1 | 0.7249 (2) | 0.4988 (3) | 0.48821 (18) | 0.0163 (6) | |
| C2 | 0.6599 (2) | 0.5394 (3) | 0.41160 (18) | 0.0184 (6) | |
| H2A | 0.6811 | 0.6052 | 0.3815 | 0.022* | |
| C3 | 0.5646 (2) | 0.4833 (3) | 0.38005 (18) | 0.0177 (6) | |
| C4 | 0.5987 (2) | 0.3485 (3) | 0.49946 (18) | 0.0164 (6) | |
| C5 | 0.6942 (2) | 0.4040 (3) | 0.53340 (18) | 0.0167 (6) | |
| H5A | 0.7379 | 0.3778 | 0.5865 | 0.020* | |
| C6 | 0.4891 (2) | 0.5170 (3) | 0.29786 (19) | 0.0244 (7) | |
| H6A | 0.4711 | 0.4472 | 0.2610 | 0.037* | |
| H6B | 0.5230 | 0.5759 | 0.2664 | 0.037* | |
| H6C | 0.4237 | 0.5499 | 0.3123 | 0.037* | |
| C7 | 0.5605 (2) | 0.2412 (3) | 0.5387 (2) | 0.0255 (7) | |
| H7A | 0.4840 | 0.2490 | 0.5389 | 0.038* | |
| H7B | 0.6003 | 0.2319 | 0.5988 | 0.038* | |
| H7C | 0.5722 | 0.1721 | 0.5042 | 0.038* | |
| C8 | 0.8979 (2) | 0.5106 (3) | 0.57885 (18) | 0.0164 (6) | |
| C9 | 0.9320 (2) | 0.5738 (3) | 0.65427 (19) | 0.0197 (6) | |
| H9A | 0.9006 | 0.6475 | 0.6631 | 0.024* | |
| C10 | 1.0135 (2) | 0.5270 (3) | 0.71692 (19) | 0.0204 (6) | |
| C11 | 1.0248 (2) | 0.3602 (3) | 0.62686 (19) | 0.0184 (6) | |
| C12 | 0.9443 (2) | 0.4049 (3) | 0.56316 (19) | 0.0178 (6) | |
| H12A | 0.9212 | 0.3640 | 0.5097 | 0.021* | |
| C13 | 1.0564 (3) | 0.5851 (3) | 0.8025 (2) | 0.0358 (9) | |
| H13A | 1.0534 | 0.5298 | 0.8501 | 0.054* | |
| H13B | 1.0131 | 0.6547 | 0.8088 | 0.054* | |
| H13C | 1.1308 | 0.6089 | 0.8046 | 0.054* | |
| C14 | 1.0780 (3) | 0.2449 (3) | 0.6199 (2) | 0.0268 (7) | |
| H14A | 1.1537 | 0.2503 | 0.6483 | 0.040* | |
| H14B | 1.0725 | 0.2247 | 0.5581 | 0.040* | |
| H14C | 1.0429 | 0.1839 | 0.6485 | 0.040* | |
| C15 | 0.2854 (2) | 0.0901 (3) | 0.3686 (2) | 0.0548 (13) | |
| C16 | 1.3531 (3) | 0.3933 (3) | 0.9209 (3) | 0.0378 (9) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0170 (4) | 0.0248 (4) | 0.0308 (4) | −0.0001 (3) | 0.0029 (3) | −0.0091 (3) |
| S2 | 0.0188 (4) | 0.0357 (5) | 0.0208 (4) | −0.0053 (3) | −0.0038 (3) | 0.0133 (3) |
| F1 | 0.0720 (13) | 0.0293 (11) | 0.0614 (17) | −0.0171 (9) | −0.0252 (11) | 0.0380 (11) |
| F2 | 0.0720 (13) | 0.0293 (11) | 0.0614 (17) | −0.0171 (9) | −0.0252 (11) | 0.0380 (11) |
| F3 | 0.0720 (13) | 0.0293 (11) | 0.0614 (17) | −0.0171 (9) | −0.0252 (11) | 0.0380 (11) |
| F1' | 0.0720 (13) | 0.0293 (11) | 0.0614 (17) | −0.0171 (9) | −0.0252 (11) | 0.0380 (11) |
| F2' | 0.0720 (13) | 0.0293 (11) | 0.0614 (17) | −0.0171 (9) | −0.0252 (11) | 0.0380 (11) |
| F3' | 0.0720 (13) | 0.0293 (11) | 0.0614 (17) | −0.0171 (9) | −0.0252 (11) | 0.0380 (11) |
| F4 | 0.0295 (12) | 0.0722 (17) | 0.0517 (14) | −0.0213 (12) | −0.0126 (10) | 0.0118 (12) |
| F5 | 0.0547 (14) | 0.0371 (12) | 0.0327 (11) | 0.0026 (10) | −0.0012 (10) | −0.0034 (9) |
| F6 | 0.0656 (17) | 0.0273 (13) | 0.084 (2) | −0.0100 (12) | −0.0240 (14) | 0.0171 (12) |
| O1 | 0.0126 (9) | 0.0218 (11) | 0.0226 (11) | −0.0023 (8) | −0.0029 (8) | 0.0070 (9) |
| O2 | 0.050 (4) | 0.060 (7) | 0.096 (8) | 0.011 (4) | 0.041 (4) | −0.024 (6) |
| O2' | 0.035 (6) | 0.039 (8) | 0.047 (8) | 0.003 (4) | 0.017 (5) | −0.011 (5) |
| O3 | 0.0352 (13) | 0.0293 (13) | 0.0318 (13) | −0.0027 (11) | −0.0139 (10) | −0.0049 (10) |
| O4 | 0.0194 (11) | 0.0242 (12) | 0.0279 (12) | −0.0032 (9) | −0.0019 (9) | −0.0056 (9) |
| O5 | 0.0270 (12) | 0.0356 (13) | 0.0202 (11) | 0.0024 (10) | −0.0002 (9) | 0.0045 (10) |
| O6 | 0.0331 (14) | 0.071 (2) | 0.0257 (13) | −0.0148 (13) | −0.0037 (10) | 0.0244 (13) |
| O7 | 0.0202 (11) | 0.0377 (14) | 0.0354 (13) | −0.0016 (10) | −0.0024 (10) | 0.0132 (11) |
| N1 | 0.0118 (11) | 0.0201 (13) | 0.0165 (12) | −0.0003 (10) | −0.0004 (9) | −0.0007 (10) |
| N2 | 0.0136 (11) | 0.0239 (14) | 0.0152 (12) | 0.0018 (10) | −0.0004 (9) | 0.0021 (10) |
| C1 | 0.0118 (13) | 0.0184 (14) | 0.0181 (14) | 0.0011 (11) | 0.0013 (11) | −0.0007 (11) |
| C2 | 0.0161 (13) | 0.0225 (16) | 0.0168 (14) | 0.0015 (12) | 0.0040 (11) | 0.0058 (12) |
| C3 | 0.0162 (14) | 0.0213 (15) | 0.0157 (13) | 0.0042 (12) | 0.0035 (11) | 0.0019 (12) |
| C4 | 0.0155 (13) | 0.0184 (15) | 0.0148 (13) | 0.0021 (11) | 0.0020 (11) | 0.0012 (11) |
| C5 | 0.0147 (13) | 0.0213 (15) | 0.0130 (13) | 0.0023 (11) | 0.0003 (10) | 0.0024 (11) |
| C6 | 0.0175 (14) | 0.0340 (18) | 0.0191 (15) | 0.0005 (13) | −0.0026 (12) | 0.0071 (13) |
| C7 | 0.0218 (15) | 0.0262 (17) | 0.0250 (16) | −0.0054 (13) | −0.0037 (12) | 0.0070 (13) |
| C8 | 0.0104 (12) | 0.0207 (15) | 0.0175 (14) | −0.0027 (11) | 0.0012 (10) | 0.0050 (11) |
| C9 | 0.0162 (14) | 0.0205 (15) | 0.0228 (15) | 0.0015 (12) | 0.0049 (11) | −0.0001 (12) |
| C10 | 0.0172 (14) | 0.0264 (16) | 0.0172 (14) | −0.0002 (12) | 0.0028 (11) | −0.0020 (12) |
| C11 | 0.0143 (13) | 0.0212 (15) | 0.0190 (14) | −0.0019 (12) | 0.0012 (11) | 0.0005 (12) |
| C12 | 0.0138 (13) | 0.0236 (16) | 0.0150 (13) | −0.0028 (12) | 0.0002 (11) | −0.0018 (12) |
| C13 | 0.0367 (19) | 0.042 (2) | 0.0243 (17) | 0.0095 (16) | −0.0037 (15) | −0.0122 (15) |
| C14 | 0.0226 (15) | 0.0240 (17) | 0.0303 (17) | 0.0034 (13) | −0.0033 (13) | −0.0024 (14) |
| C15 | 0.040 (2) | 0.025 (2) | 0.080 (3) | −0.0091 (18) | −0.033 (2) | 0.004 (2) |
| C16 | 0.0332 (19) | 0.032 (2) | 0.041 (2) | −0.0067 (16) | −0.0092 (16) | 0.0111 (16) |
Geometric parameters (Å, º)
| S1—O2 | 1.422 (6) | C1—C2 | 1.392 (4) |
| S1—O3 | 1.435 (2) | C2—C3 | 1.373 (4) |
| S1—O4 | 1.448 (2) | C2—H2A | 0.9500 |
| S1—O2' | 1.459 (9) | C3—C6 | 1.493 (4) |
| S1—C15 | 1.797 (4) | C4—C5 | 1.380 (4) |
| S2—O5 | 1.432 (2) | C4—C7 | 1.488 (4) |
| S2—O6 | 1.439 (2) | C5—H5A | 0.9500 |
| S2—O7 | 1.448 (2) | C6—H6A | 0.9800 |
| S2—C16 | 1.825 (4) | C6—H6B | 0.9800 |
| F1—C15 | 1.352 (4) | C6—H6C | 0.9800 |
| F2—C15 | 1.368 (4) | C7—H7A | 0.9800 |
| F3—C15 | 1.393 (4) | C7—H7B | 0.9800 |
| F1'—C15 | 1.364 (4) | C7—H7C | 0.9800 |
| F2'—C15 | 1.353 (4) | C8—C9 | 1.374 (4) |
| F3'—C15 | 1.314 (4) | C8—C12 | 1.382 (4) |
| F4—C16 | 1.335 (4) | C9—C10 | 1.384 (4) |
| F5—C16 | 1.334 (4) | C9—H9A | 0.9500 |
| F6—C16 | 1.332 (4) | C10—C13 | 1.493 (4) |
| O1—C1 | 1.364 (3) | C11—C12 | 1.376 (4) |
| O1—C8 | 1.389 (3) | C11—C14 | 1.489 (4) |
| N1—C3 | 1.351 (4) | C12—H12A | 0.9500 |
| N1—C4 | 1.354 (4) | C13—H13A | 0.9800 |
| N1—H1N | 0.86 (2) | C13—H13B | 0.9800 |
| N2—C10 | 1.340 (4) | C13—H13C | 0.9800 |
| N2—C11 | 1.350 (4) | C14—H14A | 0.9800 |
| N2—H2N | 0.87 (2) | C14—H14B | 0.9800 |
| C1—C5 | 1.386 (4) | C14—H14C | 0.9800 |
| O2—S1—O3 | 120.0 (12) | H7A—C7—H7C | 109.5 |
| O2—S1—O4 | 114.2 (5) | H7B—C7—H7C | 109.5 |
| O3—S1—O4 | 114.57 (14) | C9—C8—C12 | 122.1 (3) |
| O3—S1—O2' | 108.6 (9) | C9—C8—O1 | 117.9 (3) |
| O4—S1—O2' | 112.7 (8) | C12—C8—O1 | 119.9 (3) |
| O2—S1—C15 | 98.1 (16) | C8—C9—C10 | 118.1 (3) |
| O3—S1—C15 | 102.94 (14) | C8—C9—H9A | 121.0 |
| O4—S1—C15 | 102.87 (14) | C10—C9—H9A | 121.0 |
| O2'—S1—C15 | 114.8 (13) | N2—C10—C9 | 118.6 (3) |
| O5—S2—O6 | 115.54 (17) | N2—C10—C13 | 117.8 (3) |
| O5—S2—O7 | 115.02 (14) | C9—C10—C13 | 123.6 (3) |
| O6—S2—O7 | 113.47 (15) | N2—C11—C12 | 118.2 (3) |
| O5—S2—C16 | 103.86 (15) | N2—C11—C14 | 118.0 (3) |
| O6—S2—C16 | 103.92 (17) | C12—C11—C14 | 123.8 (3) |
| O7—S2—C16 | 102.88 (17) | C11—C12—C8 | 118.4 (3) |
| C1—O1—C8 | 119.3 (2) | C11—C12—H12A | 120.8 |
| C3—N1—C4 | 123.6 (2) | C8—C12—H12A | 120.8 |
| C3—N1—H1N | 119 (2) | C10—C13—H13A | 109.5 |
| C4—N1—H1N | 117 (2) | C10—C13—H13B | 109.5 |
| C10—N2—C11 | 124.5 (3) | H13A—C13—H13B | 109.5 |
| C10—N2—H2N | 120 (2) | C10—C13—H13C | 109.5 |
| C11—N2—H2N | 114 (2) | H13A—C13—H13C | 109.5 |
| O1—C1—C5 | 123.4 (2) | H13B—C13—H13C | 109.5 |
| O1—C1—C2 | 115.6 (3) | C11—C14—H14A | 109.5 |
| C5—C1—C2 | 121.0 (3) | C11—C14—H14B | 109.5 |
| C3—C2—C1 | 119.2 (3) | H14A—C14—H14B | 109.5 |
| C3—C2—H2A | 120.4 | C11—C14—H14C | 109.5 |
| C1—C2—H2A | 120.4 | H14A—C14—H14C | 109.5 |
| N1—C3—C2 | 118.6 (3) | H14B—C14—H14C | 109.5 |
| N1—C3—C6 | 117.2 (3) | F3'—C15—F2' | 112.1 (4) |
| C2—C3—C6 | 124.2 (3) | F3'—C15—F1' | 113.4 (4) |
| N1—C4—C5 | 119.1 (3) | F2'—C15—F1' | 104.6 (4) |
| N1—C4—C7 | 117.4 (3) | F1—C15—F2 | 103.8 (4) |
| C5—C4—C7 | 123.4 (3) | F1—C15—F3 | 101.0 (4) |
| C4—C5—C1 | 118.4 (3) | F2—C15—F3 | 102.9 (4) |
| C4—C5—H5A | 120.8 | F3'—C15—S1 | 120.6 (3) |
| C1—C5—H5A | 120.8 | F1—C15—S1 | 122.0 (3) |
| C3—C6—H6A | 109.5 | F2'—C15—S1 | 102.7 (3) |
| C3—C6—H6B | 109.5 | F1'—C15—S1 | 101.5 (3) |
| H6A—C6—H6B | 109.5 | F2—C15—S1 | 121.0 (3) |
| C3—C6—H6C | 109.5 | F3—C15—S1 | 102.5 (3) |
| H6A—C6—H6C | 109.5 | F6—C16—F5 | 107.7 (3) |
| H6B—C6—H6C | 109.5 | F6—C16—F4 | 107.9 (3) |
| C4—C7—H7A | 109.5 | F5—C16—F4 | 107.7 (3) |
| C4—C7—H7B | 109.5 | F6—C16—S2 | 111.2 (2) |
| H7A—C7—H7B | 109.5 | F5—C16—S2 | 111.3 (2) |
| C4—C7—H7C | 109.5 | F4—C16—S2 | 110.9 (3) |
| C8—O1—C1—C5 | 15.1 (4) | O2'—S1—C15—F3' | −178.5 (8) |
| C8—O1—C1—C2 | −165.8 (3) | O2—S1—C15—F1 | −63.4 (9) |
| O1—C1—C2—C3 | −180.0 (3) | O3—S1—C15—F1 | 60.0 (4) |
| C5—C1—C2—C3 | −0.9 (4) | O4—S1—C15—F1 | 179.4 (4) |
| C4—N1—C3—C2 | −1.3 (4) | O2'—S1—C15—F1 | −57.7 (9) |
| C4—N1—C3—C6 | 178.2 (3) | O2—S1—C15—F2' | 50.2 (9) |
| C1—C2—C3—N1 | 0.6 (4) | O3—S1—C15—F2' | 173.7 (4) |
| C1—C2—C3—C6 | −178.8 (3) | O4—S1—C15—F2' | −67.0 (4) |
| C3—N1—C4—C5 | 2.1 (4) | O2'—S1—C15—F2' | 55.9 (9) |
| C3—N1—C4—C7 | −176.1 (3) | O2—S1—C15—F1' | −57.8 (8) |
| N1—C4—C5—C1 | −2.2 (4) | O3—S1—C15—F1' | 65.6 (3) |
| C7—C4—C5—C1 | 175.8 (3) | O4—S1—C15—F1' | −175.0 (3) |
| O1—C1—C5—C4 | −179.3 (3) | O2'—S1—C15—F1' | −52.2 (8) |
| C2—C1—C5—C4 | 1.6 (4) | O2—S1—C15—F2 | 71.3 (9) |
| C1—O1—C8—C9 | −121.9 (3) | O3—S1—C15—F2 | −165.3 (4) |
| C1—O1—C8—C12 | 61.5 (3) | O4—S1—C15—F2 | −45.9 (4) |
| C12—C8—C9—C10 | −1.9 (4) | O2'—S1—C15—F2 | 76.9 (9) |
| O1—C8—C9—C10 | −178.4 (2) | O2—S1—C15—F3 | −175.1 (8) |
| C11—N2—C10—C9 | 0.1 (4) | O3—S1—C15—F3 | −51.7 (3) |
| C11—N2—C10—C13 | 179.5 (3) | O4—S1—C15—F3 | 67.7 (3) |
| C8—C9—C10—N2 | 0.7 (4) | O2'—S1—C15—F3 | −169.4 (8) |
| C8—C9—C10—C13 | −178.6 (3) | O5—S2—C16—F6 | −178.0 (3) |
| C10—N2—C11—C12 | 0.3 (4) | O6—S2—C16—F6 | 60.8 (3) |
| C10—N2—C11—C14 | −178.4 (3) | O7—S2—C16—F6 | −57.8 (3) |
| N2—C11—C12—C8 | −1.4 (4) | O5—S2—C16—F5 | −57.9 (3) |
| C14—C11—C12—C8 | 177.2 (3) | O6—S2—C16—F5 | −179.2 (3) |
| C9—C8—C12—C11 | 2.3 (4) | O7—S2—C16—F5 | 62.3 (3) |
| O1—C8—C12—C11 | 178.7 (2) | O5—S2—C16—F4 | 62.0 (3) |
| O2—S1—C15—F3' | 175.9 (9) | O6—S2—C16—F4 | −59.3 (3) |
| O3—S1—C15—F3' | −60.7 (4) | O7—S2—C16—F4 | −177.8 (2) |
| O4—S1—C15—F3' | 58.7 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O4 | 0.86 (2) | 1.93 (2) | 2.783 (3) | 171 (3) |
| N2—H2N···O7 | 0.87 (2) | 1.97 (2) | 2.826 (3) | 169 (3) |
| C2—H2A···O6i | 0.95 | 2.36 | 3.170 (4) | 142 |
| C6—H6B···O6i | 0.98 | 2.50 | 3.383 (4) | 149 |
| C7—H7B···O3ii | 0.98 | 2.47 | 3.421 (4) | 164 |
| C9—H9A···O3iii | 0.95 | 2.44 | 3.293 (4) | 149 |
| C12—H12A···O5iv | 0.95 | 2.26 | 3.168 (4) | 160 |
| C14—H14A···O6 | 0.98 | 2.52 | 3.436 (4) | 155 |
Symmetry codes: (i) −x+2, −y+1, −z+1; (ii) x+1/2, −y+1/2, z+1/2; (iii) −x+1, −y+1, −z+1; (iv) x−1/2, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FF2121).
References
- Bruker (2005). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Constable, E. C., Cargill Thompson, A. M. W., Harveson, P., Macko, L. & Zehnder, M. (1995). Chem. Eur. J. 1, 360–367.
- Dunne, S. J., von Nagy-Felsobuki, E. I. & Mackay, M. F. (1996). Acta Cryst. C52, 2040–2042.
- Heijden, S. P. N. van der, Griffith, E. A. H., Chandler, W. D. & Robertson, B. E. (1975). Can. J. Chem. 53, 2084–2092.
- Maas, G. (1985). J. Chem. Soc. Perkin Trans. 2, pp. 1985–1988.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S1600536813027505/ff2121sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813027505/ff2121Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813027505/ff2121Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report


