Abstract
In the title compound, C13H15NO3, the pyrrolidine ring makes a dihedral angle of 4.69 (9)° with the 3-methoxy-phenyl ring. In the crystal, hydrogen-bonded chains running along [101] are generated by connecting neighbouring molecules via C—H⋯O hydrogen bonds. Parallel chains are linked by further C—H⋯O hydrogen bonds, forming a three-dimensional structure.
Related literature
For the bioactivity of pyrrolidine-2,5-dione derivatives, see: Obniska et al. (2012 ▶); Ha et al. (2011 ▶); Kaminski et al. (2011 ▶). For related structures, see: Khorasani & Fernandes (2012 ▶); Mayes et al. (2008 ▶).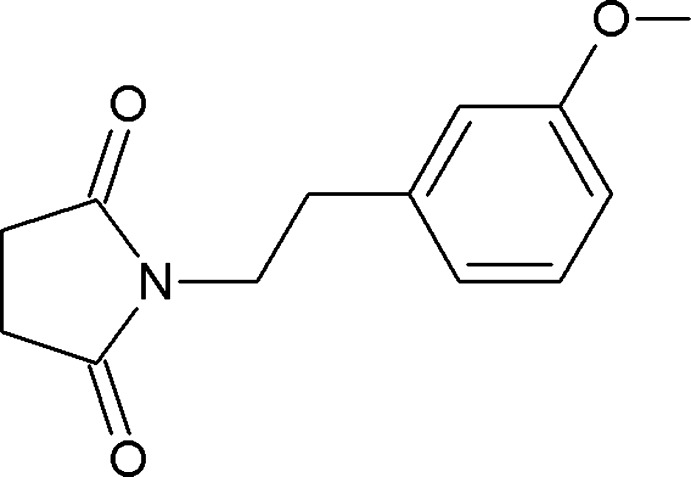
Experimental
Crystal data
C13H15NO3
M r = 233.26
Monoclinic,

a = 12.8719 (9) Å
b = 12.5878 (8) Å
c = 7.4523 (5) Å
β = 90.831 (3)°
V = 1207.36 (14) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 293 K
0.40 × 0.35 × 0.20 mm
Data collection
Bruker SMART APEXII area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2008 ▶) T min = 0.964, T max = 0.982
5692 measured reflections
2615 independent reflections
2328 reflections with I > 2σ(I)
R int = 0.024
Refinement
R[F 2 > 2σ(F 2)] = 0.035
wR(F 2) = 0.097
S = 1.02
2615 reflections
156 parameters
2 restraints
H-atom parameters constrained
Δρmax = 0.12 e Å−3
Δρmin = −0.16 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813023751/su2640sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813023751/su2640Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813023751/su2640Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C1—H1B⋯O3i | 0.96 | 2.54 | 3.418 (3) | 151 |
| C8—H8B⋯O1ii | 0.97 | 2.54 | 3.469 (2) | 161 |
| C12—H12A⋯O2iii | 0.97 | 2.57 | 3.456 (3) | 152 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
We are grateful to Dr C. R. Ramanathan, Department of Chemistry, Pondicherry University for support of this research. The authors thank the TBI X-ray facility, CAS in Crystallography and Biophysics, University of Madras, India, for the data collection. ZF and DV acknowledge the UGC (SAP–CAS) for departmental facilities. ZF also thanks the UGC for a meritorious fellowship.
supplementary crystallographic information
1. Comment
Pyrrolidine-2,5-dione derivates are an important class of heterocylic compounds with essential applications in medicinal chemistry and organic synthesis. They exhibit numerous bioactivities, for example anticonvulsant (Obniska et al., 2012; Kaminski et al., 2011) and tyrosinase inhibitory activity (Ha et al., 2011). In the field of organic chemistry derivates, like 1-bromopyrrolidine-2,5-dione (NBS), are the most commonly used halogenation reagents. In view of the different applications of this class of compounds, we have synthesized the title derivative and report herein on its crystal structure.
In the title compound, Fig. 1, the pyrrolidine ring (N1/C10—C13) makes a dihedral angle of 4.69 (9)° with the benzene ring (C2—C7).
In the crystal, hydrogen-bonded chains running along [101] are generated by connecting neighbouring molecules via C—H···O hydrogen bonds (Table 1 and Fig. 2). Parallel chains are linked by further C—H···O hydrogen bonds forming a three-dimensional structure (Table 1 and Fig. 2).
2. Experimental
3-methoxy phenethylamine (1.51 g, 10 mmol) and succinic anhydride (1.2 g, 12 mmol) were stirred at room temperature in dry ethyl acetate for 30 min. Ethyl acetate was removed under reduced pressure, and the resulting residue was dissolved in toluene. Acetyl chloride (5 equiv) was then added and the mixture refluxed for 1 h. The reaction mixture was washed with aqueous Na2CO3 and dried over anhydrous Na2SO4, filtered and concentrated under reduced pressure followed by silica gel column purification using hexane ethyl acetate (30:70) as eluent to afford the title compound as a colourless solid. Single crystals suitable for X-ray diffraction analysis were obtained by slow evaporation of a solution of the title compound in ethanol at room temperature.
3. Refinement
The H atoms were placed in calculated positions and treated as riding atoms: C—H = 0.93 Å to 0.97 Å, with Uiso(H) = 1.5Ueq(C-methyl) and = 1.2Ueq(C) for other H atoms.
Figures
Fig. 1.

The molecular structure of the title molecule, with atom labelling. Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.

The crystal packing of the title compound viewed along the c axis. Hydrogen bonds are shown as dashed lines (see Table 1 for details; H atoms not involved in hydrogen bonding have been omitted for clarity).
Crystal data
| C13H15NO3 | F(000) = 496 |
| Mr = 233.26 | Dx = 1.283 Mg m−3 |
| Monoclinic, Cc | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: C -2yc | Cell parameters from 2615 reflections |
| a = 12.8719 (9) Å | θ = 2.3–28.4° |
| b = 12.5878 (8) Å | µ = 0.09 mm−1 |
| c = 7.4523 (5) Å | T = 293 K |
| β = 90.831 (3)° | Block, colourless |
| V = 1207.36 (14) Å3 | 0.40 × 0.35 × 0.20 mm |
| Z = 4 |
Data collection
| Bruker SMART APEXII area-detector diffractometer | 2615 independent reflections |
| Radiation source: fine-focus sealed tube | 2328 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.024 |
| ω and φ scans | θmax = 28.4°, θmin = 2.3° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −17→17 |
| Tmin = 0.964, Tmax = 0.982 | k = −15→16 |
| 5692 measured reflections | l = −9→9 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.035 | H-atom parameters constrained |
| wR(F2) = 0.097 | w = 1/[σ2(Fo2) + (0.0544P)2 + 0.1677P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max < 0.001 |
| 2615 reflections | Δρmax = 0.12 e Å−3 |
| 156 parameters | Δρmin = −0.16 e Å−3 |
| 2 restraints | Extinction correction: SHELXL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0149 (15) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.95763 (19) | 0.43725 (18) | 1.0655 (3) | 0.0832 (6) | |
| H1A | 0.9806 | 0.4494 | 0.9452 | 0.125* | |
| H1B | 1.0043 | 0.4716 | 1.1487 | 0.125* | |
| H1C | 0.8890 | 0.4657 | 1.0789 | 0.125* | |
| C2 | 0.90109 (11) | 0.26275 (13) | 0.9852 (2) | 0.0485 (3) | |
| C3 | 0.91325 (14) | 0.15484 (15) | 1.0131 (2) | 0.0633 (4) | |
| H3 | 0.9582 | 0.1300 | 1.1025 | 0.076* | |
| C4 | 0.85800 (17) | 0.08476 (14) | 0.9071 (3) | 0.0695 (5) | |
| H4 | 0.8652 | 0.0121 | 0.9261 | 0.083* | |
| C5 | 0.79213 (14) | 0.12096 (13) | 0.7730 (2) | 0.0609 (4) | |
| H5 | 0.7552 | 0.0726 | 0.7026 | 0.073* | |
| C6 | 0.78069 (11) | 0.22844 (13) | 0.74273 (18) | 0.0472 (3) | |
| C7 | 0.83541 (11) | 0.29992 (12) | 0.85029 (19) | 0.0450 (3) | |
| H7 | 0.8280 | 0.3726 | 0.8317 | 0.054* | |
| C8 | 0.70939 (12) | 0.26805 (15) | 0.5942 (2) | 0.0564 (4) | |
| H8A | 0.6893 | 0.3408 | 0.6193 | 0.068* | |
| H8B | 0.6468 | 0.2251 | 0.5907 | 0.068* | |
| C9 | 0.76102 (13) | 0.26346 (15) | 0.4132 (2) | 0.0565 (4) | |
| H9A | 0.8161 | 0.3159 | 0.4094 | 0.068* | |
| H9B | 0.7920 | 0.1939 | 0.3972 | 0.068* | |
| C10 | 0.63151 (13) | 0.20320 (14) | 0.1850 (2) | 0.0573 (4) | |
| C11 | 0.56115 (15) | 0.25110 (19) | 0.0462 (3) | 0.0737 (6) | |
| H11A | 0.4891 | 0.2352 | 0.0712 | 0.088* | |
| H11B | 0.5774 | 0.2247 | −0.0724 | 0.088* | |
| C12 | 0.58151 (16) | 0.36961 (19) | 0.0592 (3) | 0.0777 (6) | |
| H12A | 0.6066 | 0.3969 | −0.0540 | 0.093* | |
| H12B | 0.5185 | 0.4073 | 0.0896 | 0.093* | |
| C13 | 0.66250 (15) | 0.38270 (13) | 0.2046 (2) | 0.0609 (4) | |
| N1 | 0.68700 (9) | 0.28356 (10) | 0.26754 (15) | 0.0475 (3) | |
| O1 | 0.95629 (10) | 0.32628 (11) | 1.10049 (17) | 0.0690 (3) | |
| O2 | 0.70187 (17) | 0.46337 (11) | 0.2584 (2) | 0.0967 (5) | |
| O3 | 0.64082 (15) | 0.11124 (11) | 0.2245 (2) | 0.0906 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0866 (14) | 0.0787 (13) | 0.0843 (14) | −0.0200 (11) | −0.0031 (11) | −0.0264 (11) |
| C2 | 0.0412 (7) | 0.0623 (8) | 0.0421 (7) | 0.0004 (7) | 0.0000 (6) | −0.0038 (7) |
| C3 | 0.0629 (10) | 0.0683 (10) | 0.0585 (10) | 0.0098 (8) | −0.0075 (8) | 0.0138 (8) |
| C4 | 0.0825 (12) | 0.0484 (9) | 0.0773 (12) | 0.0001 (8) | −0.0032 (10) | 0.0119 (8) |
| C5 | 0.0636 (10) | 0.0534 (9) | 0.0658 (11) | −0.0099 (7) | −0.0003 (8) | −0.0074 (7) |
| C6 | 0.0405 (7) | 0.0586 (8) | 0.0425 (7) | −0.0008 (6) | 0.0019 (6) | −0.0018 (6) |
| C7 | 0.0429 (7) | 0.0486 (7) | 0.0437 (7) | 0.0016 (6) | 0.0014 (6) | −0.0002 (6) |
| C8 | 0.0444 (7) | 0.0769 (10) | 0.0478 (8) | 0.0049 (7) | −0.0064 (6) | −0.0044 (8) |
| C9 | 0.0434 (7) | 0.0776 (11) | 0.0483 (8) | 0.0044 (7) | −0.0084 (6) | 0.0034 (8) |
| C10 | 0.0570 (9) | 0.0649 (11) | 0.0502 (8) | −0.0133 (7) | 0.0058 (7) | −0.0053 (7) |
| C11 | 0.0508 (9) | 0.1229 (19) | 0.0473 (8) | −0.0130 (10) | −0.0052 (7) | 0.0007 (10) |
| C12 | 0.0687 (12) | 0.1047 (16) | 0.0596 (10) | 0.0264 (11) | 0.0000 (9) | 0.0204 (10) |
| C13 | 0.0717 (11) | 0.0584 (9) | 0.0529 (10) | 0.0049 (8) | 0.0093 (8) | 0.0062 (7) |
| N1 | 0.0448 (6) | 0.0538 (7) | 0.0439 (7) | −0.0009 (5) | −0.0041 (5) | −0.0004 (5) |
| O1 | 0.0654 (7) | 0.0838 (9) | 0.0573 (7) | −0.0055 (6) | −0.0185 (5) | −0.0104 (6) |
| O2 | 0.1454 (15) | 0.0532 (7) | 0.0917 (10) | −0.0170 (9) | 0.0081 (10) | −0.0025 (7) |
| O3 | 0.1184 (13) | 0.0569 (8) | 0.0962 (12) | −0.0184 (7) | −0.0033 (10) | −0.0054 (7) |
Geometric parameters (Å, º)
| C1—O1 | 1.421 (3) | C8—H8A | 0.9700 |
| C1—H1A | 0.9600 | C8—H8B | 0.9700 |
| C1—H1B | 0.9600 | C9—N1 | 1.4562 (19) |
| C1—H1C | 0.9600 | C9—H9A | 0.9700 |
| C2—O1 | 1.3654 (19) | C9—H9B | 0.9700 |
| C2—C3 | 1.383 (2) | C10—O3 | 1.200 (2) |
| C2—C7 | 1.386 (2) | C10—N1 | 1.378 (2) |
| C3—C4 | 1.376 (3) | C10—C11 | 1.492 (3) |
| C3—H3 | 0.9300 | C11—C12 | 1.517 (3) |
| C4—C5 | 1.379 (3) | C11—H11A | 0.9700 |
| C4—H4 | 0.9300 | C11—H11B | 0.9700 |
| C5—C6 | 1.379 (2) | C12—C13 | 1.502 (3) |
| C5—H5 | 0.9300 | C12—H12A | 0.9700 |
| C6—C7 | 1.390 (2) | C12—H12B | 0.9700 |
| C6—C8 | 1.512 (2) | C13—O2 | 1.201 (2) |
| C7—H7 | 0.9300 | C13—N1 | 1.368 (2) |
| C8—C9 | 1.513 (2) | ||
| O1—C1—H1A | 109.5 | H8A—C8—H8B | 107.9 |
| O1—C1—H1B | 109.5 | N1—C9—C8 | 111.51 (13) |
| H1A—C1—H1B | 109.5 | N1—C9—H9A | 109.3 |
| O1—C1—H1C | 109.5 | C8—C9—H9A | 109.3 |
| H1A—C1—H1C | 109.5 | N1—C9—H9B | 109.3 |
| H1B—C1—H1C | 109.5 | C8—C9—H9B | 109.3 |
| O1—C2—C3 | 115.10 (14) | H9A—C9—H9B | 108.0 |
| O1—C2—C7 | 124.41 (14) | O3—C10—N1 | 123.31 (18) |
| C3—C2—C7 | 120.48 (14) | O3—C10—C11 | 128.13 (18) |
| C4—C3—C2 | 119.16 (15) | N1—C10—C11 | 108.56 (15) |
| C4—C3—H3 | 120.4 | C10—C11—C12 | 104.51 (15) |
| C2—C3—H3 | 120.4 | C10—C11—H11A | 110.8 |
| C3—C4—C5 | 120.78 (16) | C12—C11—H11A | 110.9 |
| C3—C4—H4 | 119.6 | C10—C11—H11B | 110.8 |
| C5—C4—H4 | 119.6 | C12—C11—H11B | 110.9 |
| C4—C5—C6 | 120.39 (16) | H11A—C11—H11B | 108.9 |
| C4—C5—H5 | 119.8 | C13—C12—C11 | 105.73 (15) |
| C6—C5—H5 | 119.8 | C13—C12—H12A | 110.6 |
| C5—C6—C7 | 119.25 (14) | C11—C12—H12A | 110.6 |
| C5—C6—C8 | 120.36 (14) | C13—C12—H12B | 110.6 |
| C7—C6—C8 | 120.39 (15) | C11—C12—H12B | 110.6 |
| C2—C7—C6 | 119.91 (14) | H12A—C12—H12B | 108.7 |
| C2—C7—H7 | 120.0 | O2—C13—N1 | 124.25 (18) |
| C6—C7—H7 | 120.0 | O2—C13—C12 | 128.20 (19) |
| C6—C8—C9 | 111.72 (12) | N1—C13—C12 | 107.54 (16) |
| C6—C8—H8A | 109.3 | C13—N1—C10 | 113.65 (14) |
| C9—C8—H8A | 109.3 | C13—N1—C9 | 123.99 (15) |
| C6—C8—H8B | 109.3 | C10—N1—C9 | 122.31 (15) |
| C9—C8—H8B | 109.3 | C2—O1—C1 | 117.90 (14) |
| O1—C2—C3—C4 | 177.92 (17) | C10—C11—C12—C13 | 0.1 (2) |
| C7—C2—C3—C4 | −1.1 (3) | C11—C12—C13—O2 | 179.5 (2) |
| C2—C3—C4—C5 | 0.8 (3) | C11—C12—C13—N1 | 0.5 (2) |
| C3—C4—C5—C6 | 0.1 (3) | O2—C13—N1—C10 | −179.95 (18) |
| C4—C5—C6—C7 | −0.7 (3) | C12—C13—N1—C10 | −0.94 (19) |
| C4—C5—C6—C8 | 179.27 (16) | O2—C13—N1—C9 | 2.7 (3) |
| O1—C2—C7—C6 | −178.43 (15) | C12—C13—N1—C9 | −178.33 (15) |
| C3—C2—C7—C6 | 0.4 (2) | O3—C10—N1—C13 | −178.52 (19) |
| C5—C6—C7—C2 | 0.5 (2) | C11—C10—N1—C13 | 1.00 (19) |
| C8—C6—C7—C2 | −179.55 (14) | O3—C10—N1—C9 | −1.1 (2) |
| C5—C6—C8—C9 | −80.96 (19) | C11—C10—N1—C9 | 178.43 (16) |
| C7—C6—C8—C9 | 99.05 (18) | C8—C9—N1—C13 | 87.3 (2) |
| C6—C8—C9—N1 | 169.92 (15) | C8—C9—N1—C10 | −89.88 (19) |
| O3—C10—C11—C12 | 178.89 (19) | C3—C2—O1—C1 | 172.27 (18) |
| N1—C10—C11—C12 | −0.6 (2) | C7—C2—O1—C1 | −8.8 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1B···O3i | 0.96 | 2.54 | 3.418 (3) | 151 |
| C8—H8B···O1ii | 0.97 | 2.54 | 3.469 (2) | 161 |
| C12—H12A···O2iii | 0.97 | 2.57 | 3.456 (3) | 152 |
Symmetry codes: (i) x+1/2, y+1/2, z+1; (ii) x−1/2, −y+1/2, z−1/2; (iii) x, −y+1, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2640).
References
- Bruker (2008). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, U. S. A.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Ha, Y. M., Kim, J., Parkl, Y. J., Park, D., Choi, Y. J., Kim, J. M., Chung, K. W., Han, Y. K., Park, J. Y., Lee, J. Y., Moon, H. R. & Chung, H. Y. (2011). Med. Chem. Commun. 2, 542–549.
- Kaminski, K., Rzepka, S. & Obniska, J. (2011). Bioorg. Med. Chem. Lett. 21, 5800–5803. [DOI] [PubMed]
- Khorasani, S. & Fernandes, M. A. (2012). Acta Cryst. E68, o1503. [DOI] [PMC free article] [PubMed]
- Mayes, B. A., McGarry, P., Moussa, A. & Watkin, D. J. (2008). Acta Cryst. E64, o1355. [DOI] [PMC free article] [PubMed]
- Obniska, J., Rzepka, S. & Kamin’ ski, K. (2012). Bioorg. Med. Chem. 20, 4872–4880. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813023751/su2640sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813023751/su2640Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813023751/su2640Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report


