Abstract
The asymmetric unit of the title compound, C11H10N4O3, contains two independent molecules in which the benzene rings make dihedral angles of 38.3 (2) and 87.1 (2)° with respect to the triazole rings. In the crystal, the molecules are linked by C—H⋯O hydrogen bonds, forming chains along [021]. Further, weak C—O⋯π [3.865 (5) Å, 83.8 (3)°] and N—O⋯π [3.275 (5) and 3.240 (6) Å, 141.8 (4) and 102.8 (3)°] interactions are observed.
Related literature
For chemical and biological properties and pharmocological applications of 1,2,3-triazole derivative, see: Nithinchandra et al. (2012 ▶, 2013 ▶); Biagi et al. (2004 ▶); Manfredini et al. (2000 ▶); Sherement et al. (2004 ▶). For bond-length data, see: Allen et al. (1987 ▶).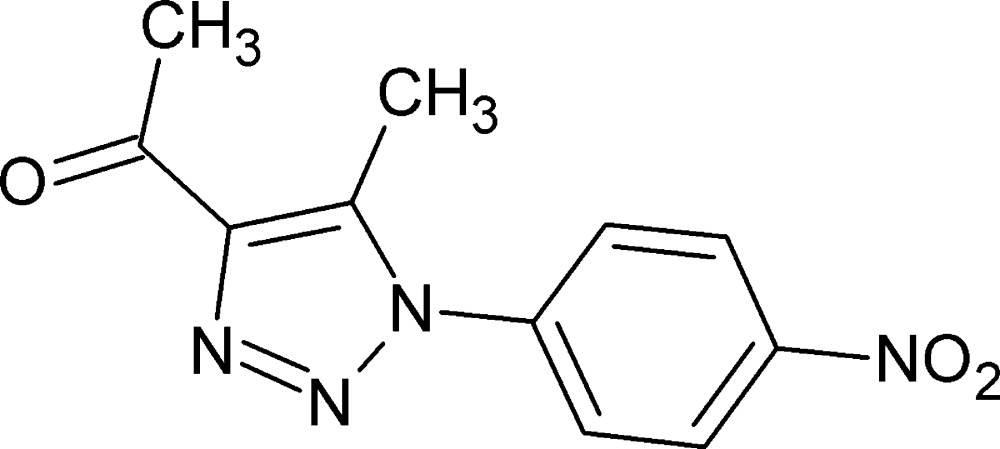
Experimental
Crystal data
C11H10N4O3
M r = 246.23
Orthorhombic,

a = 7.2786 (10) Å
b = 11.5055 (16) Å
c = 27.220 (4) Å
V = 2279.5 (6) Å3
Z = 8
Cu Kα radiation
μ = 0.91 mm−1
T = 296 K
0.23 × 0.22 × 0.21 mm
Data collection
Bruker X8 Proteum diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2013 ▶) T min = 0.818, T max = 0.831
8697 measured reflections
1910 independent reflections
1617 reflections with I > 2σ(I)
R int = 0.057
Refinement
R[F 2 > 2σ(F 2)] = 0.046
wR(F 2) = 0.138
S = 1.08
1910 reflections
329 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.17 e Å−3
Δρmin = −0.17 e Å−3
Data collection: APEX2 (Bruker, 2013 ▶); cell refinement: SAINT (Bruker, 2013 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: Mercury.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813029425/is5313sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813029425/is5313Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C5B—H5B1⋯O3A i | 0.96 | 2.51 | 3.306 (8) | 141 |
| C10B—H10B⋯O1A | 0.93 | 2.58 | 3.197 (7) | 124 |
Symmetry code: (i)  .
.
Acknowledgments
The authors are thankful to the IOE, University of Mysore, for providing the single-crystal X-ray diffraction facility. VN is grateful to the UGC for the award of an RFSMS Fellowship. RD acknowledges the UGC, New Delhi, for financial support under the Major Research Project Scheme [UGC MRP No. F.41–882/2012 (SR) dated 01/07/2012].
supplementary crystallographic information
1. Comment
1,2,3-Triazoles are attractive constructs, because of their unique chemical properties and they find many applications in organic and medicinal chemistry (Nithinchandra et al., 2013). They are found to be potent antimicrobial (Sherement et al., 2004)and antiviral agents. Some of them have exhibited antiproliferative and anti-inflammatory property (Nithinchandra et al., 2012). Also, 1,2,3-triazoles are used as DNA cleaving agents (Manfredini et al., 2000) and potassium channel activators (Biagi et al., 2004).
The asymmetric unit of of the title compound consists of two molecules A and B (Fig. 1). They show conformational difference, as evident from dihedral angles. The dihedral angle between benzene ring and triazole moiety is 38.2 (3)° in A and 87.6 (4)° in B. The values of the bond lengths are similar to the reported literature (Allen et al., 1987).
In the crystal, the molecules are linked to one another with the C—H···O hydrogen bonds (Table 1). Also, short contacts of the type C3A—O1A···Cg4(x + 1/2, y - 1, z) with a distance 3.865 (5) Å [angle 83.8 (3)°], N4A—O2A···Cg3(x - 1, y - 1, z + 1/2) with a distance 3.275 (5) Å [angle 141.8 (4)°] and N4B—O3B···Cg1(x - 1/2, y - 1, z) with a distance 3.240 (6) Å [angle 102.8 (3)°] help in crystal stabilization. These interactions form a three dimensional architecture (Fig. 2), where Cg1, Cg3 and Cg4 are the centroids of the N1A/N2A/N3A/C1A/C2A, N1B/N2B/N3B/C1B/C2B and C6B–C11B rings, respectively.
2. Experimental
1-Azido-4-nitrobenzene (0.01 mol) was treated with acetyl acetone (0.01 mol) in methanol (10 ml) and the mixture was cooled to 0 °C. Sodium methoxide (0.01 mol) was added under inert atmosphere to the above mixture and stirred at ambient temperature for 8 h. Progress of the reaction was monitored by TLC (ethyl acetate/petroleum ether, 2:3, v/v). After completion of the reaction, the mixture was poured onto ice cold water. The precipitated solid was filtered, washed with water and recrystallized from ethanol. Single crystals suitable for X-ray analysis were obtained from a 1:2 mixture of DMF and ethanol by slow evaporation.
3. Refinement
All the H atoms were fixed geometrically (C—H = 0.93–0.96 Å) and allowed to ride on their parent atoms with Uiso(H) = 1.5Ueq(C-methyl) and 1.2Ueq(C) for other H atoms. The Flack parameter x refines to 0.2 (4) with unmerged data, and the absolute structure cannot be determined reliably. The final refinement was performed with the merged data.
Figures
Fig. 1.

Asymmetric unit of the title compound with 50% probability ellipsoids.
Fig. 2.

Packing diagram of the title compound, viewed along the b axis. Dotted lines indicate hydrogen bonds and short contacts involved.
Crystal data
| C11H10N4O3 | F(000) = 1024 |
| Mr = 246.23 | Dx = 1.435 Mg m−3 |
| Orthorhombic, Pca21 | Cu Kα radiation, λ = 1.54178 Å |
| Hall symbol: P 2c -2ac | Cell parameters from 1910 reflections |
| a = 7.2786 (10) Å | θ = 3.2–64.2° |
| b = 11.5055 (16) Å | µ = 0.91 mm−1 |
| c = 27.220 (4) Å | T = 296 K |
| V = 2279.5 (6) Å3 | Block, red |
| Z = 8 | 0.23 × 0.22 × 0.21 mm |
Data collection
| Bruker X8 Proteum diffractometer | 1910 independent reflections |
| Radiation source: Bruker MicroStar microfocus rotating anode | 1617 reflections with I > 2σ(I) |
| Helios multilayer optics monochromator | Rint = 0.057 |
| Detector resolution: 10.7 pixels mm-1 | θmax = 64.2°, θmin = 3.3° |
| φ and ω scans | h = −8→8 |
| Absorption correction: multi-scan (SADABS; Bruker, 2013) | k = −5→13 |
| Tmin = 0.818, Tmax = 0.831 | l = −30→31 |
| 8697 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.046 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.138 | H-atom parameters constrained |
| S = 1.08 | w = 1/[σ2(Fo2) + (0.0932P)2] where P = (Fo2 + 2Fc2)/3 |
| 1910 reflections | (Δ/σ)max < 0.001 |
| 329 parameters | Δρmax = 0.17 e Å−3 |
| 1 restraint | Δρmin = −0.17 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > σ(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1A | 0.1681 (6) | 0.7317 (4) | 0.30010 (16) | 0.0907 (16) | |
| O2A | 0.5412 (8) | 0.8545 (4) | 0.64933 (16) | 0.1063 (18) | |
| O3A | 0.4670 (10) | 1.0325 (5) | 0.64815 (18) | 0.129 (3) | |
| N1A | 0.4033 (6) | 0.9759 (4) | 0.35049 (15) | 0.0680 (16) | |
| N2A | 0.4450 (6) | 0.9990 (4) | 0.39568 (14) | 0.0657 (14) | |
| N3A | 0.3707 (5) | 0.9122 (3) | 0.42410 (12) | 0.0522 (11) | |
| N4A | 0.4913 (7) | 0.9402 (4) | 0.62761 (15) | 0.0767 (16) | |
| C1A | 0.2812 (6) | 0.8331 (4) | 0.39621 (15) | 0.0497 (12) | |
| C2A | 0.3036 (6) | 0.8752 (4) | 0.34871 (16) | 0.0570 (14) | |
| C3A | 0.2316 (7) | 0.8288 (5) | 0.30239 (18) | 0.0670 (16) | |
| C4A | 0.2355 (10) | 0.9058 (6) | 0.2585 (2) | 0.084 (2) | |
| C5A | 0.1735 (7) | 0.7346 (4) | 0.41484 (19) | 0.0667 (17) | |
| C6A | 0.3986 (6) | 0.9188 (4) | 0.47594 (15) | 0.0503 (14) | |
| C7A | 0.3940 (7) | 1.0257 (4) | 0.49864 (17) | 0.0603 (17) | |
| C8A | 0.4254 (7) | 1.0344 (4) | 0.54869 (16) | 0.0633 (17) | |
| C9A | 0.4592 (7) | 0.9331 (4) | 0.57459 (16) | 0.0587 (14) | |
| C10A | 0.4634 (7) | 0.8257 (4) | 0.55236 (16) | 0.0600 (16) | |
| C11A | 0.4341 (6) | 0.8185 (4) | 0.50253 (16) | 0.0577 (16) | |
| O1B | −0.0706 (6) | 0.3153 (4) | 0.04016 (14) | 0.0883 (16) | |
| O2B | 0.0771 (8) | 0.4308 (5) | 0.40221 (15) | 0.110 (2) | |
| O3B | 0.1969 (7) | 0.2600 (5) | 0.40198 (14) | 0.0950 (18) | |
| N1B | 0.3210 (5) | 0.4147 (4) | 0.10651 (14) | 0.0653 (14) | |
| N2B | 0.3525 (5) | 0.4112 (4) | 0.15379 (13) | 0.0637 (14) | |
| N3B | 0.1961 (5) | 0.3690 (3) | 0.17486 (12) | 0.0538 (11) | |
| N4B | 0.1456 (6) | 0.3479 (5) | 0.38102 (15) | 0.0733 (18) | |
| C1B | 0.0636 (6) | 0.3448 (4) | 0.14185 (17) | 0.0550 (12) | |
| C2B | 0.1480 (6) | 0.3740 (4) | 0.09745 (16) | 0.0550 (12) | |
| C3B | 0.0683 (7) | 0.3710 (4) | 0.04782 (15) | 0.0597 (16) | |
| C4B | 0.1636 (9) | 0.4365 (6) | 0.00848 (19) | 0.082 (2) | |
| C5B | −0.1210 (8) | 0.3025 (5) | 0.1540 (2) | 0.0760 (19) | |
| C6B | 0.1834 (6) | 0.3626 (4) | 0.22757 (15) | 0.0533 (14) | |
| C7B | 0.2290 (8) | 0.2619 (4) | 0.25147 (17) | 0.0673 (16) | |
| C8B | 0.2177 (7) | 0.2566 (4) | 0.30211 (19) | 0.0693 (17) | |
| C9B | 0.1605 (6) | 0.3519 (5) | 0.32681 (16) | 0.0570 (14) | |
| C10B | 0.1174 (7) | 0.4558 (5) | 0.30392 (19) | 0.0703 (17) | |
| C11B | 0.1287 (8) | 0.4607 (5) | 0.25360 (18) | 0.0680 (17) | |
| H5A1 | 0.24880 | 0.66610 | 0.41530 | 0.1000* | |
| H7A | 0.36970 | 1.09220 | 0.48030 | 0.0730* | |
| H5A2 | 0.06950 | 0.72180 | 0.39380 | 0.1000* | |
| H8A | 0.42390 | 1.10620 | 0.56440 | 0.0760* | |
| H5A3 | 0.13180 | 0.75140 | 0.44750 | 0.1000* | |
| H4A1 | 0.17180 | 0.86890 | 0.23190 | 0.1260* | |
| H10A | 0.48560 | 0.75900 | 0.57070 | 0.0720* | |
| H4A2 | 0.36060 | 0.92010 | 0.24920 | 0.1260* | |
| H11A | 0.43810 | 0.74690 | 0.48680 | 0.0690* | |
| H4A3 | 0.17660 | 0.97820 | 0.26620 | 0.1260* | |
| H5B1 | −0.13290 | 0.22300 | 0.14380 | 0.1140* | |
| H5B2 | −0.21100 | 0.34910 | 0.13740 | 0.1140* | |
| H5B3 | −0.13980 | 0.30770 | 0.18890 | 0.1140* | |
| H4B1 | 0.28300 | 0.40340 | 0.00310 | 0.1230* | |
| H4B2 | 0.17640 | 0.51630 | 0.01820 | 0.1230* | |
| H4B3 | 0.09310 | 0.43210 | −0.02130 | 0.1230* | |
| H7B | 0.26730 | 0.19720 | 0.23370 | 0.0800* | |
| H8B | 0.24890 | 0.18890 | 0.31880 | 0.0830* | |
| H10B | 0.08170 | 0.52040 | 0.32210 | 0.0840* | |
| H11B | 0.10010 | 0.52910 | 0.23710 | 0.0820* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1A | 0.115 (3) | 0.074 (3) | 0.083 (2) | −0.011 (2) | −0.018 (2) | −0.0103 (19) |
| O2A | 0.146 (4) | 0.107 (3) | 0.066 (2) | 0.015 (3) | −0.021 (3) | 0.008 (2) |
| O3A | 0.217 (7) | 0.099 (3) | 0.071 (3) | −0.001 (4) | −0.007 (3) | −0.018 (2) |
| N1A | 0.081 (3) | 0.067 (3) | 0.056 (2) | −0.010 (2) | 0.0006 (19) | 0.0059 (19) |
| N2A | 0.080 (3) | 0.063 (2) | 0.054 (2) | −0.017 (2) | 0.0022 (18) | 0.0112 (18) |
| N3A | 0.0547 (19) | 0.050 (2) | 0.0518 (19) | −0.0050 (16) | 0.0006 (14) | 0.0012 (15) |
| N4A | 0.092 (3) | 0.081 (3) | 0.057 (2) | −0.005 (3) | −0.003 (2) | −0.002 (2) |
| C1A | 0.053 (2) | 0.044 (2) | 0.052 (2) | 0.0033 (18) | −0.0018 (18) | 0.0026 (17) |
| C2A | 0.062 (2) | 0.056 (3) | 0.053 (2) | 0.002 (2) | 0.000 (2) | 0.004 (2) |
| C3A | 0.073 (3) | 0.070 (3) | 0.058 (2) | 0.011 (3) | −0.005 (2) | −0.003 (2) |
| C4A | 0.107 (4) | 0.093 (4) | 0.051 (2) | 0.004 (3) | 0.001 (3) | 0.006 (3) |
| C5A | 0.075 (3) | 0.060 (3) | 0.065 (3) | −0.009 (2) | 0.002 (2) | 0.007 (2) |
| C6A | 0.051 (2) | 0.055 (3) | 0.045 (2) | −0.0043 (18) | 0.0043 (17) | 0.0041 (18) |
| C7A | 0.066 (3) | 0.055 (3) | 0.060 (3) | −0.001 (2) | −0.002 (2) | 0.005 (2) |
| C8A | 0.075 (3) | 0.058 (3) | 0.057 (3) | −0.004 (2) | 0.004 (2) | −0.006 (2) |
| C9A | 0.059 (2) | 0.069 (3) | 0.048 (2) | −0.002 (2) | 0.0011 (18) | 0.000 (2) |
| C10A | 0.066 (3) | 0.060 (3) | 0.054 (2) | −0.001 (2) | −0.001 (2) | 0.007 (2) |
| C11A | 0.064 (3) | 0.052 (3) | 0.057 (2) | 0.002 (2) | −0.0023 (19) | 0.0002 (19) |
| O1B | 0.110 (3) | 0.092 (3) | 0.063 (2) | −0.039 (2) | −0.0193 (19) | 0.0015 (18) |
| O2B | 0.139 (4) | 0.130 (4) | 0.061 (3) | 0.005 (3) | 0.017 (2) | −0.022 (2) |
| O3B | 0.105 (3) | 0.123 (4) | 0.057 (2) | −0.018 (3) | −0.003 (2) | 0.018 (2) |
| N1B | 0.069 (2) | 0.076 (3) | 0.051 (2) | −0.0069 (19) | 0.0015 (18) | 0.0005 (18) |
| N2B | 0.060 (2) | 0.084 (3) | 0.0470 (19) | −0.012 (2) | 0.0031 (16) | 0.0026 (18) |
| N3B | 0.058 (2) | 0.055 (2) | 0.0485 (19) | −0.0005 (17) | −0.0001 (15) | 0.0000 (15) |
| N4B | 0.074 (3) | 0.099 (4) | 0.047 (2) | −0.019 (3) | −0.0022 (19) | −0.004 (2) |
| C1B | 0.062 (2) | 0.050 (2) | 0.053 (2) | −0.0065 (19) | 0.0007 (18) | −0.0003 (18) |
| C2B | 0.060 (2) | 0.053 (2) | 0.052 (2) | −0.0023 (18) | 0.0045 (19) | 0.0005 (18) |
| C3B | 0.076 (3) | 0.054 (3) | 0.049 (2) | −0.003 (2) | −0.003 (2) | −0.0024 (19) |
| C4B | 0.090 (4) | 0.105 (5) | 0.050 (2) | −0.010 (3) | 0.002 (3) | 0.007 (3) |
| C5B | 0.072 (3) | 0.088 (4) | 0.068 (3) | −0.023 (3) | 0.006 (2) | 0.007 (3) |
| C6B | 0.060 (2) | 0.059 (3) | 0.041 (2) | 0.000 (2) | 0.0024 (17) | −0.0021 (18) |
| C7B | 0.090 (3) | 0.059 (3) | 0.053 (2) | 0.009 (2) | 0.004 (2) | 0.004 (2) |
| C8B | 0.084 (3) | 0.065 (3) | 0.059 (3) | 0.006 (3) | 0.004 (2) | 0.011 (2) |
| C9B | 0.055 (2) | 0.072 (3) | 0.044 (2) | −0.010 (2) | 0.0004 (17) | 0.000 (2) |
| C10B | 0.082 (3) | 0.070 (3) | 0.059 (3) | 0.005 (3) | 0.003 (2) | −0.012 (2) |
| C11B | 0.089 (3) | 0.060 (3) | 0.055 (3) | 0.009 (3) | 0.002 (2) | 0.000 (2) |
Geometric parameters (Å, º)
| O1A—C3A | 1.211 (7) | C4A—H4A2 | 0.9600 |
| O2A—N4A | 1.206 (7) | C4A—H4A3 | 0.9600 |
| O3A—N4A | 1.213 (7) | C5A—H5A3 | 0.9600 |
| O1B—C3B | 1.215 (7) | C5A—H5A2 | 0.9600 |
| O2B—N4B | 1.221 (8) | C5A—H5A1 | 0.9600 |
| O3B—N4B | 1.220 (8) | C7A—H7A | 0.9300 |
| N1A—N2A | 1.295 (6) | C8A—H8A | 0.9300 |
| N1A—C2A | 1.368 (6) | C10A—H10A | 0.9300 |
| N2A—N3A | 1.374 (6) | C11A—H11A | 0.9300 |
| N3A—C1A | 1.352 (6) | C1B—C2B | 1.397 (6) |
| N3A—C6A | 1.428 (5) | C1B—C5B | 1.467 (7) |
| N4A—C9A | 1.464 (6) | C2B—C3B | 1.471 (6) |
| N1B—N2B | 1.308 (5) | C3B—C4B | 1.482 (7) |
| N1B—C2B | 1.366 (6) | C6B—C7B | 1.370 (6) |
| N2B—N3B | 1.364 (5) | C6B—C11B | 1.391 (7) |
| N3B—C1B | 1.347 (6) | C7B—C8B | 1.382 (7) |
| N3B—C6B | 1.440 (5) | C8B—C9B | 1.352 (7) |
| N4B—C9B | 1.480 (6) | C9B—C10B | 1.384 (8) |
| C1A—C5A | 1.468 (7) | C10B—C11B | 1.373 (7) |
| C1A—C2A | 1.390 (6) | C4B—H4B1 | 0.9600 |
| C2A—C3A | 1.466 (7) | C4B—H4B2 | 0.9600 |
| C3A—C4A | 1.488 (8) | C4B—H4B3 | 0.9600 |
| C6A—C7A | 1.377 (6) | C5B—H5B1 | 0.9600 |
| C6A—C11A | 1.387 (6) | C5B—H5B2 | 0.9600 |
| C7A—C8A | 1.385 (6) | C5B—H5B3 | 0.9600 |
| C8A—C9A | 1.384 (6) | C7B—H7B | 0.9300 |
| C9A—C10A | 1.376 (6) | C8B—H8B | 0.9300 |
| C10A—C11A | 1.376 (6) | C10B—H10B | 0.9300 |
| C4A—H4A1 | 0.9600 | C11B—H11B | 0.9300 |
| N2A—N1A—C2A | 109.4 (4) | C8A—C7A—H7A | 120.00 |
| N1A—N2A—N3A | 107.1 (4) | C6A—C7A—H7A | 120.00 |
| N2A—N3A—C1A | 111.3 (3) | C9A—C8A—H8A | 121.00 |
| N2A—N3A—C6A | 117.5 (3) | C7A—C8A—H8A | 121.00 |
| C1A—N3A—C6A | 131.2 (4) | C9A—C10A—H10A | 121.00 |
| O2A—N4A—O3A | 122.3 (5) | C11A—C10A—H10A | 120.00 |
| O2A—N4A—C9A | 119.1 (4) | C6A—C11A—H11A | 120.00 |
| O3A—N4A—C9A | 118.7 (5) | C10A—C11A—H11A | 120.00 |
| N2B—N1B—C2B | 109.2 (4) | N3B—C1B—C2B | 102.3 (4) |
| N1B—N2B—N3B | 106.2 (3) | N3B—C1B—C5B | 125.0 (4) |
| N2B—N3B—C1B | 113.0 (3) | C2B—C1B—C5B | 132.7 (4) |
| N2B—N3B—C6B | 119.4 (3) | N1B—C2B—C1B | 109.4 (4) |
| C1B—N3B—C6B | 127.5 (4) | N1B—C2B—C3B | 122.5 (4) |
| O3B—N4B—C9B | 118.0 (5) | C1B—C2B—C3B | 128.0 (4) |
| O2B—N4B—O3B | 123.5 (4) | O1B—C3B—C2B | 119.9 (4) |
| O2B—N4B—C9B | 118.5 (5) | O1B—C3B—C4B | 122.3 (4) |
| N3A—C1A—C5A | 125.6 (4) | C2B—C3B—C4B | 117.9 (4) |
| C2A—C1A—C5A | 130.7 (4) | N3B—C6B—C7B | 120.1 (4) |
| N3A—C1A—C2A | 103.4 (4) | N3B—C6B—C11B | 119.0 (4) |
| N1A—C2A—C1A | 108.9 (4) | C7B—C6B—C11B | 120.9 (4) |
| N1A—C2A—C3A | 121.9 (4) | C6B—C7B—C8B | 119.8 (4) |
| C1A—C2A—C3A | 129.1 (4) | C7B—C8B—C9B | 118.6 (4) |
| O1A—C3A—C2A | 121.1 (5) | N4B—C9B—C8B | 119.5 (5) |
| O1A—C3A—C4A | 121.0 (5) | N4B—C9B—C10B | 117.3 (5) |
| C2A—C3A—C4A | 117.8 (5) | C8B—C9B—C10B | 123.1 (4) |
| N3A—C6A—C11A | 119.9 (4) | C9B—C10B—C11B | 118.1 (5) |
| N3A—C6A—C7A | 119.2 (4) | C6B—C11B—C10B | 119.5 (5) |
| C7A—C6A—C11A | 120.9 (4) | C3B—C4B—H4B1 | 109.00 |
| C6A—C7A—C8A | 120.1 (4) | C3B—C4B—H4B2 | 109.00 |
| C7A—C8A—C9A | 118.0 (4) | C3B—C4B—H4B3 | 109.00 |
| C8A—C9A—C10A | 122.4 (4) | H4B1—C4B—H4B2 | 109.00 |
| N4A—C9A—C8A | 118.9 (4) | H4B1—C4B—H4B3 | 110.00 |
| N4A—C9A—C10A | 118.7 (4) | H4B2—C4B—H4B3 | 110.00 |
| C9A—C10A—C11A | 119.0 (4) | C1B—C5B—H5B1 | 109.00 |
| C6A—C11A—C10A | 119.6 (4) | C1B—C5B—H5B2 | 109.00 |
| C3A—C4A—H4A1 | 109.00 | C1B—C5B—H5B3 | 109.00 |
| C3A—C4A—H4A2 | 109.00 | H5B1—C5B—H5B2 | 110.00 |
| H4A1—C4A—H4A2 | 110.00 | H5B1—C5B—H5B3 | 109.00 |
| H4A1—C4A—H4A3 | 109.00 | H5B2—C5B—H5B3 | 109.00 |
| C3A—C4A—H4A3 | 109.00 | C6B—C7B—H7B | 120.00 |
| H4A2—C4A—H4A3 | 109.00 | C8B—C7B—H7B | 120.00 |
| C1A—C5A—H5A1 | 109.00 | C7B—C8B—H8B | 121.00 |
| C1A—C5A—H5A2 | 109.00 | C9B—C8B—H8B | 121.00 |
| C1A—C5A—H5A3 | 109.00 | C9B—C10B—H10B | 121.00 |
| H5A1—C5A—H5A2 | 109.00 | C11B—C10B—H10B | 121.00 |
| H5A1—C5A—H5A3 | 110.00 | C6B—C11B—H11B | 120.00 |
| H5A2—C5A—H5A3 | 110.00 | C10B—C11B—H11B | 120.00 |
| C2A—N1A—N2A—N3A | −0.3 (5) | N3A—C1A—C2A—C3A | −177.7 (5) |
| N2A—N1A—C2A—C1A | 0.3 (5) | C5A—C1A—C2A—C3A | −3.4 (8) |
| N2A—N1A—C2A—C3A | 178.0 (4) | C5A—C1A—C2A—N1A | 174.2 (5) |
| N1A—N2A—N3A—C1A | 0.2 (5) | C1A—C2A—C3A—O1A | −14.0 (8) |
| N1A—N2A—N3A—C6A | −180.0 (4) | C1A—C2A—C3A—C4A | 164.8 (5) |
| N2A—N3A—C1A—C2A | 0.0 (5) | N1A—C2A—C3A—C4A | −12.5 (7) |
| N2A—N3A—C1A—C5A | −174.7 (4) | N1A—C2A—C3A—O1A | 168.8 (5) |
| C6A—N3A—C1A—C2A | −179.9 (4) | N3A—C6A—C11A—C10A | 179.1 (4) |
| C6A—N3A—C1A—C5A | 5.4 (7) | N3A—C6A—C7A—C8A | −178.3 (4) |
| N2A—N3A—C6A—C7A | 37.8 (6) | C11A—C6A—C7A—C8A | 0.1 (7) |
| N2A—N3A—C6A—C11A | −140.7 (4) | C7A—C6A—C11A—C10A | 0.6 (7) |
| C1A—N3A—C6A—C7A | −142.4 (5) | C6A—C7A—C8A—C9A | −0.6 (7) |
| C1A—N3A—C6A—C11A | 39.1 (7) | C7A—C8A—C9A—C10A | 0.3 (8) |
| O2A—N4A—C9A—C8A | −171.7 (5) | C7A—C8A—C9A—N4A | −179.5 (5) |
| O2A—N4A—C9A—C10A | 8.5 (8) | N4A—C9A—C10A—C11A | −179.7 (4) |
| O3A—N4A—C9A—C8A | 8.2 (8) | C8A—C9A—C10A—C11A | 0.4 (8) |
| O3A—N4A—C9A—C10A | −171.7 (6) | C9A—C10A—C11A—C6A | −0.9 (7) |
| C2B—N1B—N2B—N3B | −0.7 (5) | N3B—C1B—C2B—N1B | −0.9 (5) |
| N2B—N1B—C2B—C1B | 1.1 (6) | N3B—C1B—C2B—C3B | −177.2 (4) |
| N2B—N1B—C2B—C3B | 177.6 (4) | C5B—C1B—C2B—N1B | 177.0 (5) |
| N1B—N2B—N3B—C6B | −175.6 (4) | C5B—C1B—C2B—C3B | 0.7 (9) |
| N1B—N2B—N3B—C1B | 0.2 (5) | N1B—C2B—C3B—O1B | 166.6 (5) |
| N2B—N3B—C1B—C2B | 0.5 (5) | N1B—C2B—C3B—C4B | −12.9 (7) |
| N2B—N3B—C1B—C5B | −177.6 (5) | C1B—C2B—C3B—O1B | −17.6 (8) |
| C1B—N3B—C6B—C11B | −90.7 (6) | C1B—C2B—C3B—C4B | 162.9 (5) |
| C6B—N3B—C1B—C5B | −2.3 (7) | N3B—C6B—C7B—C8B | 179.6 (4) |
| N2B—N3B—C6B—C7B | −94.0 (5) | C11B—C6B—C7B—C8B | 1.1 (8) |
| C6B—N3B—C1B—C2B | 175.9 (4) | N3B—C6B—C11B—C10B | −179.5 (5) |
| C1B—N3B—C6B—C7B | 90.9 (6) | C7B—C6B—C11B—C10B | −1.1 (8) |
| N2B—N3B—C6B—C11B | 84.5 (5) | C6B—C7B—C8B—C9B | 0.3 (8) |
| O3B—N4B—C9B—C8B | 4.5 (7) | C7B—C8B—C9B—N4B | 179.4 (5) |
| O2B—N4B—C9B—C10B | 8.1 (7) | C7B—C8B—C9B—C10B | −1.8 (8) |
| O2B—N4B—C9B—C8B | −173.1 (5) | N4B—C9B—C10B—C11B | −179.4 (5) |
| O3B—N4B—C9B—C10B | −174.3 (5) | C8B—C9B—C10B—C11B | 1.9 (8) |
| N3A—C1A—C2A—N1A | −0.2 (5) | C9B—C10B—C11B—C6B | −0.4 (8) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5B—H5B1···O3Ai | 0.96 | 2.51 | 3.306 (8) | 141 |
| C10B—H10B···O1A | 0.93 | 2.58 | 3.197 (7) | 124 |
Symmetry code: (i) −x+1/2, y−1, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS5313).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Prpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Biagi, G., Calderone, V., Giorgi, I., Livi, O., Martinotti, E., Martelli, A. & Nardi, A. (2004). Farmaco, 59, 397–404. [DOI] [PubMed]
- Bruker (2013). APEX2, SAINT and SADABS Bruker AXS Inc., Madison,Wisconsin, USA.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Manfredini, S., Vicentini, C. B., Manfrini, M., Bianchi, N., Rutigliano, C., Mischiati, C. & Gambari, R. (2000). Bioorg. Med. Chem. 8, 2343–2346. [DOI] [PubMed]
- Nithinchandra, Kalluraya, B., Aamir, S. & Shabaraya, A. R. (2012). Eur. J. Med. Chem. 54, 597–604. [DOI] [PubMed]
- Nithinchandra, Kalluraya, B., Shobhitha, S. & Babu, M. (2013). J. Chem. Pharm. Res. 5, 307–313.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sherement, E. A., Tomanov, R. I., Trukhin, E. V. & Berestovitskaya, V. M. (2004). Russ. J. Org. Chem. 40, 594–595.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813029425/is5313sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813029425/is5313Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report


