Abstract
The molecular structure of the title compound, C11H13IN4O3, shows a ribofuranosyl–pyrrolo O—C—N—C torsion angle of 59.1 (3)°, with the central C—N bond length being 1.446 (3) Å. The C—I bond length is 2.072 (2) Å. The amino group is coplanar with the attached aromatic ring [C—N—C—N torsion angle = −178.8 (2)°] and forms an intramolecular N—H⋯I hydrogen bond. In the crystal, O—H⋯N and N—H⋯O hydrogen bonds link the molecules into puckered layers parallel to (001). These layers are bound to each other by secondary I⋯O interactions [3.2250 (17) Å], forming a three-dimensional framework.
Related literature
For background to the use of marine natural products as therapeutic agents, see: Kazlauskas et al. (1983 ▶); Mitchell et al. (1996 ▶); Wiesner et al. (1999 ▶); Ugarkar et al. (2000 ▶); Song et al. (2011 ▶). For the structures of related compounds, see: Seela et al. (1996 ▶, 1999 ▶, 2008 ▶).
Experimental
Crystal data
C11H13IN4O3
M r = 376.15
Orthorhombic,

a = 4.9164 (2) Å
b = 14.6490 (5) Å
c = 18.0130 (6) Å
V = 1297.30 (8) Å3
Z = 4
Mo Kα radiation
μ = 2.48 mm−1
T = 130 K
0.49 × 0.08 × 0.08 mm
Data collection
Bruker SMART APEX diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2004 ▶) T min = 0.376, T max = 0.826
12307 measured reflections
3103 independent reflections
3056 reflections with I > 2σ(I)
R int = 0.019
Refinement
R[F 2 > 2σ(F 2)] = 0.020
wR(F 2) = 0.051
S = 1.06
3103 reflections
175 parameters
H-atom parameters constrained
Δρmax = 1.05 e Å−3
Δρmin = −0.30 e Å−3
Absolute structure: Flack (1983 ▶), 1274 Friedel pairs
Absolute structure parameter: −0.015 (17)
Data collection: SMART (Bruker, 2002 ▶); cell refinement: SAINT (Bruker, 2002 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and local programs.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813027931/kq2009sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813027931/kq2009Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813027931/kq2009Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2⋯N2i | 0.84 | 1.94 | 2.759 (3) | 165 |
| O3—H3⋯N1ii | 0.84 | 2.16 | 2.907 (3) | 149 |
| N4—H4A⋯O2iii | 0.88 | 2.07 | 2.915 (3) | 160 |
| N4—H4B⋯I1 | 0.88 | 2.92 | 3.636 (2) | 139 |
Symmetry codes: (i)  ; (ii)
; (ii) 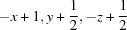 ; (iii)
; (iii)  .
.
supplementary crystallographic information
1. Comment
Marine natural products provide a rich source of chemical diversity that can be used to develop new, potentially useful therapeutic agents. Nucleosides from marine organisms show great potential as lead compounds in medicinal chemistry research. 5'-Deoxy-5-iodotubercidin (5'd-5IT, 4-amino-5-iodo-7-(5-deoxy-β-D-ribofuranosyl)pyrrolo[2,3-d]pyrimidine) was isolated from the marine red alga Hypnea vanlendiae (Kazlauskas et al., 1983) and from the marine ascidian Didemnum voeltzkowi (Mitchell et al., 1996). In vitro, all 5'-deoxytubercidin marine nucleosides show strong inhibitory activity for human adenosine kinase with 5'd-5IT being the most potent one. Therapeutic success of adenosine kinase inhibitors as active agents in animal models is documented for epilepsy and pain and as antiseizure agents (Wiesner et al., 1999; Ugarkar et al., 2000). The molecular structure is related to the derivatives studied previously (Seela et al., 1996, 1999, 2008) and shows no unexpected geometric parameters.
2. Experimental
The title compound was synthesized according to a known procedure (Song et al., 2011). Recrystallization from ethanol-water (1:1) yielded crystals suitable for X-ray analysis. Spectroscopic analysis: 1H NMR (250 MHz, DMSO-d6, δ): 1.29 (d, J = 6.16 Hz, 3H, CHCH3), 3.87–3.95 [m, 2H, H-3', H-4'], 4.41 (m, 1H, H-2'), 5.11 (s, 1H, 3'-OH), 5.33 (s, 1H, 2'-OH), 6.02 (d, J = 5.28 Hz, 1H, H-1'), 6.69 (s, 2H, 4-NH2), 7.63 (s, 1H, H-6), 8.14 (s, 1H, H-2); 13C NMR (250 MHz, DMSO-d6, δ): 19.6 (CH3, C-5'), 52.8 (C—I, C-5), 73.8 (CH, C-2'), 75.0 (CH, C-3'), 79.8 (CH, C-4'), 87.4 (CH, C-1'), 103.6 (C═C, C-4a), 127.4 (CH, C-6), 150.8 (C═C, C-7a), 152.5 (CH, C-2), 157.6 (C—NH2, C-4).
3. Refinement
Hydrogen atoms were clearly identified in difference syntheses, refined at idealized positions riding on the carbon, nitrogen or oxygen atoms with C—H 0.95–1.00, N—H 0.88, O—H 0.84 Å and with isotropic displacement parameters Uiso(H) = 1.2Ueq(C/N) or 1.5Ueq(—CH3 and —OH H atoms). All CH3 and OH hydrogen atoms were allowed to rotate but not to tip. The max. electron density residual is close (0.9 Å) to the I1 position.
Figures
Fig. 1.
Molecular structure of the title compound with anisotropic displacement parameters drawn at the 50% probability level. The intramolecular N4–H4b···I1 hydrogen bond depicted as dashed line.
Fig. 2.
Crystal packing viewed along a axis with intermolecular I···O interaction as well as hydrogen bonds as dashed lines.
Crystal data
| C11H13IN4O3 | F(000) = 736 |
| Mr = 376.15 | Dx = 1.926 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 8237 reflections |
| a = 4.9164 (2) Å | θ = 2.7–28.3° |
| b = 14.6490 (5) Å | µ = 2.48 mm−1 |
| c = 18.0130 (6) Å | T = 130 K |
| V = 1297.30 (8) Å3 | Needle, colourless |
| Z = 4 | 0.49 × 0.08 × 0.08 mm |
Data collection
| Bruker SMART APEX diffractometer | 3103 independent reflections |
| Radiation source: sealed tube | 3056 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.019 |
| φ and ω scans | θmax = 27.9°, θmin = 1.8° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2004) | h = −6→6 |
| Tmin = 0.376, Tmax = 0.826 | k = −19→19 |
| 12307 measured reflections | l = −23→22 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.020 | H-atom parameters constrained |
| wR(F2) = 0.051 | w = 1/[σ2(Fo2) + (0.0319P)2 + 0.5639P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max = 0.001 |
| 3103 reflections | Δρmax = 1.05 e Å−3 |
| 175 parameters | Δρmin = −0.30 e Å−3 |
| 0 restraints | Absolute structure: Flack (1983), 1274 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Absolute structure parameter: −0.015 (17) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| I1 | 0.89717 (3) | 0.148805 (10) | 0.080859 (9) | 0.02262 (6) | |
| O1 | 0.5927 (4) | 0.52637 (11) | 0.01354 (9) | 0.0194 (3) | |
| O2 | 1.0140 (4) | 0.58588 (12) | 0.16890 (10) | 0.0197 (4) | |
| H2 | 1.1212 | 0.5464 | 0.1858 | 0.030* | |
| O3 | 0.7492 (4) | 0.71478 (11) | 0.08382 (10) | 0.0228 (4) | |
| H3 | 0.8160 | 0.7236 | 0.1261 | 0.034* | |
| N1 | 0.2124 (5) | 0.28946 (15) | 0.26704 (12) | 0.0224 (4) | |
| N2 | 0.3108 (4) | 0.43351 (14) | 0.20816 (11) | 0.0189 (4) | |
| N3 | 0.6561 (4) | 0.42430 (13) | 0.11225 (11) | 0.0162 (4) | |
| N4 | 0.4171 (5) | 0.15312 (15) | 0.23436 (11) | 0.0243 (4) | |
| H4A | 0.3194 | 0.1255 | 0.2686 | 0.029* | |
| H4B | 0.5318 | 0.1215 | 0.2070 | 0.029* | |
| C1 | 0.8108 (5) | 0.35459 (17) | 0.08162 (13) | 0.0192 (4) | |
| H1A | 0.9408 | 0.3619 | 0.0430 | 0.023* | |
| C2 | 0.7476 (5) | 0.27404 (16) | 0.11546 (14) | 0.0191 (5) | |
| C3 | 0.5438 (5) | 0.29313 (16) | 0.17014 (13) | 0.0160 (5) | |
| C4 | 0.3910 (6) | 0.24402 (16) | 0.22372 (12) | 0.0185 (5) | |
| C5 | 0.1823 (5) | 0.37938 (18) | 0.25567 (14) | 0.0223 (5) | |
| H5A | 0.0501 | 0.4085 | 0.2860 | 0.027* | |
| C6 | 0.4924 (5) | 0.38688 (16) | 0.16605 (13) | 0.0164 (5) | |
| C7 | 0.6628 (5) | 0.51898 (15) | 0.08971 (13) | 0.0154 (4) | |
| H7A | 0.5302 | 0.5547 | 0.1202 | 0.018* | |
| C8 | 0.9424 (5) | 0.56288 (15) | 0.09505 (12) | 0.0150 (5) | |
| H8A | 1.0830 | 0.5217 | 0.0730 | 0.018* | |
| C9 | 0.9025 (5) | 0.64656 (15) | 0.04619 (12) | 0.0178 (4) | |
| H9A | 1.0796 | 0.6710 | 0.0274 | 0.021* | |
| C10 | 0.7272 (5) | 0.60780 (16) | −0.01690 (13) | 0.0182 (5) | |
| H10A | 0.5861 | 0.6538 | −0.0312 | 0.022* | |
| C11 | 0.8868 (6) | 0.58060 (18) | −0.08480 (14) | 0.0274 (5) | |
| H11A | 1.0502 | 0.5471 | −0.0697 | 0.041* | |
| H11B | 0.9398 | 0.6355 | −0.1124 | 0.041* | |
| H11C | 0.7742 | 0.5415 | −0.1165 | 0.041* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| I1 | 0.02487 (8) | 0.01568 (8) | 0.02731 (8) | 0.00340 (6) | −0.00081 (7) | −0.00292 (6) |
| O1 | 0.0223 (8) | 0.0186 (8) | 0.0174 (7) | −0.0041 (7) | −0.0021 (8) | 0.0033 (6) |
| O2 | 0.0225 (9) | 0.0172 (8) | 0.0193 (8) | 0.0048 (7) | −0.0047 (7) | −0.0027 (7) |
| O3 | 0.0320 (10) | 0.0158 (7) | 0.0207 (8) | 0.0065 (7) | −0.0005 (9) | −0.0020 (7) |
| N1 | 0.0230 (11) | 0.0239 (10) | 0.0203 (10) | −0.0025 (9) | 0.0016 (9) | 0.0048 (8) |
| N2 | 0.0217 (10) | 0.0181 (10) | 0.0170 (9) | 0.0027 (8) | 0.0005 (8) | 0.0030 (8) |
| N3 | 0.0177 (11) | 0.0120 (9) | 0.0191 (9) | 0.0006 (7) | 0.0022 (8) | 0.0007 (7) |
| N4 | 0.0286 (11) | 0.0227 (10) | 0.0216 (10) | −0.0061 (12) | 0.0010 (9) | 0.0076 (8) |
| C1 | 0.0192 (10) | 0.0170 (10) | 0.0213 (10) | 0.0002 (9) | 0.0033 (9) | −0.0001 (11) |
| C2 | 0.0207 (13) | 0.0160 (11) | 0.0206 (11) | 0.0005 (9) | −0.0014 (10) | 0.0000 (9) |
| C3 | 0.0162 (12) | 0.0145 (10) | 0.0174 (10) | −0.0010 (8) | −0.0018 (9) | 0.0019 (8) |
| C4 | 0.0209 (12) | 0.0172 (10) | 0.0175 (10) | −0.0019 (10) | −0.0055 (11) | 0.0043 (8) |
| C5 | 0.0231 (12) | 0.0227 (11) | 0.0210 (12) | 0.0039 (10) | 0.0030 (10) | 0.0017 (9) |
| C6 | 0.0167 (10) | 0.0174 (11) | 0.0149 (10) | −0.0008 (9) | −0.0026 (9) | 0.0042 (9) |
| C7 | 0.0150 (10) | 0.0130 (9) | 0.0182 (11) | −0.0019 (8) | −0.0009 (9) | 0.0020 (8) |
| C8 | 0.0154 (12) | 0.0135 (10) | 0.0161 (11) | 0.0006 (8) | 0.0007 (8) | −0.0024 (8) |
| C9 | 0.0205 (10) | 0.0116 (9) | 0.0212 (10) | −0.0014 (12) | 0.0032 (9) | −0.0001 (8) |
| C10 | 0.0233 (12) | 0.0136 (10) | 0.0177 (11) | −0.0011 (9) | 0.0018 (10) | 0.0027 (9) |
| C11 | 0.0375 (14) | 0.0256 (12) | 0.0190 (11) | −0.0042 (12) | 0.0058 (15) | 0.0001 (10) |
Geometric parameters (Å, º)
| I1—C2 | 2.072 (2) | C1—C2 | 1.364 (3) |
| O1—C7 | 1.419 (3) | C1—H1A | 0.9500 |
| O1—C10 | 1.470 (3) | C2—C3 | 1.433 (4) |
| O2—C8 | 1.417 (3) | C3—C6 | 1.398 (3) |
| O2—H2 | 0.8400 | C3—C4 | 1.419 (3) |
| O3—C9 | 1.423 (3) | C5—H5A | 0.9500 |
| O3—H3 | 0.8400 | C7—C8 | 1.521 (3) |
| N1—C5 | 1.341 (3) | C7—H7A | 1.0000 |
| N1—C4 | 1.350 (3) | C8—C9 | 1.522 (3) |
| N2—C5 | 1.327 (3) | C8—H8A | 1.0000 |
| N2—C6 | 1.356 (3) | C9—C10 | 1.535 (3) |
| N3—C6 | 1.374 (3) | C9—H9A | 1.0000 |
| N3—C1 | 1.388 (3) | C10—C11 | 1.507 (3) |
| N3—C7 | 1.446 (3) | C10—H10A | 1.0000 |
| N4—C4 | 1.351 (3) | C11—H11A | 0.9800 |
| N4—H4A | 0.8800 | C11—H11B | 0.9800 |
| N4—H4B | 0.8800 | C11—H11C | 0.9800 |
| C7—O1—C10 | 108.28 (17) | O1—C7—C8 | 104.37 (18) |
| C8—O2—H2 | 109.5 | N3—C7—C8 | 114.11 (19) |
| C9—O3—H3 | 109.5 | O1—C7—H7A | 109.5 |
| C5—N1—C4 | 117.9 (2) | N3—C7—H7A | 109.5 |
| C5—N2—C6 | 111.9 (2) | C8—C7—H7A | 109.5 |
| C6—N3—C1 | 107.93 (19) | O2—C8—C7 | 112.61 (19) |
| C6—N3—C7 | 126.5 (2) | O2—C8—C9 | 112.55 (18) |
| C1—N3—C7 | 125.6 (2) | C7—C8—C9 | 100.81 (18) |
| C4—N4—H4A | 120.0 | O2—C8—H8A | 110.2 |
| C4—N4—H4B | 120.0 | C7—C8—H8A | 110.2 |
| H4A—N4—H4B | 120.0 | C9—C8—H8A | 110.2 |
| C2—C1—N3 | 109.5 (2) | O3—C9—C8 | 111.00 (18) |
| C2—C1—H1A | 125.2 | O3—C9—C10 | 108.4 (2) |
| N3—C1—H1A | 125.2 | C8—C9—C10 | 101.68 (18) |
| C1—C2—C3 | 107.3 (2) | O3—C9—H9A | 111.8 |
| C1—C2—I1 | 123.41 (19) | C8—C9—H9A | 111.8 |
| C3—C2—I1 | 128.88 (18) | C10—C9—H9A | 111.8 |
| C6—C3—C4 | 116.0 (2) | O1—C10—C11 | 108.81 (19) |
| C6—C3—C2 | 106.4 (2) | O1—C10—C9 | 106.05 (18) |
| C4—C3—C2 | 137.6 (2) | C11—C10—C9 | 114.0 (2) |
| N1—C4—N4 | 117.7 (2) | O1—C10—H10A | 109.3 |
| N1—C4—C3 | 119.2 (2) | C11—C10—H10A | 109.3 |
| N4—C4—C3 | 123.1 (2) | C9—C10—H10A | 109.3 |
| N2—C5—N1 | 129.3 (2) | C10—C11—H11A | 109.5 |
| N2—C5—H5A | 115.4 | C10—C11—H11B | 109.5 |
| N1—C5—H5A | 115.4 | H11A—C11—H11B | 109.5 |
| N2—C6—N3 | 125.4 (2) | C10—C11—H11C | 109.5 |
| N2—C6—C3 | 125.7 (2) | H11A—C11—H11C | 109.5 |
| N3—C6—C3 | 108.9 (2) | H11B—C11—H11C | 109.5 |
| O1—C7—N3 | 109.84 (18) | ||
| C6—N3—C1—C2 | −0.2 (3) | C2—C3—C6—N2 | 179.6 (2) |
| C7—N3—C1—C2 | −178.7 (2) | C4—C3—C6—N3 | 179.7 (2) |
| N3—C1—C2—C3 | −0.1 (3) | C2—C3—C6—N3 | −0.4 (3) |
| N3—C1—C2—I1 | 173.35 (17) | C10—O1—C7—N3 | −151.81 (19) |
| C1—C2—C3—C6 | 0.3 (3) | C10—O1—C7—C8 | −29.1 (2) |
| I1—C2—C3—C6 | −172.66 (19) | C6—N3—C7—O1 | −119.2 (2) |
| C1—C2—C3—C4 | −179.9 (3) | C1—N3—C7—O1 | 59.1 (3) |
| I1—C2—C3—C4 | 7.2 (5) | C6—N3—C7—C8 | 124.0 (2) |
| C5—N1—C4—N4 | −178.8 (2) | C1—N3—C7—C8 | −57.7 (3) |
| C5—N1—C4—C3 | 1.9 (4) | O1—C7—C8—O2 | 162.92 (18) |
| C6—C3—C4—N1 | −0.8 (3) | N3—C7—C8—O2 | −77.2 (2) |
| C2—C3—C4—N1 | 179.3 (3) | O1—C7—C8—C9 | 42.8 (2) |
| C6—C3—C4—N4 | 179.9 (2) | N3—C7—C8—C9 | 162.66 (19) |
| C2—C3—C4—N4 | 0.1 (5) | O2—C8—C9—O3 | −44.0 (3) |
| C6—N2—C5—N1 | 1.1 (4) | C7—C8—C9—O3 | 76.2 (2) |
| C4—N1—C5—N2 | −2.2 (4) | O2—C8—C9—C10 | −159.10 (19) |
| C5—N2—C6—N3 | −179.8 (2) | C7—C8—C9—C10 | −38.9 (2) |
| C5—N2—C6—C3 | 0.2 (4) | C7—O1—C10—C11 | 126.7 (2) |
| C1—N3—C6—N2 | −179.6 (2) | C7—O1—C10—C9 | 3.7 (2) |
| C7—N3—C6—N2 | −1.1 (4) | O3—C9—C10—O1 | −94.2 (2) |
| C1—N3—C6—C3 | 0.3 (3) | C8—C9—C10—O1 | 22.8 (2) |
| C7—N3—C6—C3 | 178.9 (2) | O3—C9—C10—C11 | 146.1 (2) |
| C4—C3—C6—N2 | −0.3 (4) | C8—C9—C10—C11 | −96.9 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···N2i | 0.84 | 1.94 | 2.759 (3) | 165 |
| O3—H3···N1ii | 0.84 | 2.16 | 2.907 (3) | 149 |
| N4—H4A···O2iii | 0.88 | 2.07 | 2.915 (3) | 160 |
| N4—H4B···I1 | 0.88 | 2.92 | 3.636 (2) | 139 |
Symmetry codes: (i) x+1, y, z; (ii) −x+1, y+1/2, −z+1/2; (iii) −x+1, y−1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: KQ2009).
References
- Bruker (2002). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Kazlauskas, R., Muphy, P. T., Wells, R. J., Baird-Lambert, J. A. & Jamieson, D. D. (1983). Aust. J. Chem. 36, 165–170.
- Mitchell, S. S., Pomerantz, S. C., Conception, G. P. & Ireland, C. M. (1996). J. Nat. Prod. 59, 1000–1001. [DOI] [PubMed]
- Seela, F., Ming, X., Budow, S., Eickmeier, H. & Reuter, H. (2008). Acta Cryst. C64, o293–o295. [DOI] [PubMed]
- Seela, F., Zulauf, M., Reuter, H. & Kastner, G. (1999). Acta Cryst. C55, 1560–1562. [DOI] [PubMed]
- Seela, F., Zulauf, M., Rosemeyer, H. & Reuter, H. (1996). J. Chem. Soc. Perkin Trans. 2, pp. 2373–2376.
- Sheldrick, G. M. (2004). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Song, Y., Ding, H. X., Dou, Y. H., Yang, R. C., Sun, Q., Xiao, Q. & Ju, Y. (2011). Synthesis, 9, 1442–1446.
- Ugarkar, B. G., DaRe, J. M., Kopcho, J. J., Browne, C. E., Schanzer, J. M., Wiesner, J. B. & Erion, M. D. (2000). J. Med. Chem. 43, 2883–2893. [DOI] [PubMed]
- Wiesner, J. B., Ugarkar, B. G., Castellino, A. J., Barankiewicz, J., Dumas, D. P., Gruber, H. E., Foster, A. C. & Erion, M. D. (1999). J. Pharmacol. Exp. Ther. 289, 1669–1677. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813027931/kq2009sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813027931/kq2009Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813027931/kq2009Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




