Abstract
The crystal structure of the title compound, C26H39BO2, which contains no strong hydrogen bond donors, displays only long C—H⋯O contacts between inversion-related pairs of molecules. The structure contains layers rich in oxygen and boron parallel to the ac plane. The dioxaborinane ring adopts an envelope conformation with the C atom attached to the two methyl groups as the flap .
Related literature
For the synthesis and applications of allylboronic esters, see: Lombardo et al. (2002 ▶); Carosi & Hall (2007 ▶); Althaus et al. (2010 ▶); Fandrick et al. (2010 ▶); Clary et al. (2011 ▶); Hesse et al. (2012 ▶); Incerti-Pradillos et al. (2013 ▶). For the X-ray structure of a boronic ester, see: Sopková-de Oliveira Santos et al. (2003 ▶).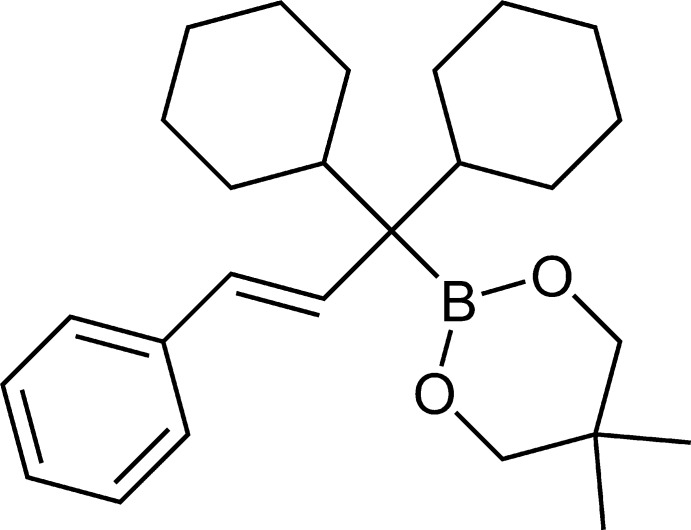
Experimental
Crystal data
C26H39BO2
M r = 394.38
Triclinic,

a = 9.4967 (3) Å
b = 11.2837 (2) Å
c = 12.0297 (4) Å
α = 109.897 (2)°
β = 96.388 (2)°
γ = 102.048 (2)°
V = 1161.90 (6) Å3
Z = 2
Mo Kα radiation
μ = 0.07 mm−1
T = 150 K
0.38 × 0.30 × 0.28 mm
Data collection
Nonius KappaCCD diffractometer
Absorption correction: multi-scan (DENZO/SCALEPACK; Otwinowski & Minor, 1997 ▶) T min = 0.975, T max = 0.981
8277 measured reflections
5287 independent reflections
4303 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.050
wR(F 2) = 0.128
S = 1.03
5287 reflections
264 parameters
H-atom parameters constrained
Δρmax = 0.32 e Å−3
Δρmin = −0.20 e Å−3
Data collection: COLLECT (Nonius, 2000 ▶); cell refinement: SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: DENZO (Otwinowski & Minor, 1997 ▶) and SCALEPACK; program(s) used to solve structure: SIR92 (Altomare et al., 1994) ▶; program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP99 for Windows (Farrugia, 2012 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶) and CHEMDRAW Ultra (Cambridge Soft, 2001 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S1600536813021739/go2094sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813021739/go2094Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813021739/go2094Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C22—H22B⋯O2i | 0.99 | 2.69 | 3.5773 (18) | 150 |
Symmetry code: (i)  .
.
Acknowledgments
The authors would like to extend their appreciation to the Deanship of Scientific Research at King Saud University for its funding for this research through the research group project RGP-VPP-239.
supplementary crystallographic information
1. Comment
The title compound I, a useful synthetic intermediate, was synthesized by the reaction of (E)-dicyclohexylstyrylborane with the anion of dichloromethyl methyl ether followed by esterification with 2,2-dimethyl-1,3-propanediol. Allylboronic esters have been synthesized from the reaction of lithiated carbamates with vinylboranes [Althaus et al. (2010)], and from the reaction of primary allyl halides with pinacolborane and magnesium [Incerti-Pradillos et al. (2013), Clary et al. (2011)]. Allylboronic esters are important synthetic intermediates, which have been shown to react with aldehydes to give homoallylic alcohols [Lombardo et al. (2002)], with the control of the newly-generated stereogenic centre possible through use of a chiral catalyst [Carosi et al. (2007)]. Allylboronic esters take part in a zinc alkoxide catalysed reaction with ketones to give the corresponding homoallylic alcohol products [Fandrick et al. (2010)], and also take part in a proto-deboronation reaction which has been used to synthesize the pheromone of the Californian red scale beetle [Hesse et al. (2012)]. For the X-ray structure of a boronic ester, see: Sopková-de Oliveira Santos et al. (2003).
In the molecule (Figure 1), the two cyclohexyl groups assume a chair conformation and an envelope conformation is observed for the dioxaborinane ring. The phenylallyl group is not planar as the plane through the double bond makes an angle of 20.84 ° with the phenyl group. There are no strong hydrogen bond donors in the structure. Long contacts of C—H···O type occur between pairs of molecules to form loosely bound dimers (Figure 2). The dimers are stacked along the a-axis to form a structure with layers rich in oxygen and boron parallel to the ac plane (Figure 3).
2. Refinement
H atoms were positioned geometrically and refined using a riding model with Uiso(H) = 1.2 times Ueq for the atom they are bonded to except for the methyl groups where 1.5 times Ueq was used with free rotation about the C—C bond. Of the low angle reflections not included in the refinement only (0 0 1) and (0 1 0) were omitted due to low intensities consistent with being obscured by the beamstop. The rest were eliminated automatically during data processing possibly as overloads.
Figures
Fig. 1.
A molecule showing atom labels and 50% probability displacement ellipsoids for non-H atoms.
Fig. 2.
A pair of molecules showing C—H···O interactions as dotted lines.
Fig. 3.
Molecular packing in the crystal structure showing oxygen and boron rich layers.
Crystal data
| C26H39BO2 | Z = 2 |
| Mr = 394.38 | F(000) = 432 |
| Triclinic, P1 | Dx = 1.127 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.4967 (3) Å | Cell parameters from 4303 reflections |
| b = 11.2837 (2) Å | θ = 2.2–27.5° |
| c = 12.0297 (4) Å | µ = 0.07 mm−1 |
| α = 109.897 (2)° | T = 150 K |
| β = 96.388 (2)° | Block, colourless |
| γ = 102.048 (2)° | 0.38 × 0.30 × 0.28 mm |
| V = 1161.90 (6) Å3 |
Data collection
| Nonius KappaCCD diffractometer | 5287 independent reflections |
| Radiation source: fine-focus sealed tube | 4303 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.027 |
| CCD slices, ω and phi scans | θmax = 27.5°, θmin = 2.1° |
| Absorption correction: multi-scan (DENZO/SCALEPACK; Otwinowski & Minor, 1997) | h = −12→12 |
| Tmin = 0.975, Tmax = 0.981 | k = −14→14 |
| 8277 measured reflections | l = −15→12 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.128 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0488P)2 + 0.4651P] where P = (Fo2 + 2Fc2)/3 |
| 5287 reflections | (Δ/σ)max = 0.004 |
| 264 parameters | Δρmax = 0.32 e Å−3 |
| 0 restraints | Δρmin = −0.20 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.24096 (15) | 0.23892 (13) | 0.62817 (11) | 0.0235 (3) | |
| C2 | 0.22913 (18) | 0.35166 (15) | 0.71836 (13) | 0.0348 (3) | |
| H2 | 0.2828 | 0.4349 | 0.7232 | 0.042* | |
| C3 | 0.14007 (19) | 0.34351 (17) | 0.80085 (14) | 0.0411 (4) | |
| H3 | 0.1319 | 0.4211 | 0.8607 | 0.049* | |
| C4 | 0.06330 (17) | 0.22337 (17) | 0.79648 (14) | 0.0382 (4) | |
| H4 | 0.0029 | 0.2179 | 0.8534 | 0.046* | |
| C5 | 0.07497 (17) | 0.11101 (16) | 0.70869 (14) | 0.0348 (3) | |
| H5 | 0.0228 | 0.0280 | 0.7055 | 0.042* | |
| C6 | 0.16250 (16) | 0.11876 (13) | 0.62506 (13) | 0.0281 (3) | |
| H6 | 0.1689 | 0.0407 | 0.5648 | 0.034* | |
| C7 | 0.33581 (15) | 0.25145 (13) | 0.54118 (12) | 0.0255 (3) | |
| H7 | 0.4098 | 0.3312 | 0.5644 | 0.031* | |
| C8 | 0.32693 (14) | 0.16134 (12) | 0.43362 (11) | 0.0221 (3) | |
| H8 | 0.2530 | 0.0818 | 0.4117 | 0.027* | |
| C9 | 0.42193 (14) | 0.17080 (12) | 0.34118 (11) | 0.0205 (3) | |
| C10 | 0.58492 (14) | 0.24810 (12) | 0.40612 (11) | 0.0218 (3) | |
| H10 | 0.5813 | 0.3293 | 0.4725 | 0.026* | |
| C11 | 0.65750 (15) | 0.16852 (13) | 0.46484 (12) | 0.0260 (3) | |
| H11A | 0.5978 | 0.1466 | 0.5210 | 0.031* | |
| H11B | 0.6589 | 0.0856 | 0.4014 | 0.031* | |
| C12 | 0.81407 (16) | 0.24166 (14) | 0.53369 (13) | 0.0330 (3) | |
| H12A | 0.8121 | 0.3195 | 0.6031 | 0.040* | |
| H12B | 0.8576 | 0.1844 | 0.5655 | 0.040* | |
| C13 | 0.90896 (17) | 0.28434 (15) | 0.45299 (15) | 0.0380 (4) | |
| H13A | 0.9200 | 0.2064 | 0.3883 | 0.046* | |
| H13B | 1.0079 | 0.3364 | 0.5010 | 0.046* | |
| C14 | 0.83863 (17) | 0.36592 (15) | 0.39717 (15) | 0.0361 (4) | |
| H14A | 0.8996 | 0.3908 | 0.3430 | 0.043* | |
| H14B | 0.8349 | 0.4471 | 0.4618 | 0.043* | |
| C15 | 0.68323 (15) | 0.29066 (13) | 0.32573 (13) | 0.0283 (3) | |
| H15A | 0.6875 | 0.2125 | 0.2577 | 0.034* | |
| H15B | 0.6400 | 0.3466 | 0.2918 | 0.034* | |
| C16 | 0.35441 (15) | 0.23557 (12) | 0.25946 (12) | 0.0229 (3) | |
| H16 | 0.4152 | 0.2331 | 0.1963 | 0.027* | |
| C17 | 0.19670 (16) | 0.16006 (14) | 0.19300 (13) | 0.0309 (3) | |
| H17A | 0.1928 | 0.0671 | 0.1500 | 0.037* | |
| H17B | 0.1315 | 0.1645 | 0.2522 | 0.037* | |
| C18 | 0.14178 (19) | 0.21631 (17) | 0.10238 (15) | 0.0404 (4) | |
| H18A | 0.2011 | 0.2042 | 0.0387 | 0.048* | |
| H18B | 0.0384 | 0.1683 | 0.0635 | 0.048* | |
| C19 | 0.1523 (2) | 0.36161 (18) | 0.16395 (16) | 0.0436 (4) | |
| H19A | 0.1255 | 0.3970 | 0.1021 | 0.052* | |
| H19B | 0.0814 | 0.3726 | 0.2187 | 0.052* | |
| C20 | 0.30661 (19) | 0.43826 (15) | 0.23583 (14) | 0.0364 (4) | |
| H20A | 0.3073 | 0.5301 | 0.2804 | 0.044* | |
| H20B | 0.3754 | 0.4379 | 0.1797 | 0.044* | |
| C21 | 0.35828 (17) | 0.37906 (13) | 0.32525 (13) | 0.0287 (3) | |
| H21A | 0.4597 | 0.4290 | 0.3689 | 0.034* | |
| H21B | 0.2940 | 0.3855 | 0.3852 | 0.034* | |
| C22 | 0.51831 (16) | −0.10974 (13) | 0.09454 (13) | 0.0290 (3) | |
| H22A | 0.6131 | −0.1146 | 0.1338 | 0.035* | |
| H22B | 0.5259 | −0.1158 | 0.0115 | 0.035* | |
| C23 | 0.39668 (15) | −0.22485 (12) | 0.08888 (12) | 0.0256 (3) | |
| C24 | 0.37886 (18) | −0.20513 (13) | 0.21769 (12) | 0.0301 (3) | |
| H24A | 0.2915 | −0.2720 | 0.2158 | 0.036* | |
| H24B | 0.4659 | −0.2183 | 0.2607 | 0.036* | |
| C25 | 0.25412 (17) | −0.23168 (15) | 0.01216 (14) | 0.0363 (4) | |
| H25A | 0.1768 | −0.3060 | 0.0092 | 0.055* | |
| H25B | 0.2688 | −0.2427 | −0.0697 | 0.055* | |
| H25C | 0.2250 | −0.1507 | 0.0477 | 0.055* | |
| C26 | 0.44291 (19) | −0.35104 (14) | 0.03502 (14) | 0.0373 (4) | |
| H26A | 0.5375 | −0.3438 | 0.0822 | 0.056* | |
| H26B | 0.4524 | −0.3656 | −0.0486 | 0.056* | |
| H26C | 0.3685 | −0.4246 | 0.0369 | 0.056* | |
| B1 | 0.42494 (16) | 0.02685 (14) | 0.25703 (13) | 0.0210 (3) | |
| O1 | 0.36243 (11) | −0.07729 (8) | 0.28350 (8) | 0.0261 (2) | |
| O2 | 0.49222 (11) | 0.01427 (9) | 0.16035 (8) | 0.0273 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0250 (7) | 0.0294 (6) | 0.0191 (6) | 0.0096 (5) | 0.0060 (5) | 0.0110 (5) |
| C2 | 0.0404 (9) | 0.0318 (7) | 0.0276 (7) | 0.0065 (6) | 0.0118 (6) | 0.0056 (6) |
| C3 | 0.0444 (9) | 0.0484 (9) | 0.0257 (8) | 0.0151 (8) | 0.0147 (7) | 0.0040 (7) |
| C4 | 0.0306 (8) | 0.0644 (10) | 0.0285 (8) | 0.0168 (7) | 0.0142 (6) | 0.0232 (7) |
| C5 | 0.0303 (8) | 0.0443 (8) | 0.0394 (8) | 0.0094 (6) | 0.0119 (6) | 0.0263 (7) |
| C6 | 0.0299 (7) | 0.0298 (7) | 0.0296 (7) | 0.0115 (6) | 0.0099 (6) | 0.0138 (6) |
| C7 | 0.0291 (7) | 0.0248 (6) | 0.0240 (7) | 0.0063 (5) | 0.0094 (5) | 0.0102 (5) |
| C8 | 0.0238 (7) | 0.0240 (6) | 0.0219 (6) | 0.0079 (5) | 0.0073 (5) | 0.0108 (5) |
| C9 | 0.0243 (6) | 0.0216 (6) | 0.0188 (6) | 0.0083 (5) | 0.0072 (5) | 0.0092 (5) |
| C10 | 0.0229 (6) | 0.0225 (6) | 0.0210 (6) | 0.0072 (5) | 0.0066 (5) | 0.0081 (5) |
| C11 | 0.0286 (7) | 0.0282 (6) | 0.0230 (7) | 0.0092 (5) | 0.0049 (5) | 0.0106 (5) |
| C12 | 0.0309 (8) | 0.0338 (7) | 0.0318 (8) | 0.0121 (6) | 0.0013 (6) | 0.0085 (6) |
| C13 | 0.0264 (8) | 0.0372 (8) | 0.0450 (9) | 0.0078 (6) | 0.0061 (7) | 0.0093 (7) |
| C14 | 0.0276 (8) | 0.0352 (8) | 0.0442 (9) | 0.0035 (6) | 0.0120 (7) | 0.0145 (7) |
| C15 | 0.0285 (7) | 0.0301 (7) | 0.0295 (7) | 0.0067 (6) | 0.0111 (6) | 0.0140 (6) |
| C16 | 0.0265 (7) | 0.0262 (6) | 0.0219 (6) | 0.0114 (5) | 0.0088 (5) | 0.0123 (5) |
| C17 | 0.0295 (8) | 0.0356 (7) | 0.0295 (7) | 0.0114 (6) | 0.0042 (6) | 0.0136 (6) |
| C18 | 0.0355 (9) | 0.0569 (10) | 0.0353 (8) | 0.0190 (8) | 0.0016 (7) | 0.0228 (7) |
| C19 | 0.0482 (10) | 0.0627 (11) | 0.0443 (9) | 0.0370 (9) | 0.0182 (8) | 0.0340 (8) |
| C20 | 0.0511 (10) | 0.0379 (8) | 0.0391 (8) | 0.0276 (7) | 0.0211 (7) | 0.0244 (7) |
| C21 | 0.0378 (8) | 0.0276 (7) | 0.0282 (7) | 0.0158 (6) | 0.0107 (6) | 0.0142 (5) |
| C22 | 0.0338 (8) | 0.0268 (7) | 0.0280 (7) | 0.0129 (6) | 0.0142 (6) | 0.0067 (5) |
| C23 | 0.0303 (7) | 0.0246 (6) | 0.0224 (7) | 0.0103 (5) | 0.0080 (5) | 0.0068 (5) |
| C24 | 0.0475 (9) | 0.0219 (6) | 0.0245 (7) | 0.0128 (6) | 0.0112 (6) | 0.0096 (5) |
| C25 | 0.0369 (9) | 0.0403 (8) | 0.0265 (7) | 0.0126 (7) | 0.0036 (6) | 0.0054 (6) |
| C26 | 0.0498 (10) | 0.0284 (7) | 0.0339 (8) | 0.0173 (7) | 0.0133 (7) | 0.0064 (6) |
| B1 | 0.0235 (7) | 0.0238 (7) | 0.0179 (7) | 0.0080 (5) | 0.0047 (5) | 0.0094 (5) |
| O1 | 0.0400 (6) | 0.0207 (4) | 0.0199 (5) | 0.0090 (4) | 0.0112 (4) | 0.0082 (4) |
| O2 | 0.0363 (6) | 0.0229 (4) | 0.0255 (5) | 0.0100 (4) | 0.0153 (4) | 0.0083 (4) |
Geometric parameters (Å, º)
| C1—C6 | 1.3902 (19) | C16—C17 | 1.531 (2) |
| C1—C2 | 1.3983 (18) | C16—C21 | 1.5316 (17) |
| C1—C7 | 1.4777 (17) | C16—H16 | 1.0000 |
| C2—C3 | 1.388 (2) | C17—C18 | 1.532 (2) |
| C2—H2 | 0.9500 | C17—H17A | 0.9900 |
| C3—C4 | 1.379 (2) | C17—H17B | 0.9900 |
| C3—H3 | 0.9500 | C18—C19 | 1.528 (2) |
| C4—C5 | 1.381 (2) | C18—H18A | 0.9900 |
| C4—H4 | 0.9500 | C18—H18B | 0.9900 |
| C5—C6 | 1.3879 (19) | C19—C20 | 1.524 (3) |
| C5—H5 | 0.9500 | C19—H19A | 0.9900 |
| C6—H6 | 0.9500 | C19—H19B | 0.9900 |
| C7—C8 | 1.3264 (18) | C20—C21 | 1.5338 (19) |
| C7—H7 | 0.9500 | C20—H20A | 0.9900 |
| C8—C9 | 1.5251 (16) | C20—H20B | 0.9900 |
| C8—H8 | 0.9500 | C21—H21A | 0.9900 |
| C9—C16 | 1.5672 (17) | C21—H21B | 0.9900 |
| C9—C10 | 1.5717 (18) | C22—O2 | 1.4424 (15) |
| C9—B1 | 1.6034 (18) | C22—C23 | 1.5238 (19) |
| C10—C11 | 1.5369 (18) | C22—H22A | 0.9900 |
| C10—C15 | 1.5386 (17) | C22—H22B | 0.9900 |
| C10—H10 | 1.0000 | C23—C24 | 1.5227 (18) |
| C11—C12 | 1.525 (2) | C23—C25 | 1.523 (2) |
| C11—H11A | 0.9900 | C23—C26 | 1.5285 (18) |
| C11—H11B | 0.9900 | C24—O1 | 1.4408 (15) |
| C12—C13 | 1.523 (2) | C24—H24A | 0.9900 |
| C12—H12A | 0.9900 | C24—H24B | 0.9900 |
| C12—H12B | 0.9900 | C25—H25A | 0.9800 |
| C13—C14 | 1.524 (2) | C25—H25B | 0.9800 |
| C13—H13A | 0.9900 | C25—H25C | 0.9800 |
| C13—H13B | 0.9900 | C26—H26A | 0.9800 |
| C14—C15 | 1.528 (2) | C26—H26B | 0.9800 |
| C14—H14A | 0.9900 | C26—H26C | 0.9800 |
| C14—H14B | 0.9900 | B1—O1 | 1.3565 (17) |
| C15—H15A | 0.9900 | B1—O2 | 1.3682 (16) |
| C15—H15B | 0.9900 | ||
| C6—C1—C2 | 117.89 (12) | C17—C16—H16 | 106.8 |
| C6—C1—C7 | 122.74 (12) | C21—C16—H16 | 106.8 |
| C2—C1—C7 | 119.37 (12) | C9—C16—H16 | 106.8 |
| C3—C2—C1 | 120.89 (14) | C18—C17—C16 | 111.14 (12) |
| C3—C2—H2 | 119.6 | C18—C17—H17A | 109.4 |
| C1—C2—H2 | 119.6 | C16—C17—H17A | 109.4 |
| C4—C3—C2 | 120.37 (14) | C18—C17—H17B | 109.4 |
| C4—C3—H3 | 119.8 | C16—C17—H17B | 109.4 |
| C2—C3—H3 | 119.8 | H17A—C17—H17B | 108.0 |
| C3—C4—C5 | 119.46 (13) | C19—C18—C17 | 111.29 (13) |
| C3—C4—H4 | 120.3 | C19—C18—H18A | 109.4 |
| C5—C4—H4 | 120.3 | C17—C18—H18A | 109.4 |
| C4—C5—C6 | 120.36 (14) | C19—C18—H18B | 109.4 |
| C4—C5—H5 | 119.8 | C17—C18—H18B | 109.4 |
| C6—C5—H5 | 119.8 | H18A—C18—H18B | 108.0 |
| C5—C6—C1 | 121.03 (13) | C20—C19—C18 | 111.51 (12) |
| C5—C6—H6 | 119.5 | C20—C19—H19A | 109.3 |
| C1—C6—H6 | 119.5 | C18—C19—H19A | 109.3 |
| C8—C7—C1 | 125.85 (12) | C20—C19—H19B | 109.3 |
| C8—C7—H7 | 117.1 | C18—C19—H19B | 109.3 |
| C1—C7—H7 | 117.1 | H19A—C19—H19B | 108.0 |
| C7—C8—C9 | 127.42 (12) | C19—C20—C21 | 111.18 (13) |
| C7—C8—H8 | 116.3 | C19—C20—H20A | 109.4 |
| C9—C8—H8 | 116.3 | C21—C20—H20A | 109.4 |
| C8—C9—C16 | 109.58 (10) | C19—C20—H20B | 109.4 |
| C8—C9—C10 | 110.43 (10) | C21—C20—H20B | 109.4 |
| C16—C9—C10 | 111.93 (10) | H20A—C20—H20B | 108.0 |
| C8—C9—B1 | 109.36 (10) | C16—C21—C20 | 110.77 (12) |
| C16—C9—B1 | 108.39 (10) | C16—C21—H21A | 109.5 |
| C10—C9—B1 | 107.06 (10) | C20—C21—H21A | 109.5 |
| C11—C10—C15 | 109.13 (11) | C16—C21—H21B | 109.5 |
| C11—C10—C9 | 110.54 (10) | C20—C21—H21B | 109.5 |
| C15—C10—C9 | 115.16 (11) | H21A—C21—H21B | 108.1 |
| C11—C10—H10 | 107.2 | O2—C22—C23 | 112.27 (10) |
| C15—C10—H10 | 107.2 | O2—C22—H22A | 109.2 |
| C9—C10—H10 | 107.2 | C23—C22—H22A | 109.2 |
| C12—C11—C10 | 112.66 (11) | O2—C22—H22B | 109.2 |
| C12—C11—H11A | 109.1 | C23—C22—H22B | 109.2 |
| C10—C11—H11A | 109.1 | H22A—C22—H22B | 107.9 |
| C12—C11—H11B | 109.1 | C24—C23—C25 | 111.00 (12) |
| C10—C11—H11B | 109.1 | C24—C23—C22 | 107.48 (11) |
| H11A—C11—H11B | 107.8 | C25—C23—C22 | 110.22 (12) |
| C13—C12—C11 | 111.29 (12) | C24—C23—C26 | 108.90 (12) |
| C13—C12—H12A | 109.4 | C25—C23—C26 | 110.06 (12) |
| C11—C12—H12A | 109.4 | C22—C23—C26 | 109.12 (11) |
| C13—C12—H12B | 109.4 | O1—C24—C23 | 112.89 (11) |
| C11—C12—H12B | 109.4 | O1—C24—H24A | 109.0 |
| H12A—C12—H12B | 108.0 | C23—C24—H24A | 109.0 |
| C12—C13—C14 | 110.08 (12) | O1—C24—H24B | 109.0 |
| C12—C13—H13A | 109.6 | C23—C24—H24B | 109.0 |
| C14—C13—H13A | 109.6 | H24A—C24—H24B | 107.8 |
| C12—C13—H13B | 109.6 | C23—C25—H25A | 109.5 |
| C14—C13—H13B | 109.6 | C23—C25—H25B | 109.5 |
| H13A—C13—H13B | 108.2 | H25A—C25—H25B | 109.5 |
| C13—C14—C15 | 111.34 (12) | C23—C25—H25C | 109.5 |
| C13—C14—H14A | 109.4 | H25A—C25—H25C | 109.5 |
| C15—C14—H14A | 109.4 | H25B—C25—H25C | 109.5 |
| C13—C14—H14B | 109.4 | C23—C26—H26A | 109.5 |
| C15—C14—H14B | 109.4 | C23—C26—H26B | 109.5 |
| H14A—C14—H14B | 108.0 | H26A—C26—H26B | 109.5 |
| C14—C15—C10 | 111.18 (12) | C23—C26—H26C | 109.5 |
| C14—C15—H15A | 109.4 | H26A—C26—H26C | 109.5 |
| C10—C15—H15A | 109.4 | H26B—C26—H26C | 109.5 |
| C14—C15—H15B | 109.4 | O1—B1—O2 | 122.35 (11) |
| C10—C15—H15B | 109.4 | O1—B1—C9 | 119.72 (11) |
| H15A—C15—H15B | 108.0 | O2—B1—C9 | 117.93 (11) |
| C17—C16—C21 | 108.63 (11) | B1—O1—C24 | 120.49 (10) |
| C17—C16—C9 | 112.83 (11) | B1—O2—C22 | 119.36 (10) |
| C21—C16—C9 | 114.57 (11) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C22—H22B···O2i | 0.99 | 2.69 | 3.5773 (18) | 150 |
Symmetry code: (i) −x+1, −y, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: GO2094).
References
- Althaus, M., Mahmood, A., Suárez, J. R., Thomas, S. P. & Aggarwal, V. K. (2010). J. Am. Chem. Soc. 132, 4025–4028. [DOI] [PubMed]
- Altomare, A., Cascarano, G., Giacovazzo, C., Guagliardi, A., Burla, M. C., Polidori, G. & Camalli, M. (1994). J. Appl. Cryst. 27, 435.
- Cambridge Soft (2001). CHEMDRAW Ultra Cambridge Soft Corporation, Cambridge, Massachusetts, USA.
- Carosi, L. & Hall, D. G. (2007). Angew. Chem. Int. Ed. 46, 5913–5915. [DOI] [PubMed]
- Clary, J. W., Rettenmaier, T. J., Snelling, R., Bryks, W., Banwell, J., Wipke, W. T. & Singaram, B. (2011). J. Org. Chem. 76, 9602–9610. [DOI] [PubMed]
- Fandrick, K. R., Fandrick, D. R., Gao, J. J., Reeves, J. T., Tan, Z., Li, W., Song, J. J., Lu, B., Yee, N. K. & Senanayake, C. H. (2010). Org. Lett. 12, 3748–3751. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Hesse, M. J., Butts, C. P., Willis, C. L. & Aggarwal, V. K. (2012). Angew. Chem. Int. Ed. 51, 12444–12448. [DOI] [PubMed]
- Incerti-Pradillos, C. A., Kabeshov, M. A. & Malkov, A. V. (2013). Angew. Chem. Int. Ed. 52, 5338–5341. [DOI] [PubMed]
- Lombardo, M., Morganti, S., Tozzi, M. & Trombini, C. (2002). Eur. J. Org. Chem. pp. 2823–2830.
- Nonius (2000). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sopková-de Oliveira Santos, J., Lancelot, J.-C., Bouillon, A. & Rault, S. (2003). Acta Cryst. C59, o111–o113. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S1600536813021739/go2094sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813021739/go2094Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813021739/go2094Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





