Abstract
The title compound, C21H25NO3 [systematic name: (3aS,9aR,10aR,10bS,E)-3-[(E)-4-(4-aminobenzylidene)-6,9a-dimethyl-3a,4,5,8,9,9a,10a,10b-octahydrooxireno[2′,3′:9,10]cyclodeca[1,2-b]furan-2(3H)-one] was obtained from the reaction of parthenolide [synonym: 4,5-epoxygermacra-1(10),11(13)-dieno-12,6-lactone] with 4-iodoaniline under Heck reaction conditions. It was identified as the E-isomer (conformation about the exocyclic methylidene C=C bond; the conformation about the C=C bond in the ten-membered ring is also E). The molecule is built up from fused ten-, five- (lactone) and three-membered (epoxide) rings with a 4-aminophenyl group as a substituent. The ten-membered ring displays an approximate chair–chair conformation, while the lactone ring has an envelope conformation with the C atom bonded to the ring O atom as the flap. The dihedral angle between the benzene ring of the 4-aminophenyl moiety and the lactone ring mean plane is 23.50 (8)°. In the crystal, molecules are linked via N—H⋯O hydrogen bonds, between the amine group and the lactone and epoxide ring O atoms, forming chains propagating along the b-axis direction. Adjacent chains are linked via C—H⋯O interactions, forming an undulating two-dimensional network lying parallel to the plane (001). The absolute structure of the molecule in the crystal was confirmed by resonance scattering [Flack parameter = 0.03 (3)].
Related literature
For the biological activity of parthenolide, see: Hall et al. (1979 ▶). For the biological activity of parthenolide derivatives similar to the title compound, see: Hanson et al. (1970 ▶); Hehner et al. (1998 ▶); Kupchan et al. (1971 ▶); Nasim et al. (2011 ▶); Neelakantan et al. (2009 ▶); Oka et al. (2007 ▶); Ralstin et al. (2006 ▶); Rodriguez et al. (1976 ▶); Sun et al. (2006 ▶). For the synthesis and crystal structures of similar molecules, see: Han et al. (2009 ▶).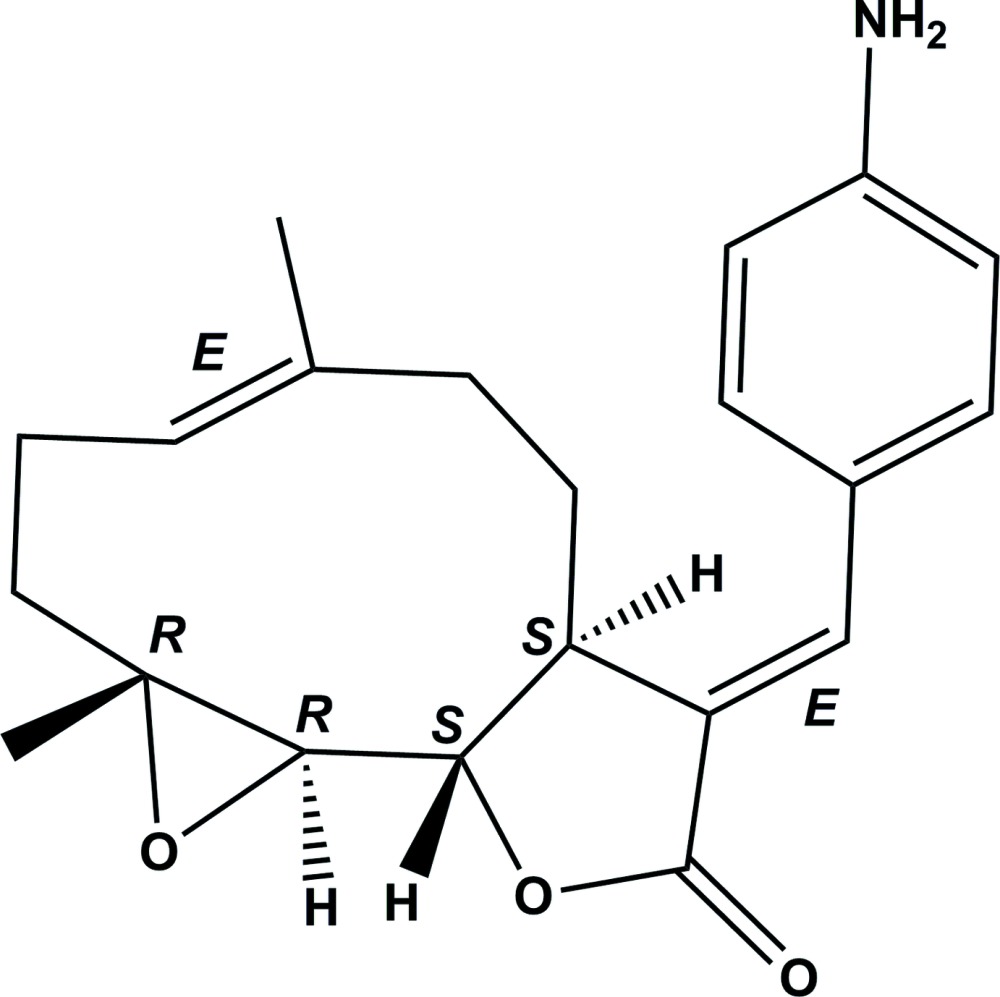
Experimental
Crystal data
C21H25NO3
M r = 339.42
Orthorhombic,
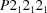
a = 8.5619 (1) Å
b = 11.8846 (2) Å
c = 17.7457 (3) Å
V = 1805.71 (5) Å3
Z = 4
Cu Kα radiation
μ = 0.66 mm−1
T = 90 K
0.20 × 0.16 × 0.12 mm
Data collection
Bruker X8 Proteum diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.843, T max = 0.956
25400 measured reflections
3296 independent reflections
3267 reflections with I > 2σ(I)
R int = 0.033
Refinement
R[F 2 > 2σ(F 2)] = 0.027
wR(F 2) = 0.070
S = 1.06
3296 reflections
234 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.13 e Å−3
Δρmin = −0.14 e Å−3
Absolute structure: Flack x parameter determined using 1383 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▶)
Absolute structure parameter: 0.03 (3)
Data collection: APEX2 (Bruker, 2006 ▶); cell refinement: SAINT (Bruker, 2006 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL2013 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813028730/su2647sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813028730/su2647Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1A⋯O1i | 0.94 (2) | 2.25 (3) | 3.133 (2) | 156 (2) |
| N1—H2B⋯O2i | 0.90 (2) | 2.57 (2) | 3.077 (2) | 116 (2) |
| C2—H2A⋯O3ii | 0.99 | 2.63 | 3.372 (2) | 132 |
Symmetry codes: (i)  ; (ii)
; (ii) 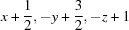 .
.
Acknowledgments
This work was supported by NIH/NCI [grant No. CA158275].
supplementary crystallographic information
1. Comment
In recent years the parthenolide molecule [systematic name: (1aR,7aS,10aS,10bS,Z)-1a,5-dimethyl-8-methylene- 2,3,6,7,7a,8,10a,10b-octahydrooxireno[2',3':9,10]cyclodeca [1,2-b]furan-9(1aH)-one] and several structurally related sesquiterpene lactone analogues have been extensively studied due to their potent anti-tumor and cytotoxic properties (Oka et al., 2007; Ralstin et al., 2006; Sun et al., 2006). The presence of an exo-methylene-γ-lactone functionality in these molecules is an important structural feature because of its exceptional reactivity toward nucleophilic functional groups (Rodriguez et al., 1976). The exo-methylene-γ-lactone moiety was reported to be essential for significant cytotoxic activity (Kupchan et al., 1971), for inhibition of nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB; Hehner et al., 1998) and for the antitumor activity of these sesquiterpene lactones (Hanson et al., 1970).
Parthenolide is a sesquiterpene lactone of the germacranolide class which occurs naturally in the plant feverfew (Tanacetum parthenium), and is believed to be the active chemical constituent responsible for the plant's biological activity (Hall et al., 1979). It has become a key intermediate for the synthesis of several novel antileukemic compounds over the past decade. From our research on antileukemia analogues of parthenolide, we have improved the poor water solubility properties of parthenolide and related sesquiterpenes by incorporating amino moieties at the exocyclic methylidene carbon via Michael addition (Neelakantan et al., 2009). Also, in a recent communication we have reported the synthesis and antileukemic activity of a melampolide sesquiterpene lactone, melampomagnolide B (Nasim et al., 2011).
The current study focuses on the synthesis of the title E-olefinic analogue of parthenolide which was obtained from the reaction of parthenolide with 4-iodoaniline utilizing Heck chemistry (Han et al., 2009). In order to obtain detailed information on the structural conformation of the title compound and to establish the geometry of the exocyclic double bond, a single-crystal X-ray structure determination has been carried out.
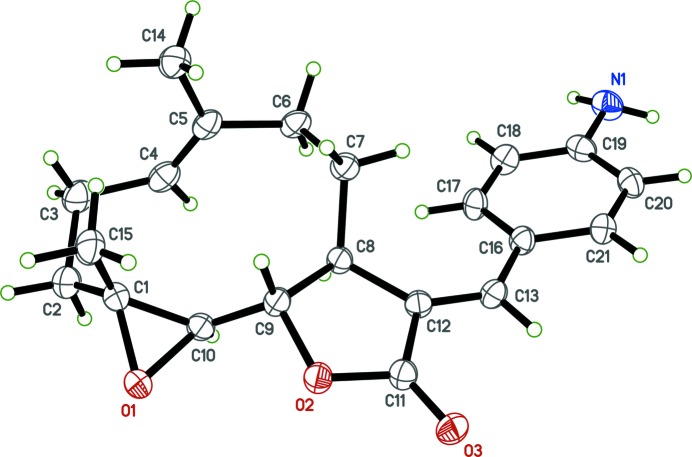
The title molecule, Fig. 1, contains an E-exocyclic olefinic bond C12═C13. The ten-membered ring displays an approximate chair-chair conformation, while the lactone ring has an envelope conformation with atom C9 as the flap. The dihedral angle between the benzene ring of the 4-aminophenyl moiety and the lactone ring mean plane is 23.50 (8) °.
The crystal structures of the structurally related 3-trifluoromethylphenyl and 2-trifluoromethylphenyl derivatives of parthenolide have been reported previously (Han et al., 2009). These molecules also have an E-conformation about the exocyclic olefinic double bond, and their absolute stereochemistries were also determined by resonance scattering. The lactone rings of these compounds have envelope conformations with the C-atom bonded to the O-atom as the flap, as in the title compound. The dihedral angles between the lactone ring mean plane and the aromatic rings of the 3-trifluoromethylphenyl and 2-trifluoromethylphenyl moieties are 40.52 (12) and 48.07 (12) °, respectively, compared to 23.50 (8) ° in the title compound.
Significant deviations from the ideal bond angle geometry around the carbon atoms, C12, C13 and C11, that are in the sp2 state and involved in the double bonds are observed; the C12═C13–C16, C13═C12–C8 and O3═ C11–C12 bond angles with the respective values of 131.43 (14), 132.77 (14), and 129.80 (14)°, deviate from the ideal value of 120°.
In the crystal, molecules are linked via N—H···O hydrogen bonds (Fig. 2 and Table 1), between the amine group and the lactone and epoxide ring O atoms, forming chains propagating along the b-axis. Adjacent chains are linked via C—H···O interactions (Table 1) forming undulating two-dimensional networks lying parallel to the plane (001).
2. Experimental
A mixture of parthenolide [Chemtek, Worcester, MA, USA](50 mg, 0.20 mmol), triethylamine (60 mg, 0.61 mmol), and 4-iodoaniline (48.56 mg, 0.22 mmol) in dimethylformamide (0.1 ml) was treated with palladium(II) acetate (0.5 mg, 0.002 mmol) and then stirred at 353 K for 24 h. Han et al. (2009) reported the synthesis of similar chiral molecules. The reaction mixture was cooled to room temperature, water (8 ml) was added, and the mixture was extracted with ethyl acetate (10 ml × 3). The separated organics were dried over Na2SO4 and concentrated under reduced pressure. The obtained crude residue was purified using silica flash chromatography (7:3, hexanes/EtOAc) to afford the title compound, which was recrystallized from a mixture of hexanes and ethyl acetate(1:1) as yellow needle-like crystals (40 mg, 58 % yield; M.p. = 512–514 K). Spectroscopic data for the title compound are available in the archived CIF.
3. Refinement
All the H-atoms were located in difference electron density maps. The NH2 H atoms were refined with Uiso(H) = 1.5Ueq(N). The C-bound H atoms were placed in idealized positions and refined as riding atoms: C-H = 0.98, 0.99, 1.00 and 0.95 Å for CH3, CH2, CH and Csp2H H atoms, respectively, with Uiso(H) = 1.5Ueq(C-methyl) and = 1.2Ueq(C) for other H atoms.
Figures
Fig. 1.
The molecular structure of the title molecule, with atom labelling. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
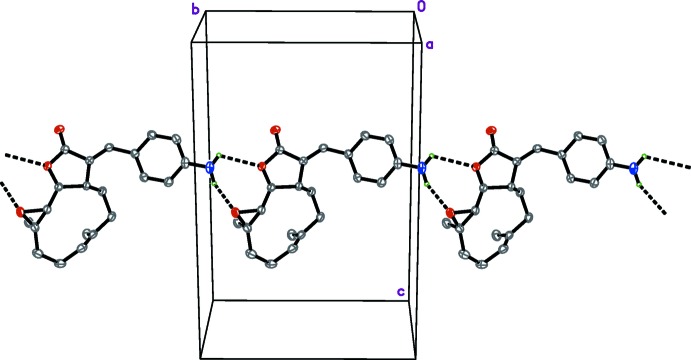
A partial crystal packing plot of the title compound showing the N-H···O hydrogen bonds that join the molecules into chains parallel to the b axis (see Table 1 for details; H atoms not involved in these hydrogen-bonds have been omitted to enhance clarity).
Crystal data
| C21H25NO3 | Dx = 1.249 Mg m−3 |
| Mr = 339.42 | Melting point = 512–514 K |
| Orthorhombic, P212121 | Cu Kα radiation, λ = 1.54178 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 9832 reflections |
| a = 8.5619 (1) Å | θ = 5.2–68.0° |
| b = 11.8846 (2) Å | µ = 0.66 mm−1 |
| c = 17.7457 (3) Å | T = 90 K |
| V = 1805.71 (5) Å3 | Block, colourless |
| Z = 4 | 0.20 × 0.16 × 0.12 mm |
| F(000) = 728 |
Data collection
| Bruker X8 Proteum diffractometer | 3296 independent reflections |
| Radiation source: fine-focus rotating anode | 3267 reflections with I > 2σ(I) |
| Detector resolution: 5.6 pixels mm-1 | Rint = 0.033 |
| φ and ω scans | θmax = 68.2°, θmin = 5.7° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −10→10 |
| Tmin = 0.843, Tmax = 0.956 | k = −11→14 |
| 25400 measured reflections | l = −21→15 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.027 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.070 | w = 1/[σ2(Fo2) + (0.0401P)2 + 0.303P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max < 0.001 |
| 3296 reflections | Δρmax = 0.13 e Å−3 |
| 234 parameters | Δρmin = −0.14 e Å−3 |
| 0 restraints | Absolute structure: Flack parameter determined using 1383 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| Primary atom site location: structure-invariant direct methods | Absolute structure parameter: 0.03 (3) |
Special details
| Experimental. Spectroscopic data for the title compound: 1H NMR (400 MHz, CDCl3): δ 7.54-7.55 (d, J=2.8 Hz, 1H), 7.23-7.26 (d, J=8.4 Hz, 2H), 6.68-6.70 (d, J=8.0 Hz, 2H), 5.28-5.30 (d, J=11.2 Hz, 1H), 4.02 (brs, 2H), 3.91-3.95 (t, J=8.0Hz, 1H), 3.25 (m, 1H), 2.82-2.84 (d, J=8.8 Hz, 1H), 2.28-2.45 (m, 1H), 2.14-2.24 (m, 5H), 1.69 (s, 3H), 1.61 (s, 1H), 1.43-1.45 (m, 1H), 1.31(s, 3H); 13C NMR (100 MHz,CDCl3) δ 171.63, 148.15, 138.64, 134.79, 132.17, 125.12, 124.03, 123.29, 114.42, 82.71, 66.62, 61.58, 46.98, 41.88, 36.10, 29.33, 24.33, 17.49, 17.36 ppm. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.69671 (14) | 0.83851 (9) | 0.57032 (6) | 0.0269 (3) | |
| O2 | 0.57735 (14) | 0.71820 (9) | 0.43765 (6) | 0.0247 (3) | |
| O3 | 0.57901 (15) | 0.65318 (10) | 0.31945 (6) | 0.0276 (3) | |
| N1 | 0.48608 (19) | −0.03270 (12) | 0.45455 (10) | 0.0323 (3) | |
| H1A | 0.539 (3) | −0.055 (2) | 0.4989 (14) | 0.049* | |
| H2B | 0.495 (3) | −0.077 (2) | 0.4135 (14) | 0.049* | |
| C1 | 0.61245 (19) | 0.79003 (14) | 0.63430 (9) | 0.0255 (4) | |
| C2 | 0.7040 (2) | 0.76874 (16) | 0.70559 (9) | 0.0321 (4) | |
| H1 | 0.7143 | 0.8397 | 0.7344 | 0.038* | |
| H2A | 0.8102 | 0.7420 | 0.6926 | 0.038* | |
| C3 | 0.6202 (2) | 0.67944 (15) | 0.75440 (9) | 0.0325 (4) | |
| H3A | 0.6881 | 0.6579 | 0.7972 | 0.039* | |
| H3B | 0.5227 | 0.7117 | 0.7752 | 0.039* | |
| C4 | 0.5821 (2) | 0.57669 (14) | 0.70835 (9) | 0.0277 (4) | |
| H4 | 0.6688 | 0.5325 | 0.6928 | 0.033* | |
| C5 | 0.4420 (2) | 0.53993 (13) | 0.68657 (8) | 0.0248 (3) | |
| C6 | 0.4284 (2) | 0.44729 (13) | 0.62863 (8) | 0.0259 (3) | |
| H6A | 0.3432 | 0.3957 | 0.6438 | 0.031* | |
| H6B | 0.5268 | 0.4035 | 0.6282 | 0.031* | |
| C7 | 0.39551 (19) | 0.49041 (13) | 0.54839 (8) | 0.0237 (3) | |
| H7A | 0.3090 | 0.5455 | 0.5508 | 0.028* | |
| H7B | 0.3597 | 0.4263 | 0.5172 | 0.028* | |
| C8 | 0.53600 (18) | 0.54638 (12) | 0.50866 (8) | 0.0198 (3) | |
| H8 | 0.6346 | 0.5178 | 0.5321 | 0.024* | |
| C9 | 0.53593 (18) | 0.67758 (12) | 0.51252 (8) | 0.0209 (3) | |
| H9 | 0.4293 | 0.7050 | 0.5263 | 0.025* | |
| C10 | 0.65311 (18) | 0.72056 (12) | 0.56849 (8) | 0.0219 (3) | |
| H10 | 0.7423 | 0.6679 | 0.5775 | 0.026* | |
| C11 | 0.56586 (19) | 0.63346 (13) | 0.38588 (8) | 0.0225 (3) | |
| C12 | 0.54277 (18) | 0.52539 (12) | 0.42479 (8) | 0.0207 (3) | |
| C13 | 0.53493 (19) | 0.43138 (13) | 0.38311 (8) | 0.0223 (3) | |
| H13 | 0.5363 | 0.4441 | 0.3303 | 0.027* | |
| C14 | 0.2884 (2) | 0.58698 (15) | 0.71301 (10) | 0.0321 (4) | |
| H14A | 0.3071 | 0.6429 | 0.7526 | 0.048* | |
| H14B | 0.2236 | 0.5260 | 0.7331 | 0.048* | |
| H14C | 0.2345 | 0.6227 | 0.6706 | 0.048* | |
| C15 | 0.4537 (2) | 0.84179 (14) | 0.64419 (9) | 0.0301 (4) | |
| H15A | 0.4639 | 0.9142 | 0.6702 | 0.045* | |
| H15B | 0.3878 | 0.7913 | 0.6741 | 0.045* | |
| H15C | 0.4057 | 0.8537 | 0.5947 | 0.045* | |
| C16 | 0.52461 (18) | 0.31328 (13) | 0.40450 (9) | 0.0228 (3) | |
| C17 | 0.5641 (2) | 0.27104 (12) | 0.47583 (8) | 0.0253 (3) | |
| H17 | 0.6003 | 0.3214 | 0.5136 | 0.030* | |
| C18 | 0.5517 (2) | 0.15768 (13) | 0.49238 (9) | 0.0276 (3) | |
| H18 | 0.5777 | 0.1314 | 0.5414 | 0.033* | |
| C19 | 0.50076 (19) | 0.08115 (13) | 0.43735 (10) | 0.0266 (3) | |
| C20 | 0.46429 (19) | 0.12208 (14) | 0.36578 (9) | 0.0266 (3) | |
| H20 | 0.4306 | 0.0715 | 0.3276 | 0.032* | |
| C21 | 0.47670 (19) | 0.23526 (13) | 0.35002 (9) | 0.0245 (3) | |
| H21 | 0.4521 | 0.2611 | 0.3008 | 0.029* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0341 (6) | 0.0208 (5) | 0.0258 (6) | −0.0064 (5) | 0.0045 (5) | −0.0028 (5) |
| O2 | 0.0368 (6) | 0.0174 (5) | 0.0199 (5) | −0.0020 (5) | 0.0000 (5) | 0.0017 (4) |
| O3 | 0.0358 (6) | 0.0279 (6) | 0.0190 (5) | −0.0027 (5) | −0.0006 (5) | 0.0043 (4) |
| N1 | 0.0364 (8) | 0.0191 (7) | 0.0415 (8) | −0.0011 (6) | 0.0020 (7) | 0.0013 (6) |
| C1 | 0.0310 (8) | 0.0222 (7) | 0.0232 (8) | −0.0057 (6) | 0.0029 (6) | −0.0029 (6) |
| C2 | 0.0339 (9) | 0.0375 (9) | 0.0248 (8) | −0.0065 (8) | −0.0004 (7) | −0.0055 (7) |
| C3 | 0.0343 (9) | 0.0430 (10) | 0.0201 (8) | −0.0023 (8) | −0.0030 (7) | 0.0005 (7) |
| C4 | 0.0320 (8) | 0.0297 (8) | 0.0212 (7) | 0.0066 (7) | 0.0028 (7) | 0.0048 (6) |
| C5 | 0.0342 (9) | 0.0220 (7) | 0.0183 (7) | 0.0041 (7) | 0.0033 (7) | 0.0048 (6) |
| C6 | 0.0346 (9) | 0.0194 (7) | 0.0236 (7) | 0.0020 (7) | 0.0069 (7) | 0.0046 (6) |
| C7 | 0.0269 (8) | 0.0203 (7) | 0.0238 (8) | −0.0003 (6) | 0.0024 (6) | 0.0014 (6) |
| C8 | 0.0237 (8) | 0.0164 (7) | 0.0191 (7) | 0.0006 (6) | 0.0002 (6) | 0.0006 (5) |
| C9 | 0.0266 (8) | 0.0175 (7) | 0.0185 (7) | 0.0021 (6) | 0.0016 (6) | 0.0008 (5) |
| C10 | 0.0261 (7) | 0.0177 (7) | 0.0220 (7) | −0.0004 (6) | 0.0016 (6) | 0.0014 (6) |
| C11 | 0.0256 (8) | 0.0206 (7) | 0.0212 (7) | 0.0012 (6) | −0.0012 (6) | −0.0002 (6) |
| C12 | 0.0229 (7) | 0.0194 (7) | 0.0197 (7) | −0.0002 (6) | −0.0007 (6) | 0.0014 (6) |
| C13 | 0.0254 (8) | 0.0233 (8) | 0.0183 (7) | 0.0023 (6) | −0.0002 (6) | 0.0009 (6) |
| C14 | 0.0346 (9) | 0.0279 (8) | 0.0340 (9) | 0.0007 (8) | 0.0070 (7) | −0.0037 (7) |
| C15 | 0.0374 (9) | 0.0242 (8) | 0.0287 (8) | 0.0011 (7) | 0.0045 (7) | −0.0054 (7) |
| C16 | 0.0260 (8) | 0.0196 (7) | 0.0229 (7) | 0.0015 (6) | 0.0009 (6) | −0.0024 (6) |
| C17 | 0.0325 (8) | 0.0198 (7) | 0.0235 (7) | 0.0036 (7) | −0.0022 (7) | −0.0033 (6) |
| C18 | 0.0329 (9) | 0.0235 (7) | 0.0265 (8) | 0.0046 (7) | −0.0001 (7) | 0.0014 (6) |
| C19 | 0.0242 (7) | 0.0192 (7) | 0.0365 (9) | 0.0005 (6) | 0.0031 (7) | −0.0004 (7) |
| C20 | 0.0256 (8) | 0.0229 (8) | 0.0314 (8) | −0.0004 (6) | −0.0019 (7) | −0.0060 (6) |
| C21 | 0.0265 (8) | 0.0242 (8) | 0.0229 (7) | 0.0016 (6) | −0.0011 (6) | −0.0031 (6) |
Geometric parameters (Å, º)
| O1—C10 | 1.4510 (18) | C8—C12 | 1.5101 (19) |
| O1—C1 | 1.4634 (19) | C8—C9 | 1.561 (2) |
| O2—C11 | 1.3667 (18) | C8—H8 | 1.0000 |
| O2—C9 | 1.4573 (17) | C9—C10 | 1.501 (2) |
| O3—C11 | 1.2072 (18) | C9—H9 | 1.0000 |
| N1—C19 | 1.393 (2) | C10—H10 | 1.0000 |
| N1—H1A | 0.94 (2) | C11—C12 | 1.472 (2) |
| N1—H2B | 0.90 (2) | C12—C13 | 1.342 (2) |
| C1—C10 | 1.472 (2) | C13—C16 | 1.457 (2) |
| C1—C15 | 1.502 (2) | C13—H13 | 0.9500 |
| C1—C2 | 1.510 (2) | C14—H14A | 0.9800 |
| C2—C3 | 1.547 (2) | C14—H14B | 0.9800 |
| C2—H1 | 0.9900 | C14—H14C | 0.9800 |
| C2—H2A | 0.9900 | C15—H15A | 0.9800 |
| C3—C4 | 1.505 (2) | C15—H15B | 0.9800 |
| C3—H3A | 0.9900 | C15—H15C | 0.9800 |
| C3—H3B | 0.9900 | C16—C21 | 1.401 (2) |
| C4—C5 | 1.334 (3) | C16—C17 | 1.403 (2) |
| C4—H4 | 0.9500 | C17—C18 | 1.383 (2) |
| C5—C14 | 1.504 (2) | C17—H17 | 0.9500 |
| C5—C6 | 1.511 (2) | C18—C19 | 1.404 (2) |
| C6—C7 | 1.539 (2) | C18—H18 | 0.9500 |
| C6—H6A | 0.9900 | C19—C20 | 1.395 (2) |
| C6—H6B | 0.9900 | C20—C21 | 1.378 (2) |
| C7—C8 | 1.545 (2) | C20—H20 | 0.9500 |
| C7—H7A | 0.9900 | C21—H21 | 0.9500 |
| C7—H7B | 0.9900 | ||
| C10—O1—C1 | 60.67 (10) | C10—C9—C8 | 111.63 (12) |
| C11—O2—C9 | 110.56 (11) | O2—C9—H9 | 109.7 |
| C19—N1—H1A | 114.0 (15) | C10—C9—H9 | 109.7 |
| C19—N1—H2B | 112.6 (15) | C8—C9—H9 | 109.7 |
| H1A—N1—H2B | 118 (2) | O1—C10—C1 | 60.08 (9) |
| O1—C1—C10 | 59.25 (9) | O1—C10—C9 | 121.02 (12) |
| O1—C1—C15 | 112.05 (13) | C1—C10—C9 | 123.89 (14) |
| C10—C1—C15 | 122.43 (14) | O1—C10—H10 | 113.8 |
| O1—C1—C2 | 117.39 (14) | C1—C10—H10 | 113.8 |
| C10—C1—C2 | 116.62 (15) | C9—C10—H10 | 113.8 |
| C15—C1—C2 | 116.15 (14) | O3—C11—O2 | 120.44 (14) |
| C1—C2—C3 | 110.08 (14) | O3—C11—C12 | 129.80 (14) |
| C1—C2—H1 | 109.6 | O2—C11—C12 | 109.72 (12) |
| C3—C2—H1 | 109.6 | C13—C12—C11 | 118.35 (13) |
| C1—C2—H2A | 109.6 | C13—C12—C8 | 132.77 (14) |
| C3—C2—H2A | 109.6 | C11—C12—C8 | 108.87 (12) |
| H1—C2—H2A | 108.2 | C12—C13—C16 | 131.43 (14) |
| C4—C3—C2 | 110.68 (13) | C12—C13—H13 | 114.3 |
| C4—C3—H3A | 109.5 | C16—C13—H13 | 114.3 |
| C2—C3—H3A | 109.5 | C5—C14—H14A | 109.5 |
| C4—C3—H3B | 109.5 | C5—C14—H14B | 109.5 |
| C2—C3—H3B | 109.5 | H14A—C14—H14B | 109.5 |
| H3A—C3—H3B | 108.1 | C5—C14—H14C | 109.5 |
| C5—C4—C3 | 128.15 (15) | H14A—C14—H14C | 109.5 |
| C5—C4—H4 | 115.9 | H14B—C14—H14C | 109.5 |
| C3—C4—H4 | 115.9 | C1—C15—H15A | 109.5 |
| C4—C5—C14 | 125.04 (14) | C1—C15—H15B | 109.5 |
| C4—C5—C6 | 120.33 (15) | H15A—C15—H15B | 109.5 |
| C14—C5—C6 | 114.58 (15) | C1—C15—H15C | 109.5 |
| C5—C6—C7 | 113.64 (12) | H15A—C15—H15C | 109.5 |
| C5—C6—H6A | 108.8 | H15B—C15—H15C | 109.5 |
| C7—C6—H6A | 108.8 | C21—C16—C17 | 117.15 (14) |
| C5—C6—H6B | 108.8 | C21—C16—C13 | 118.40 (14) |
| C7—C6—H6B | 108.8 | C17—C16—C13 | 124.42 (14) |
| H6A—C6—H6B | 107.7 | C18—C17—C16 | 121.43 (15) |
| C6—C7—C8 | 115.03 (13) | C18—C17—H17 | 119.3 |
| C6—C7—H7A | 108.5 | C16—C17—H17 | 119.3 |
| C8—C7—H7A | 108.5 | C17—C18—C19 | 120.49 (15) |
| C6—C7—H7B | 108.5 | C17—C18—H18 | 119.8 |
| C8—C7—H7B | 108.5 | C19—C18—H18 | 119.8 |
| H7A—C7—H7B | 107.5 | N1—C19—C20 | 121.19 (15) |
| C12—C8—C7 | 114.12 (13) | N1—C19—C18 | 120.33 (16) |
| C12—C8—C9 | 102.02 (11) | C20—C19—C18 | 118.47 (15) |
| C7—C8—C9 | 114.19 (12) | C21—C20—C19 | 120.52 (15) |
| C12—C8—H8 | 108.7 | C21—C20—H20 | 119.7 |
| C7—C8—H8 | 108.7 | C19—C20—H20 | 119.7 |
| C9—C8—H8 | 108.7 | C20—C21—C16 | 121.91 (15) |
| O2—C9—C10 | 109.13 (12) | C20—C21—H21 | 119.0 |
| O2—C9—C8 | 106.91 (11) | C16—C21—H21 | 119.0 |
| C10—O1—C1—C15 | −115.61 (15) | C8—C9—C10—O1 | 166.95 (12) |
| C10—O1—C1—C2 | 106.19 (17) | O2—C9—C10—C1 | 121.71 (15) |
| O1—C1—C2—C3 | −158.44 (14) | C8—C9—C10—C1 | −120.33 (15) |
| C10—C1—C2—C3 | −91.06 (18) | C9—O2—C11—O3 | 171.96 (15) |
| C15—C1—C2—C3 | 65.05 (19) | C9—O2—C11—C12 | −9.99 (16) |
| C1—C2—C3—C4 | 51.03 (19) | O3—C11—C12—C13 | 0.4 (3) |
| C2—C3—C4—C5 | −110.84 (19) | O2—C11—C12—C13 | −177.46 (14) |
| C3—C4—C5—C14 | −8.7 (3) | O3—C11—C12—C8 | 179.14 (17) |
| C3—C4—C5—C6 | 168.64 (15) | O2—C11—C12—C8 | 1.33 (18) |
| C4—C5—C6—C7 | −98.81 (18) | C7—C8—C12—C13 | −50.8 (2) |
| C14—C5—C6—C7 | 78.79 (18) | C9—C8—C12—C13 | −174.49 (17) |
| C5—C6—C7—C8 | 73.72 (17) | C7—C8—C12—C11 | 130.63 (13) |
| C6—C7—C8—C12 | 144.91 (13) | C9—C8—C12—C11 | 6.96 (17) |
| C6—C7—C8—C9 | −98.26 (15) | C11—C12—C13—C16 | 174.88 (16) |
| C11—O2—C9—C10 | 135.27 (13) | C8—C12—C13—C16 | −3.6 (3) |
| C11—O2—C9—C8 | 14.39 (16) | C12—C13—C16—C21 | 163.27 (17) |
| C12—C8—C9—O2 | −12.50 (15) | C12—C13—C16—C17 | −18.8 (3) |
| C7—C8—C9—O2 | −136.12 (12) | C21—C16—C17—C18 | −2.0 (2) |
| C12—C8—C9—C10 | −131.78 (13) | C13—C16—C17—C18 | −179.96 (16) |
| C7—C8—C9—C10 | 104.60 (15) | C16—C17—C18—C19 | 0.9 (3) |
| C1—O1—C10—C9 | 113.87 (16) | C17—C18—C19—N1 | −179.03 (16) |
| C15—C1—C10—O1 | 98.02 (16) | C17—C18—C19—C20 | 0.4 (2) |
| C2—C1—C10—O1 | −107.50 (15) | N1—C19—C20—C21 | 178.82 (16) |
| O1—C1—C10—C9 | −109.26 (15) | C18—C19—C20—C21 | −0.6 (2) |
| C15—C1—C10—C9 | −11.2 (2) | C19—C20—C21—C16 | −0.5 (2) |
| C2—C1—C10—C9 | 143.25 (15) | C17—C16—C21—C20 | 1.8 (2) |
| O2—C9—C10—O1 | 49.00 (18) | C13—C16—C21—C20 | 179.88 (15) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O1i | 0.94 (2) | 2.25 (3) | 3.133 (2) | 156 (2) |
| N1—H2B···O2i | 0.90 (2) | 2.57 (2) | 3.077 (2) | 116 (2) |
| C2—H2A···O3ii | 0.99 | 2.63 | 3.372 (2) | 132 |
Symmetry codes: (i) x, y−1, z; (ii) x+1/2, −y+3/2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2647).
References
- Bruker (2006). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Hall, I. H., Lee, K. H., Starnes, C. O., Sumida, Y., Wu, R. Y., Waddell, T. G., Cochran, J. W. & Gerhart, K. G. (1979). J. Pharm. Sci. 68, 537–542. [DOI] [PubMed]
- Han, C., Barrios, F. J., Riofski, M. V. & Colby, D. A. (2009). J. Org. Chem. 74, 7176–7179. [DOI] [PubMed]
- Hanson, R. L., Lardy, H. A. & Kupchan, S. M. (1970). Science, 168, 378–380. [DOI] [PubMed]
- Hehner, S. P., Heinrich, M., Bork, P. M., Vogt, M., Ratter, F., Lehmann, V., Osthoff, K. S., Dröge, W. & Schmitz, M. L. (1998). J. Biol. Chem. 273, 1288–1297. [DOI] [PubMed]
- Kupchan, S. M., Eakin, M. A. & Thomas, A. M. (1971). J. Med. Chem. 14, 1147–1152. [DOI] [PubMed]
- Nasim, S., Pei, S. S., Hagen, F. K., Jordan, C. T. & Crooks, P. A. (2011). Bioorg. Med. Chem. 19, 1515–1519. [DOI] [PubMed]
- Neelakantan, S., Nasim, S., Guzman, M. L., Jordan, C. T. & Crooks, P. A. (2009). Bioorg. Med. Chem. Lett. 19, 4346–4349. [DOI] [PubMed]
- Oka, D., Nishimura, K., Shiba, M., Nakai, Y., Arai, Y., Nakayama, M., Takayama, H., Inoue, H., Okuyama, A. & Nonomura, N. (2007). Int. J. Cancer, 120, 2576–2581. [DOI] [PubMed]
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Ralstin, M. C., Gage, E. A., Yip-Schneider, M. T., Klein, P. J., Wiebke, E. A. & Schmidt, C. M. (2006). Mol. Cancer Res. 4, 387–399. [DOI] [PubMed]
- Rodriguez, E., Towers, G. H. & Mitchell, J. C. (1976). Phytochemistry, 15, 1573–1580.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122.
- Sun, H.-X., Zheng, Q.-F. & Tu, J. (2006). Bioorg. Med. Chem. 14, 1189–1198. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813028730/su2647sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813028730/su2647Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report