Abstract
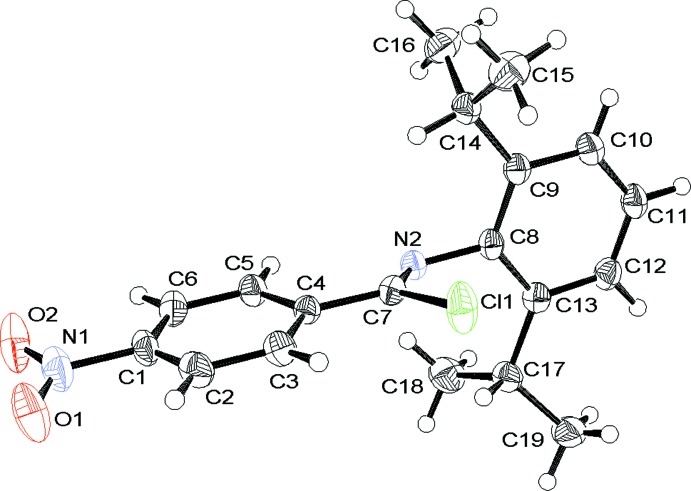
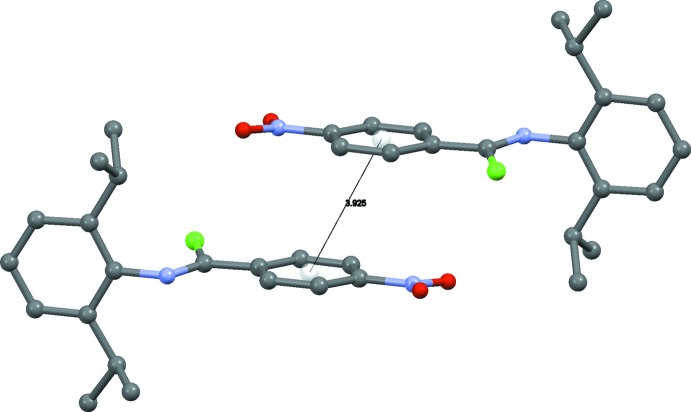
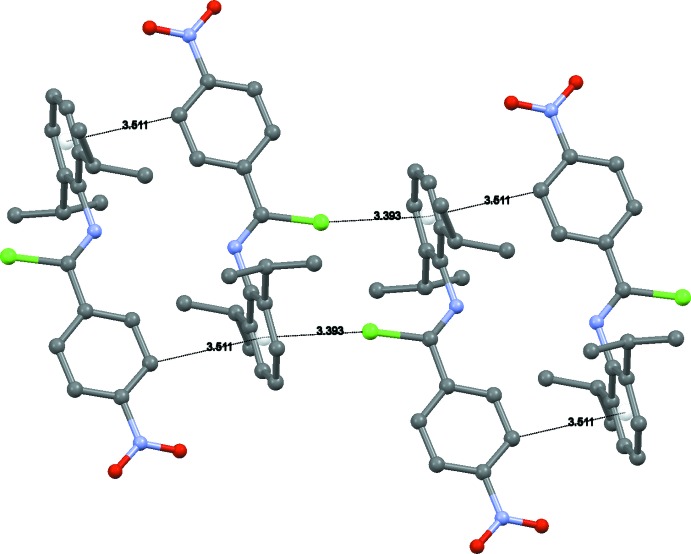
In the title compound, C19H21ClN2O2, the aromatic rings are approximately perpendicular to each other, subtending a dihedral angle of 87.7 (1)°. In the crystal, the 4-nitrophenyl groups of pairs of neighbouring molecules are parallel and oriented head-to-tail with a ring centroid–centroid distance of 3.9247 (12) Å, leading to a π–π interaction between the pair. The faces of each phenyl ring of the 2,6-diisopropylphenyl group interact with two different groups, viz. a chloro group of an adjacent molecule on one side and the edge of the 4-nitrophenyl ring of a second molecule on the other side.
Related literature
For the synthesis and applications of imidoyl chlorides, see: Pelter et al. (1975 ▶); Manley & Bilodeau (2002 ▶); Cunico & Pandey (2005 ▶); Raussukana et al. (2006 ▶); Zheng & Alper (2008 ▶); Kuszpit et al. (2011 ▶). For a related structure of an imidoyl chloride, see: Seidelmann et al. (1998 ▶).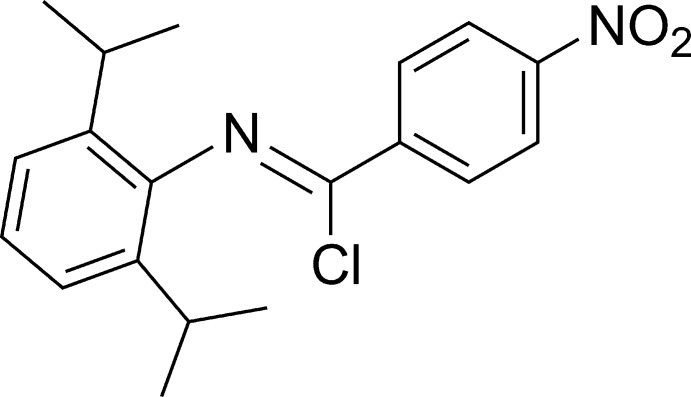
Experimental
Crystal data
C19H21ClN2O2
M r = 344.83
Triclinic,
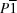
a = 8.2988 (4) Å
b = 10.4667 (3) Å
c = 10.9665 (3) Å
α = 75.568 (2)°
β = 85.411 (2)°
γ = 74.145 (2)°
V = 887.33 (6) Å3
Z = 2
Mo Kα radiation
μ = 0.23 mm−1
T = 150 K
0.35 × 0.20 × 0.15 mm
Data collection
Nonius KappaCCD diffractometer
Absorption correction: multi-scan (DENZO/SCALEPACK; Otwinowski & Minor, 1997 ▶) T min = 0.924, T max = 0.967
6021 measured reflections
4232 independent reflections
3108 reflections with I > 2σ(I)
R int = 0.034
Refinement
R[F 2 > 2σ(F 2)] = 0.062
wR(F 2) = 0.173
S = 1.06
4232 reflections
222 parameters
H-atom parameters constrained
Δρmax = 0.32 e Å−3
Δρmin = −0.41 e Å−3
Data collection: COLLECT (Nonius, 2000 ▶); cell refinement: DENZO/SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: DENZO/SCALEPACK; program(s) used to solve structure: SIR92 (Altomare et al., 1993 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP99 for Windows (Farrugia, 2012 ▶) and Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S1600536813020862/is5293sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813020862/is5293Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813020862/is5293Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 and Cg2 are the centroids of the C1–C6 and C8–C13 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C6—H6⋯Cg2i | 0.95 | 2.67 | 3.511 (2) | 147 |
| C16—H16B⋯Cg1ii | 0.98 | 2.79 | 3.663 (3) | 149 |
Symmetry codes: (i) 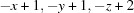 ; (ii)
; (ii) 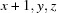 .
.
Acknowledgments
The authors would like to extend their appreciation to the Deanship of Scientific Research at King Saud University for its funding for this research through the research group project RGP-VPP-239.
supplementary crystallographic information
1. Comment
The title compound I, a useful synthetic intermediate, was synthesized in good yield by the reaction of N-(2,6-diisopropylphenyl)-4-nitrobenzamide with phosphorus pentachloride. Imidoyl chlorides are useful reactive intermediates in syntheses of ketones from trialkylcyanoborates (Pelter et al., 1975), of highly substituted 2-imidazolines via a ring-expansion reaction with aziridines (Kuszpit et al., 2011), and by in situ reaction with pyridine-1-oxides to give 2-aminopyridine amides (Manley et al., 2002). They have also been used as precursors to α-iminoamides (Cunico et al., 2005), isoquinolin-1(2H)-ones via a palladium-catalyzed reaction with diethyl(2-iodoaryl)malonates (Zheng et al., 2008), and 1,3-oxathiolanones and benzoxathianones by reaction with mercaptocarboxylic acids (Raussukana et al., 2006). The X-ray crystal structure of N-(diethylaminothiocarbonyl)ferrocenecarbimidoyl chloride has been reported (Seidelmann et al., 1998).
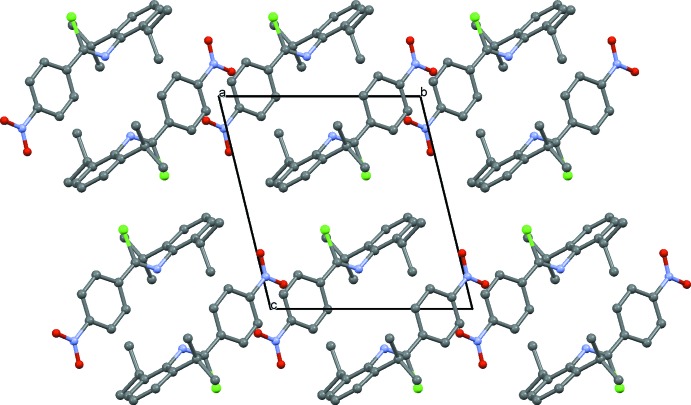
In the molecule (Fig. 1), the aromatic rings of the 2,6-diisopropylphenyl and 4-nitrophenyl groups are approximately perpendicular to each other; the dihedral angle between the least-squares planes through the rings is 87.7 (1)°. The molecule has no strong hydrogen bond donor and the crystal structure is shown in Figure 2. The 4-nitrophenyl groups of neighboring molecules are parallel and oriented head-to-tail with a ring centroid-centroid distance of 3.9247 (12) Å, leading to a π–π interaction (Fig. 3). One face of the phenyl ring of the 2,6-diisopropylphenyl group interacts with the chloro group of an adjacent molecule (C7—Cl1···Cg2) and the other face of the same ring interacts with the edge of the 4-nitrophenyl ring of a second molecule (C6—H6···Cg2; Fig. 4); Cg2 is the centroid of the C8–C13 ring. Another interaction, C16—H16B···Cg1, is also observed; Cg1 is the centroid of the C1–C6 ring.
2. Experimental
Synthesis ofN-(2,6-diisopropylphenyl)-4-nitrobenzimidoyl chloride (I)
An oven dried two necked 100 ml flask equipped with a magnetic stirrer, septum-capped reflux condenser and septum was flushed with N2 and phosphorus pentachloride (4.42 g, 21 mmol) and dry toluene (40 ml) were added. The mixture was stirred for 5 min then N-(2,6-diisopropylphenyl)-4-nitrobenzamide (6.90 g, 21 mmol) was quickly added to the flask under a fast stream of N2, and the septum replaced by a stopper. The mixture was heated to reflux for 2 h, whereupon it became homogeneous and gas evolution was observed. Phosphorus oxychloride and toluene were removed under reduced pressure and the crude product was quickly extracted with hot diethyl ether (3 × 80 ml). The diethyl ether washings were evaporated under a fast stream of N2 overnight, during which process bright yellow prisms of N-(2,6-diisopropylphenyl)-4-nitrobenzimidoyl chloride (6.93 g, 95%) separated; m.p. 144–146 °C. HREI+–MS m/z: calcd for C19H21N2O235Cl 344.1292, found 344.1301.
3. Refinement
H atoms were positioned geometrically (C—H = 0.95–1.00 Å) and refined using a riding model with Uiso(H) = 1.2Ueq(C) or 1.5Ueq(methyl C), allowing for free rotation of the methyl groups about the C—C bond.
Figures
Fig. 1.
Molecular structure of the title compound, showing atom labels and 50% probability displacement ellipsoids for non-H atoms.
Fig. 2.
A packing view of the title compound along the a axis.
Fig. 3.
A pair of molecules showing the ring centroid-centroid distance for parallel 4-nitrobenzyl groups.
Fig. 4.
A segment showing edge-to-face and chloro-to-face contacts in the crystal structure.
Crystal data
| C19H21ClN2O2 | Z = 2 |
| Mr = 344.83 | F(000) = 364 |
| Triclinic, P1 | Dx = 1.291 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.2988 (4) Å | Cell parameters from 3108 reflections |
| b = 10.4667 (3) Å | θ = 2.8–28.3° |
| c = 10.9665 (3) Å | µ = 0.23 mm−1 |
| α = 75.568 (2)° | T = 150 K |
| β = 85.411 (2)° | Block, yellow |
| γ = 74.145 (2)° | 0.35 × 0.20 × 0.15 mm |
| V = 887.33 (6) Å3 |
Data collection
| Nonius KappaCCD diffractometer | 4232 independent reflections |
| Radiation source: fine-focus sealed tube | 3108 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.034 |
| ω and φ scans | θmax = 28.3°, θmin = 2.8° |
| Absorption correction: multi-scan (DENZO/SCALEPACK; Otwinowski & Minor, 1997) | h = −11→10 |
| Tmin = 0.924, Tmax = 0.967 | k = −13→13 |
| 6021 measured reflections | l = −14→14 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.062 | H-atom parameters constrained |
| wR(F2) = 0.173 | w = 1/[σ2(Fo2) + (0.0765P)2 + 0.589P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max = 0.005 |
| 4232 reflections | Δρmax = 0.32 e Å−3 |
| 222 parameters | Δρmin = −0.41 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.094 (10) |
Special details
| Experimental. 1H (400 MHz; CDCl3) δ: 8.25 (2 H, d, J = 8.4 Hz), 8.17 (2 H, d, J = 8.4 Hz), 7.05–7.12 (2 H, m), 6.97–7.04 (1 H, m), 2.66 (2 H, app. sept, J = 6.9 Hz), 1.11 (6 H, d, J = 6.6 Hz), 1.05 (6 H, d, J = 6.6 Hz) – the two 6 H doublets coalesced at 50 °C; 13C (125 MHz; CDCl3) δ: 149.9 (s), 143.4 (s), 141.8 (s), 140.1 (s), 136.3 (s), 130.3 (d), 125.5 (d), 123.7 (d), 123.3 (d), 28.8 (d), 23.3 (q), 22.8 (q); vmax (thin film/cm-1): 3017, 2966, 2929, 2871, 1662, 1605, 1529, 1349, 1216, 1168, 1461. |
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.2830 (3) | 0.0882 (2) | 1.0555 (2) | 0.0319 (5) | |
| C2 | 0.2874 (3) | 0.0814 (2) | 0.9309 (2) | 0.0343 (5) | |
| H2 | 0.2496 | 0.0132 | 0.9075 | 0.041* | |
| C3 | 0.3483 (3) | 0.1763 (2) | 0.8410 (2) | 0.0299 (5) | |
| H3 | 0.3551 | 0.1720 | 0.7552 | 0.036* | |
| C4 | 0.3996 (2) | 0.27816 (19) | 0.87595 (19) | 0.0234 (4) | |
| C5 | 0.3944 (3) | 0.2816 (2) | 1.0028 (2) | 0.0277 (5) | |
| H5 | 0.4310 | 0.3500 | 1.0269 | 0.033* | |
| C6 | 0.3360 (3) | 0.1857 (2) | 1.0938 (2) | 0.0320 (5) | |
| H6 | 0.3326 | 0.1873 | 1.1803 | 0.038* | |
| C7 | 0.4587 (3) | 0.3856 (2) | 0.78235 (18) | 0.0237 (4) | |
| C8 | 0.5521 (3) | 0.5859 (2) | 0.72503 (18) | 0.0239 (4) | |
| C9 | 0.7254 (3) | 0.5618 (2) | 0.70425 (19) | 0.0259 (4) | |
| C10 | 0.7851 (3) | 0.6667 (2) | 0.6259 (2) | 0.0306 (5) | |
| H10 | 0.9022 | 0.6527 | 0.6104 | 0.037* | |
| C11 | 0.6771 (3) | 0.7907 (2) | 0.5702 (2) | 0.0317 (5) | |
| H11 | 0.7201 | 0.8614 | 0.5180 | 0.038* | |
| C12 | 0.5055 (3) | 0.8114 (2) | 0.5911 (2) | 0.0306 (5) | |
| H12 | 0.4321 | 0.8961 | 0.5513 | 0.037* | |
| C13 | 0.4386 (3) | 0.7111 (2) | 0.6689 (2) | 0.0267 (5) | |
| C14 | 0.8468 (3) | 0.4269 (2) | 0.7645 (2) | 0.0307 (5) | |
| H14 | 0.7792 | 0.3633 | 0.8100 | 0.037* | |
| C15 | 0.9545 (3) | 0.3613 (3) | 0.6651 (3) | 0.0427 (6) | |
| H15A | 0.8819 | 0.3544 | 0.6022 | 0.064* | |
| H15B | 1.0214 | 0.2697 | 0.7057 | 0.064* | |
| H15C | 1.0293 | 0.4178 | 0.6239 | 0.064* | |
| C16 | 0.9550 (3) | 0.4460 (3) | 0.8613 (2) | 0.0406 (6) | |
| H16A | 1.0282 | 0.5032 | 0.8185 | 0.061* | |
| H16B | 1.0235 | 0.3566 | 0.9054 | 0.061* | |
| H16C | 0.8827 | 0.4905 | 0.9222 | 0.061* | |
| C17 | 0.2510 (3) | 0.7304 (2) | 0.6917 (2) | 0.0308 (5) | |
| H17 | 0.2250 | 0.6450 | 0.6838 | 0.037* | |
| C18 | 0.2005 (3) | 0.7468 (3) | 0.8252 (3) | 0.0446 (6) | |
| H18A | 0.2680 | 0.6693 | 0.8861 | 0.067* | |
| H18B | 0.0816 | 0.7497 | 0.8397 | 0.067* | |
| H18C | 0.2195 | 0.8320 | 0.8353 | 0.067* | |
| C19 | 0.1440 (3) | 0.8492 (3) | 0.5950 (3) | 0.0438 (6) | |
| H19A | 0.1614 | 0.9356 | 0.6032 | 0.066* | |
| H19B | 0.0255 | 0.8510 | 0.6101 | 0.066* | |
| H19C | 0.1766 | 0.8370 | 0.5099 | 0.066* | |
| N1 | 0.2200 (3) | −0.0141 (2) | 1.1512 (2) | 0.0457 (6) | |
| N2 | 0.4892 (2) | 0.48451 (17) | 0.81255 (16) | 0.0246 (4) | |
| O1 | 0.1607 (3) | −0.0925 (2) | 1.1148 (2) | 0.0716 (7) | |
| O2 | 0.2325 (3) | −0.0162 (2) | 1.2616 (2) | 0.0651 (6) | |
| Cl1 | 0.48157 (10) | 0.36574 (7) | 0.62743 (5) | 0.0461 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0304 (11) | 0.0202 (10) | 0.0391 (12) | −0.0059 (8) | 0.0081 (9) | 0.0008 (9) |
| C2 | 0.0350 (12) | 0.0224 (10) | 0.0473 (14) | −0.0115 (9) | 0.0001 (10) | −0.0074 (9) |
| C3 | 0.0334 (11) | 0.0270 (11) | 0.0312 (11) | −0.0100 (9) | 0.0017 (9) | −0.0085 (9) |
| C4 | 0.0235 (10) | 0.0192 (9) | 0.0265 (10) | −0.0061 (7) | 0.0001 (8) | −0.0031 (8) |
| C5 | 0.0318 (11) | 0.0257 (10) | 0.0269 (10) | −0.0105 (8) | 0.0008 (9) | −0.0056 (8) |
| C6 | 0.0360 (12) | 0.0272 (11) | 0.0284 (11) | −0.0063 (9) | 0.0046 (9) | −0.0023 (9) |
| C7 | 0.0267 (10) | 0.0229 (9) | 0.0205 (9) | −0.0056 (8) | −0.0003 (8) | −0.0043 (7) |
| C8 | 0.0308 (11) | 0.0219 (9) | 0.0210 (9) | −0.0102 (8) | 0.0014 (8) | −0.0056 (7) |
| C9 | 0.0302 (11) | 0.0233 (10) | 0.0249 (10) | −0.0092 (8) | 0.0009 (8) | −0.0050 (8) |
| C10 | 0.0328 (11) | 0.0293 (11) | 0.0304 (11) | −0.0125 (9) | 0.0025 (9) | −0.0045 (9) |
| C11 | 0.0396 (12) | 0.0267 (11) | 0.0296 (11) | −0.0154 (9) | 0.0027 (9) | −0.0017 (8) |
| C12 | 0.0359 (12) | 0.0241 (10) | 0.0296 (11) | −0.0086 (9) | −0.0027 (9) | −0.0010 (8) |
| C13 | 0.0312 (11) | 0.0247 (10) | 0.0260 (10) | −0.0095 (8) | 0.0000 (8) | −0.0070 (8) |
| C14 | 0.0301 (11) | 0.0264 (11) | 0.0326 (11) | −0.0076 (9) | 0.0018 (9) | −0.0018 (9) |
| C15 | 0.0432 (14) | 0.0384 (13) | 0.0435 (14) | −0.0016 (11) | −0.0010 (11) | −0.0143 (11) |
| C16 | 0.0391 (13) | 0.0400 (13) | 0.0367 (13) | −0.0010 (10) | −0.0049 (11) | −0.0070 (10) |
| C17 | 0.0302 (11) | 0.0239 (10) | 0.0377 (12) | −0.0081 (8) | −0.0002 (9) | −0.0051 (9) |
| C18 | 0.0389 (14) | 0.0480 (15) | 0.0462 (15) | −0.0086 (11) | 0.0087 (11) | −0.0156 (12) |
| C19 | 0.0318 (12) | 0.0367 (13) | 0.0554 (16) | −0.0076 (10) | −0.0063 (11) | 0.0028 (11) |
| N1 | 0.0472 (13) | 0.0264 (10) | 0.0566 (15) | −0.0112 (9) | 0.0153 (11) | −0.0006 (10) |
| N2 | 0.0272 (9) | 0.0228 (8) | 0.0239 (8) | −0.0092 (7) | 0.0009 (7) | −0.0035 (7) |
| O1 | 0.0921 (18) | 0.0478 (12) | 0.0828 (17) | −0.0473 (12) | 0.0134 (14) | −0.0024 (11) |
| O2 | 0.0942 (17) | 0.0485 (12) | 0.0469 (12) | −0.0290 (12) | 0.0254 (12) | 0.0019 (9) |
| Cl1 | 0.0793 (5) | 0.0479 (4) | 0.0234 (3) | −0.0365 (3) | 0.0067 (3) | −0.0110 (2) |
Geometric parameters (Å, º)
| C1—C6 | 1.379 (3) | C12—C13 | 1.391 (3) |
| C1—C2 | 1.382 (3) | C12—H12 | 0.9500 |
| C1—N1 | 1.477 (3) | C13—C17 | 1.522 (3) |
| C2—C3 | 1.386 (3) | C14—C16 | 1.526 (3) |
| C2—H2 | 0.9500 | C14—C15 | 1.529 (3) |
| C3—C4 | 1.393 (3) | C14—H14 | 1.0000 |
| C3—H3 | 0.9500 | C15—H15A | 0.9800 |
| C4—C5 | 1.397 (3) | C15—H15B | 0.9800 |
| C4—C7 | 1.485 (3) | C15—H15C | 0.9800 |
| C5—C6 | 1.388 (3) | C16—H16A | 0.9800 |
| C5—H5 | 0.9500 | C16—H16B | 0.9800 |
| C6—H6 | 0.9500 | C16—H16C | 0.9800 |
| C7—N2 | 1.254 (3) | C17—C18 | 1.529 (4) |
| C7—Cl1 | 1.752 (2) | C17—C19 | 1.533 (3) |
| C8—C9 | 1.400 (3) | C17—H17 | 1.0000 |
| C8—C13 | 1.414 (3) | C18—H18A | 0.9800 |
| C8—N2 | 1.427 (3) | C18—H18B | 0.9800 |
| C9—C10 | 1.396 (3) | C18—H18C | 0.9800 |
| C9—C14 | 1.520 (3) | C19—H19A | 0.9800 |
| C10—C11 | 1.385 (3) | C19—H19B | 0.9800 |
| C10—H10 | 0.9500 | C19—H19C | 0.9800 |
| C11—C12 | 1.390 (3) | N1—O2 | 1.218 (3) |
| C11—H11 | 0.9500 | N1—O1 | 1.221 (3) |
| C6—C1—C2 | 122.8 (2) | C9—C14—C15 | 111.36 (19) |
| C6—C1—N1 | 118.8 (2) | C16—C14—C15 | 111.3 (2) |
| C2—C1—N1 | 118.4 (2) | C9—C14—H14 | 107.7 |
| C1—C2—C3 | 118.5 (2) | C16—C14—H14 | 107.7 |
| C1—C2—H2 | 120.7 | C15—C14—H14 | 107.7 |
| C3—C2—H2 | 120.7 | C14—C15—H15A | 109.5 |
| C2—C3—C4 | 120.3 (2) | C14—C15—H15B | 109.5 |
| C2—C3—H3 | 119.9 | H15A—C15—H15B | 109.5 |
| C4—C3—H3 | 119.9 | C14—C15—H15C | 109.5 |
| C3—C4—C5 | 119.64 (19) | H15A—C15—H15C | 109.5 |
| C3—C4—C7 | 122.22 (19) | H15B—C15—H15C | 109.5 |
| C5—C4—C7 | 118.14 (18) | C14—C16—H16A | 109.5 |
| C6—C5—C4 | 120.5 (2) | C14—C16—H16B | 109.5 |
| C6—C5—H5 | 119.7 | H16A—C16—H16B | 109.5 |
| C4—C5—H5 | 119.7 | C14—C16—H16C | 109.5 |
| C1—C6—C5 | 118.2 (2) | H16A—C16—H16C | 109.5 |
| C1—C6—H6 | 120.9 | H16B—C16—H16C | 109.5 |
| C5—C6—H6 | 120.9 | C13—C17—C18 | 111.42 (19) |
| N2—C7—C4 | 121.94 (18) | C13—C17—C19 | 113.50 (19) |
| N2—C7—Cl1 | 122.36 (16) | C18—C17—C19 | 110.2 (2) |
| C4—C7—Cl1 | 115.70 (15) | C13—C17—H17 | 107.1 |
| C9—C8—C13 | 122.16 (19) | C18—C17—H17 | 107.1 |
| C9—C8—N2 | 118.85 (17) | C19—C17—H17 | 107.1 |
| C13—C8—N2 | 118.84 (18) | C17—C18—H18A | 109.5 |
| C10—C9—C8 | 117.86 (19) | C17—C18—H18B | 109.5 |
| C10—C9—C14 | 120.20 (19) | H18A—C18—H18B | 109.5 |
| C8—C9—C14 | 121.94 (18) | C17—C18—H18C | 109.5 |
| C11—C10—C9 | 121.3 (2) | H18A—C18—H18C | 109.5 |
| C11—C10—H10 | 119.3 | H18B—C18—H18C | 109.5 |
| C9—C10—H10 | 119.3 | C17—C19—H19A | 109.5 |
| C10—C11—C12 | 119.7 (2) | C17—C19—H19B | 109.5 |
| C10—C11—H11 | 120.2 | H19A—C19—H19B | 109.5 |
| C12—C11—H11 | 120.2 | C17—C19—H19C | 109.5 |
| C11—C12—C13 | 121.6 (2) | H19A—C19—H19C | 109.5 |
| C11—C12—H12 | 119.2 | H19B—C19—H19C | 109.5 |
| C13—C12—H12 | 119.2 | O2—N1—O1 | 124.1 (2) |
| C12—C13—C8 | 117.3 (2) | O2—N1—C1 | 118.0 (2) |
| C12—C13—C17 | 122.60 (19) | O1—N1—C1 | 117.9 (2) |
| C8—C13—C17 | 120.03 (18) | C7—N2—C8 | 122.75 (18) |
| C9—C14—C16 | 110.82 (19) |
Hydrogen-bond geometry (Å, º)
Cg1 and Cg2 are the centroids of the C1–C6 and C8–C13 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C6—H6···Cg2i | 0.95 | 2.67 | 3.511 (2) | 147 |
| C16—H16B···Cg1ii | 0.98 | 2.79 | 3.663 (3) | 149 |
Symmetry codes: (i) −x+1, −y+1, −z+2; (ii) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS5293).
References
- Altomare, A., Cascarano, G., Giacovazzo, C. & Guagliardi, A. (1993). J. Appl. Cryst. 26, 343–350.
- Cunico, R. F. & Pandey, R. K. (2005). J. Org. Chem. 70, 5344–5346. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Kuszpit, M. R., Wulff, W. D. & Tepe, J. J. (2011). J. Org. Chem. 76, 2913–2919. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- Manley, P. J. & Bilodeau, M. T. (2002). Org. Lett. 4, 3127–3129. [DOI] [PubMed]
- Nonius (2000). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Pelter, A., Smith, K., Hutchings, M. G. & Rowe, K. (1975). J. Chem. Soc. Perkin Trans. 1, pp. 129–138.
- Raussukana, Y. V., Khomenko, E. A., Onys’ko, P. P. & Sinitsa, A. D. (2006). Synthesis, pp. 3195–3198.
- Seidelmann, O., Beyer, L., Lessmann, F. & Richter, R. (1998). Inorg. Chem. Commun. 1, 472–474.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Zheng, Z. & Alper, H. (2008). Org. Lett. 10, 4903–4906. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S1600536813020862/is5293sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813020862/is5293Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813020862/is5293Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report