Abstract
The asymmetric unit of the title co-crystal, 2C14H13N2 +·C10H4O8 2−·2C14H12N2·C10H6O8, comprises a 2,9-dimethyl-1,10-phenanthrolin-1-ium cation (Me2PhenH+) and a 2,9-dimethyl-1,10-phenanthroline molecule (Me2Phen), each in a general position, and half each of a 2,5-dicarboxybenzene-1,4-dicarboxylate dianion (LH2 2−) and a benzene-1,2,4,5-tetracarboxylic acid molecule (LH4), each being disposed about a centre of inversion. Small twists are evident in the dianion [the C—C—C—O torsion angles are 168.41 (18) and 16.2 (3)°], whereas a major twist is found for one carboxylic acid group in the neutral molecule [C—C—C—O = 66.3 (2) and 18.2 (3)°]. The most prominent feature of the crystal packing is the formation of linear supramolecular chains along [001] mediated by charge-assisted O—H⋯O− hydrogen bonding between alternating LH4 and LH2 2−. These are connected to the Me2PhenH+ and Me2Phen species by N—H⋯O and O—H⋯N hydrogen bonds, respectively. A three-dimensional architecture is formed by C—H⋯O and π–π interactions [inter-centroid distance = 3.5337 (17) Å].
Related literature
For salt formation with benzene-1,2,4,5-tetracarboxylic acid, see: Arman & Tiekink (2013 ▶). For a co-crystal involving 2,9-dimethyl-1,10-phenanthroline, see: Arman et al. (2010 ▶). For the structure of a 2,9-dimethyl-1,10-phenanthrolin-1-ium carboxylate salt, see: Derikvand & Olmstead (2011 ▶).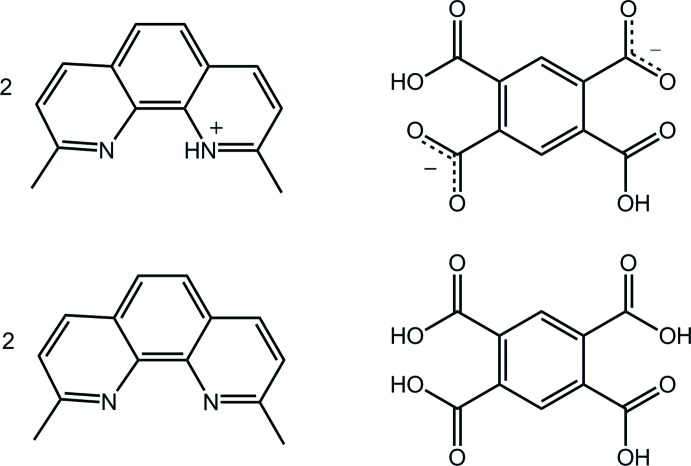
Experimental
Crystal data
2C14H13N2 +·C10H4O8 2−·2C14H12N2·C10H6O8
M r = 1341.32
Monoclinic,

a = 11.798 (4) Å
b = 13.893 (4) Å
c = 19.163 (6) Å
β = 92.216 (5)°
V = 3138.8 (16) Å3
Z = 2
Mo Kα radiation
μ = 0.10 mm−1
T = 98 K
0.48 × 0.37 × 0.09 mm
Data collection
Rigaku AFC12/SATURN724 diffractometer
Absorption correction: multi-scan (ABSCOR; Higashi, 1995 ▶) T min = 0.723, T max = 1.000
22006 measured reflections
7181 independent reflections
6007 reflections with I > 2σ(I)
R int = 0.056
Refinement
R[F 2 > 2σ(F 2)] = 0.062
wR(F 2) = 0.148
S = 1.11
7181 reflections
467 parameters
4 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.29 e Å−3
Δρmin = −0.25 e Å−3
Data collection: CrystalClear (Molecular Structure Corporation & Rigaku, 2005 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEPII (Johnson, 1976 ▶) and DIAMOND (Brandenburg, 2006 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813022691/xu5731sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813022691/xu5731Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813022691/xu5731Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O4—H1o⋯O1 | 0.85 (2) | 1.55 (2) | 2.403 (2) | 176 (3) |
| O6—H2o⋯O2i | 0.85 (1) | 1.74 (1) | 2.577 (2) | 168 (2) |
| O8—H3o⋯N4ii | 0.85 (2) | 1.79 (2) | 2.636 (2) | 173 (2) |
| N1—H1n⋯O3iii | 0.89 (2) | 2.41 (2) | 3.257 (2) | 161 (2) |
| N1—H1n⋯O4iii | 0.89 (2) | 2.35 (2) | 2.957 (2) | 126 (2) |
| C13—H13⋯O7iv | 0.95 | 2.28 | 3.225 (3) | 171 |
| C28—H28⋯O5v | 0.95 | 2.40 | 3.320 (3) | 162 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Acknowledgments
We gratefully thank the Ministry of Higher Education (Malaysia) for funding structural studies through the High-Impact Research scheme (UM.C/HIR-MOHE/SC/03).
supplementary crystallographic information
1. Comment
In continuation of on-going structural studies of salts/co-crystals formed between carboxylic acids and various pyridyl derivatives (Arman et al., 2010; Arman & Tiekink, 2013), the title salt co-crystal, (I), was isolated from the 2:3 co-crystallization of benzene-1,2,4,5-tetracarboxylic acid (LH4) and 2,9-dimethyl-1,10-phenanthroline (Me2Phen).
The asymmetric unit of (I) comprises a centrosymmetric, doubly deprotonated LH22- dianion, a centrosymmetric neutral LH4 molecule, a protonated Me2PhenH+ cation and a neutral Me2Phen molecule, Fig. 1, and is formulated as a combination of a 2:1 Me2Phen+:LH22- salt combined with a 2:1 Me2Phen:HL4 co-crystal. A salt formed between Me2PhenH+ and a hydrogen(S,S)-tartrate has been reported (Derikvand & Olmstead, 2011).
Small twists are evident in the LH22- dianion as seen in the C2—C1—C4—O2 and C1—C2—C5—O4 torsion angles of 168.41 (18) and 16.2 (3)°, respectively. This arrangement is stabilized by intramolecular O—H···O hydrogen bonds, Table 1. By contrast, a considerable twist is evident in LH4 with the C7—C6—C9—O6 and C6—C7—C10—O7 torsion angles being 66.3 (2) and 18.2 (3)°, respectively. Such variations in conformation have been discussed in some detail (Arman & Tiekink, 2013). The Me2Phen molecule and Me2PhenH+ cation are each planar with the r.m.s. deviation for the 16 non-hydrogen atoms being 0.037 and 0.036 Å, respectively.
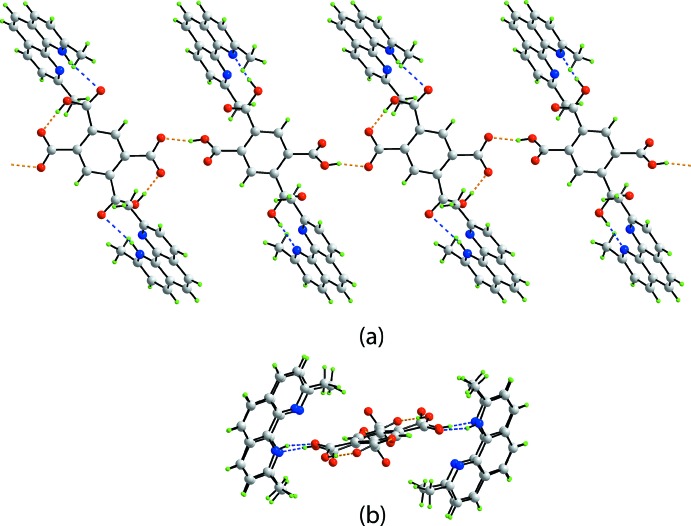
The prominent feature of the crystal packing is the formation of linear supramolecular chains along [0 0 1] comprising alternating LH4 and LH22- species connected via charge-assisted O6—H···O2 hydrogen bonding, Table 1. The hydroxyl-O8 forms an O—H···N4 hydrogen bond with the neutral Me2Phen molecules, one to either side of the carboxylic acid/carboxylate chain. The O3,O4 carboxylic acid residue accepts hydrogen bonds from the N1—H1n atom of the Me2PhenH+ cation, again, from symmetry, one to either side, leading to the supramolecular chain shown in Fig. 2a; an end-on view is shown in Fig. 2b. The Me2Phen and Me2PhenH+ cations inter-digitate along the c axis and are connected by π—π [Cg(C15–C20)···Cg(C29—C34)i = 3.5337 (17) Å for i: x, 1 + y, z] interactions between Me2PhenH+ and Me2Phen. Additional contacts are of the type C—H···O, Table 1, as illustrated in the crystal packing diagram, Fig. 3.
2. Experimental
Crystals of (I) were obtained by the co-crystallization of benzene-1,2,4,5-tetracarboxylic acid (Sigma-Aldrich), 0.06 mmol) and 2,9-dimethylphenanthroline (ACROS, 0.09 mmol) in ethanol solution. Crystals were obtained by slow evaporation.
3. Refinement
C-bound H-atoms were placed in calculated positions (C—H = 0.95–0.98 Å) and were included in the refinement in the riding model approximation with Uiso(H) set to 1.2–1.5Ueq(C). The O-and N-bound H-atoms were located in a difference Fourier map and were refined with a distance restraints of O—H = 0.84±0.01 Å and N—H = 0.88±0.01 Å, and with Uiso(H) = 1.2Ueq(N) and 1.5Ueq(O). Owing to being affected by the beam-stop, three reflections, i.e. (0 0 1), (1 0 1) and (-6 0 2), were omitted from the final cycles of refinement.
Figures
Fig. 1.
Molecular structures of the components of (I), showing atom-labelling scheme and displacement ellipsoids at the 50% probability level: (a) LH22-, (b) LH4, (c) Me2PhenH+ and (d) Me2Phen.
Fig. 2.
Views (a) side-on and (b) end-on of the supramolecular chain in (I). The O—H···O (orange), O—H···N (blue) and N—H···O (blue) hydrogen bonds are shown as dashed lines.
Fig. 3.
Unit-cell contents in (I) viewed in projection down the c axis. The C—H···O interactions are shown green dashed lines.
Crystal data
| 2C14H13N2+·C10H4O82−·2C14H12N2·C10H6O8 | F(000) = 1400 |
| Mr = 1341.32 | Dx = 1.419 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71069 Å |
| Hall symbol: -P 2yn | Cell parameters from 12762 reflections |
| a = 11.798 (4) Å | θ = 2.0–40.7° |
| b = 13.893 (4) Å | µ = 0.10 mm−1 |
| c = 19.163 (6) Å | T = 98 K |
| β = 92.216 (5)° | Prism, colourless |
| V = 3138.8 (16) Å3 | 0.48 × 0.37 × 0.09 mm |
| Z = 2 |
Data collection
| Rigaku AFC12K/SATURN724 diffractometer | 7181 independent reflections |
| Radiation source: fine-focus sealed tube | 6007 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.056 |
| ω scans | θmax = 27.5°, θmin = 2.1° |
| Absorption correction: multi-scan (ABSCOR; Higashi, 1995) | h = −15→15 |
| Tmin = 0.723, Tmax = 1.000 | k = −18→18 |
| 22006 measured reflections | l = −18→24 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.062 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.148 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.11 | w = 1/[σ2(Fo2) + (0.0555P)2 + 1.3348P] where P = (Fo2 + 2Fc2)/3 |
| 7181 reflections | (Δ/σ)max < 0.001 |
| 467 parameters | Δρmax = 0.29 e Å−3 |
| 4 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.60101 (12) | 0.56642 (11) | 0.32029 (7) | 0.0298 (3) | |
| O2 | 0.42698 (12) | 0.50799 (11) | 0.31498 (7) | 0.0271 (3) | |
| O3 | 0.80815 (11) | 0.52832 (11) | 0.50668 (7) | 0.0276 (3) | |
| O4 | 0.75764 (12) | 0.58534 (11) | 0.40310 (7) | 0.0275 (3) | |
| H1O | 0.7039 (17) | 0.5802 (19) | 0.3723 (11) | 0.041* | |
| O5 | 1.04388 (12) | 0.10773 (11) | 0.67551 (7) | 0.0279 (3) | |
| O6 | 0.93416 (12) | −0.02423 (10) | 0.68129 (6) | 0.0225 (3) | |
| H2O | 0.941 (2) | −0.0150 (17) | 0.7251 (5) | 0.034* | |
| O7 | 0.78053 (11) | 0.10681 (9) | 0.60744 (6) | 0.0217 (3) | |
| O8 | 0.69585 (11) | 0.03734 (10) | 0.51309 (7) | 0.0225 (3) | |
| H3O | 0.6399 (14) | 0.0441 (18) | 0.5394 (10) | 0.034* | |
| N1 | 0.49493 (13) | 1.05719 (11) | 0.10595 (8) | 0.0216 (3) | |
| H1N | 0.5525 (13) | 1.0368 (16) | 0.0816 (10) | 0.026* | |
| N2 | 0.59146 (14) | 0.87982 (12) | 0.11607 (8) | 0.0242 (4) | |
| N3 | 0.38546 (15) | 0.11239 (12) | 0.39446 (8) | 0.0251 (4) | |
| N4 | 0.48691 (13) | −0.06451 (12) | 0.41450 (8) | 0.0212 (3) | |
| C1 | 0.51592 (16) | 0.51633 (13) | 0.42769 (9) | 0.0183 (4) | |
| C2 | 0.61045 (15) | 0.52086 (13) | 0.47597 (9) | 0.0174 (3) | |
| C3 | 0.59041 (15) | 0.50468 (13) | 0.54626 (9) | 0.0185 (4) | |
| H3 | 0.6533 | 0.5082 | 0.5787 | 0.022* | |
| C4 | 0.51495 (16) | 0.53048 (14) | 0.34896 (9) | 0.0206 (4) | |
| C5 | 0.73412 (16) | 0.54460 (14) | 0.46160 (10) | 0.0215 (4) | |
| C6 | 0.99235 (15) | 0.02306 (12) | 0.57069 (9) | 0.0172 (3) | |
| C7 | 0.89506 (15) | 0.02660 (12) | 0.52636 (9) | 0.0170 (3) | |
| C8 | 0.90372 (15) | 0.00341 (13) | 0.45593 (9) | 0.0177 (4) | |
| H8 | 0.8380 | 0.0057 | 0.4257 | 0.021* | |
| C9 | 0.99120 (15) | 0.04220 (13) | 0.64807 (9) | 0.0191 (4) | |
| C10 | 0.78392 (15) | 0.06034 (13) | 0.55334 (9) | 0.0182 (4) | |
| C11 | 0.51318 (18) | 1.21161 (15) | 0.04831 (11) | 0.0287 (4) | |
| H11A | 0.5737 | 1.2474 | 0.0733 | 0.043* | |
| H11B | 0.4577 | 1.2569 | 0.0277 | 0.043* | |
| H11C | 0.5457 | 1.1731 | 0.0112 | 0.043* | |
| C12 | 0.45601 (16) | 1.14676 (14) | 0.09806 (10) | 0.0234 (4) | |
| C13 | 0.36476 (17) | 1.17594 (14) | 0.13837 (10) | 0.0261 (4) | |
| H13 | 0.3340 | 1.2388 | 0.1328 | 0.031* | |
| C14 | 0.32018 (17) | 1.11385 (15) | 0.18567 (10) | 0.0258 (4) | |
| H14 | 0.2590 | 1.1343 | 0.2128 | 0.031* | |
| C15 | 0.36416 (16) | 1.01985 (14) | 0.19451 (10) | 0.0220 (4) | |
| C16 | 0.45351 (16) | 0.99289 (13) | 0.15246 (10) | 0.0206 (4) | |
| C17 | 0.32532 (16) | 0.95298 (15) | 0.24508 (10) | 0.0245 (4) | |
| H17 | 0.2658 | 0.9708 | 0.2744 | 0.029* | |
| C18 | 0.37276 (17) | 0.86434 (15) | 0.25149 (10) | 0.0253 (4) | |
| H18 | 0.3459 | 0.8209 | 0.2854 | 0.030* | |
| C19 | 0.46257 (16) | 0.83521 (13) | 0.20811 (10) | 0.0225 (4) | |
| C20 | 0.50465 (16) | 0.89957 (13) | 0.15806 (10) | 0.0214 (4) | |
| C21 | 0.51552 (18) | 0.74414 (15) | 0.21224 (11) | 0.0284 (4) | |
| H21 | 0.4910 | 0.6975 | 0.2446 | 0.034* | |
| C22 | 0.60207 (18) | 0.72363 (15) | 0.16957 (11) | 0.0301 (5) | |
| H22 | 0.6371 | 0.6621 | 0.1717 | 0.036* | |
| C23 | 0.64006 (17) | 0.79384 (15) | 0.12197 (11) | 0.0271 (4) | |
| C24 | 0.73845 (19) | 0.77429 (16) | 0.07645 (12) | 0.0353 (5) | |
| H24A | 0.7311 | 0.8138 | 0.0342 | 0.053* | |
| H24B | 0.7389 | 0.7061 | 0.0635 | 0.053* | |
| H24C | 0.8095 | 0.7903 | 0.1020 | 0.053* | |
| C25 | 0.2311 (2) | 0.21602 (17) | 0.42569 (13) | 0.0394 (6) | |
| H25A | 0.1657 | 0.1888 | 0.3995 | 0.059* | |
| H25B | 0.2202 | 0.2855 | 0.4314 | 0.059* | |
| H25C | 0.2385 | 0.1854 | 0.4717 | 0.059* | |
| C26 | 0.3371 (2) | 0.19826 (15) | 0.38637 (11) | 0.0310 (5) | |
| C27 | 0.3831 (2) | 0.27008 (16) | 0.34338 (12) | 0.0391 (6) | |
| H27 | 0.3487 | 0.3318 | 0.3400 | 0.047* | |
| C28 | 0.4769 (2) | 0.24996 (18) | 0.30682 (12) | 0.0421 (6) | |
| H28 | 0.5070 | 0.2973 | 0.2769 | 0.051* | |
| C29 | 0.5297 (2) | 0.15893 (17) | 0.31328 (11) | 0.0347 (5) | |
| C30 | 0.48077 (17) | 0.09257 (15) | 0.35951 (9) | 0.0253 (4) | |
| C31 | 0.6265 (2) | 0.1311 (2) | 0.27508 (11) | 0.0434 (7) | |
| H31 | 0.6579 | 0.1756 | 0.2435 | 0.052* | |
| C32 | 0.6740 (2) | 0.0432 (2) | 0.28292 (11) | 0.0418 (6) | |
| H32 | 0.7377 | 0.0265 | 0.2566 | 0.050* | |
| C33 | 0.62883 (17) | −0.02531 (18) | 0.33100 (11) | 0.0322 (5) | |
| C34 | 0.53289 (16) | −0.00081 (15) | 0.36943 (10) | 0.0239 (4) | |
| C35 | 0.67628 (18) | −0.11709 (18) | 0.34117 (12) | 0.0379 (6) | |
| H35 | 0.7416 | −0.1353 | 0.3169 | 0.045* | |
| C36 | 0.62834 (18) | −0.18028 (17) | 0.38602 (12) | 0.0342 (5) | |
| H36 | 0.6598 | −0.2427 | 0.3929 | 0.041* | |
| C37 | 0.53169 (17) | −0.15191 (14) | 0.42193 (11) | 0.0256 (4) | |
| C38 | 0.47268 (19) | −0.22145 (15) | 0.46823 (12) | 0.0326 (5) | |
| H38A | 0.4480 | −0.1877 | 0.5099 | 0.049* | |
| H38B | 0.5250 | −0.2734 | 0.4822 | 0.049* | |
| H38C | 0.4065 | −0.2487 | 0.4428 | 0.049* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0286 (8) | 0.0431 (9) | 0.0180 (7) | −0.0085 (6) | 0.0034 (6) | 0.0057 (6) |
| O2 | 0.0271 (8) | 0.0398 (8) | 0.0145 (6) | −0.0042 (6) | 0.0010 (5) | −0.0001 (6) |
| O3 | 0.0188 (7) | 0.0367 (8) | 0.0272 (7) | 0.0001 (6) | −0.0012 (6) | 0.0025 (6) |
| O4 | 0.0218 (7) | 0.0401 (8) | 0.0208 (7) | −0.0060 (6) | 0.0042 (5) | 0.0038 (6) |
| O5 | 0.0260 (7) | 0.0373 (8) | 0.0204 (7) | −0.0079 (6) | 0.0032 (5) | −0.0083 (6) |
| O6 | 0.0290 (7) | 0.0263 (7) | 0.0122 (6) | −0.0009 (5) | 0.0024 (5) | 0.0003 (5) |
| O7 | 0.0234 (7) | 0.0229 (7) | 0.0191 (6) | −0.0006 (5) | 0.0035 (5) | −0.0034 (5) |
| O8 | 0.0170 (6) | 0.0313 (7) | 0.0193 (7) | −0.0010 (5) | 0.0029 (5) | −0.0039 (5) |
| N1 | 0.0181 (8) | 0.0210 (8) | 0.0256 (8) | 0.0010 (6) | 0.0007 (6) | −0.0010 (6) |
| N2 | 0.0229 (8) | 0.0232 (8) | 0.0264 (8) | 0.0045 (6) | −0.0002 (6) | −0.0027 (6) |
| N3 | 0.0280 (9) | 0.0243 (9) | 0.0229 (8) | 0.0006 (7) | −0.0024 (7) | 0.0001 (6) |
| N4 | 0.0174 (7) | 0.0246 (8) | 0.0215 (8) | −0.0004 (6) | −0.0010 (6) | −0.0052 (6) |
| C1 | 0.0231 (9) | 0.0168 (8) | 0.0152 (8) | 0.0017 (7) | 0.0010 (7) | 0.0004 (6) |
| C2 | 0.0190 (9) | 0.0181 (8) | 0.0152 (8) | 0.0010 (6) | 0.0022 (6) | −0.0010 (6) |
| C3 | 0.0190 (9) | 0.0212 (9) | 0.0152 (8) | 0.0024 (7) | 0.0002 (6) | 0.0006 (7) |
| C4 | 0.0229 (9) | 0.0235 (9) | 0.0155 (8) | 0.0028 (7) | 0.0027 (7) | 0.0003 (7) |
| C5 | 0.0211 (9) | 0.0223 (9) | 0.0212 (9) | −0.0010 (7) | 0.0027 (7) | −0.0035 (7) |
| C6 | 0.0208 (9) | 0.0177 (8) | 0.0133 (8) | −0.0016 (7) | 0.0036 (6) | −0.0010 (6) |
| C7 | 0.0188 (8) | 0.0166 (8) | 0.0156 (8) | −0.0012 (6) | 0.0026 (6) | 0.0005 (6) |
| C8 | 0.0164 (8) | 0.0204 (9) | 0.0160 (8) | −0.0008 (6) | −0.0018 (6) | −0.0004 (6) |
| C9 | 0.0170 (8) | 0.0252 (9) | 0.0153 (8) | 0.0024 (7) | 0.0013 (6) | −0.0013 (7) |
| C10 | 0.0205 (9) | 0.0172 (8) | 0.0172 (8) | 0.0000 (7) | 0.0020 (7) | 0.0016 (6) |
| C11 | 0.0310 (11) | 0.0250 (10) | 0.0300 (11) | 0.0021 (8) | 0.0003 (8) | 0.0029 (8) |
| C12 | 0.0221 (9) | 0.0220 (9) | 0.0257 (10) | 0.0010 (7) | −0.0033 (7) | −0.0006 (7) |
| C13 | 0.0245 (10) | 0.0218 (10) | 0.0318 (11) | 0.0062 (7) | −0.0020 (8) | −0.0048 (8) |
| C14 | 0.0216 (9) | 0.0276 (10) | 0.0283 (10) | 0.0037 (7) | 0.0022 (8) | −0.0062 (8) |
| C15 | 0.0187 (9) | 0.0249 (9) | 0.0221 (9) | 0.0004 (7) | −0.0027 (7) | −0.0041 (7) |
| C16 | 0.0173 (9) | 0.0228 (9) | 0.0213 (9) | −0.0015 (7) | −0.0029 (7) | −0.0016 (7) |
| C17 | 0.0205 (9) | 0.0316 (11) | 0.0214 (9) | −0.0034 (8) | −0.0002 (7) | −0.0025 (8) |
| C18 | 0.0268 (10) | 0.0267 (10) | 0.0222 (9) | −0.0051 (8) | −0.0023 (8) | 0.0012 (7) |
| C19 | 0.0241 (9) | 0.0206 (9) | 0.0223 (9) | −0.0021 (7) | −0.0049 (7) | −0.0026 (7) |
| C20 | 0.0212 (9) | 0.0207 (9) | 0.0222 (9) | 0.0010 (7) | −0.0036 (7) | −0.0032 (7) |
| C21 | 0.0315 (11) | 0.0226 (10) | 0.0303 (11) | −0.0020 (8) | −0.0085 (8) | 0.0010 (8) |
| C22 | 0.0328 (11) | 0.0211 (10) | 0.0357 (11) | 0.0050 (8) | −0.0072 (9) | −0.0024 (8) |
| C23 | 0.0249 (10) | 0.0258 (10) | 0.0302 (10) | 0.0035 (8) | −0.0038 (8) | −0.0056 (8) |
| C24 | 0.0323 (12) | 0.0307 (11) | 0.0432 (13) | 0.0112 (9) | 0.0034 (10) | −0.0051 (9) |
| C25 | 0.0454 (14) | 0.0277 (11) | 0.0444 (13) | 0.0117 (10) | −0.0056 (11) | −0.0029 (10) |
| C26 | 0.0396 (12) | 0.0256 (10) | 0.0268 (10) | −0.0002 (9) | −0.0113 (9) | −0.0008 (8) |
| C27 | 0.0559 (16) | 0.0267 (11) | 0.0334 (12) | −0.0054 (10) | −0.0178 (11) | 0.0059 (9) |
| C28 | 0.0616 (17) | 0.0373 (13) | 0.0263 (11) | −0.0224 (12) | −0.0150 (11) | 0.0104 (9) |
| C29 | 0.0425 (13) | 0.0411 (13) | 0.0202 (10) | −0.0199 (10) | −0.0035 (9) | 0.0016 (9) |
| C30 | 0.0283 (10) | 0.0304 (10) | 0.0168 (9) | −0.0091 (8) | −0.0024 (7) | −0.0017 (7) |
| C31 | 0.0448 (14) | 0.0651 (18) | 0.0203 (10) | −0.0326 (13) | 0.0019 (9) | −0.0003 (10) |
| C32 | 0.0334 (12) | 0.0664 (18) | 0.0265 (11) | −0.0236 (12) | 0.0105 (9) | −0.0155 (11) |
| C33 | 0.0211 (10) | 0.0505 (14) | 0.0252 (10) | −0.0123 (9) | 0.0036 (8) | −0.0151 (9) |
| C34 | 0.0183 (9) | 0.0321 (11) | 0.0215 (9) | −0.0074 (8) | 0.0015 (7) | −0.0055 (8) |
| C35 | 0.0176 (10) | 0.0558 (15) | 0.0404 (13) | −0.0023 (9) | 0.0035 (9) | −0.0252 (11) |
| C36 | 0.0228 (10) | 0.0381 (12) | 0.0412 (12) | 0.0078 (9) | −0.0039 (9) | −0.0193 (10) |
| C37 | 0.0200 (9) | 0.0268 (10) | 0.0297 (10) | 0.0030 (7) | −0.0044 (8) | −0.0090 (8) |
| C38 | 0.0321 (11) | 0.0244 (10) | 0.0408 (12) | 0.0035 (8) | −0.0062 (9) | −0.0006 (9) |
Geometric parameters (Å, º)
| O1—C4 | 1.275 (2) | C15—C16 | 1.402 (3) |
| O2—C4 | 1.244 (2) | C15—C17 | 1.431 (3) |
| O3—C5 | 1.226 (2) | C16—C20 | 1.432 (3) |
| O4—C5 | 1.295 (2) | C17—C18 | 1.356 (3) |
| O4—H1O | 0.853 (10) | C17—H17 | 0.9500 |
| O5—C9 | 1.211 (2) | C18—C19 | 1.430 (3) |
| O6—C9 | 1.320 (2) | C18—H18 | 0.9500 |
| O6—H2O | 0.850 (10) | C19—C21 | 1.412 (3) |
| O7—C10 | 1.223 (2) | C19—C20 | 1.415 (3) |
| O8—C10 | 1.309 (2) | C21—C22 | 1.363 (3) |
| O8—H3O | 0.851 (10) | C21—H21 | 0.9500 |
| N1—C12 | 1.333 (2) | C22—C23 | 1.420 (3) |
| N1—C16 | 1.366 (2) | C22—H22 | 0.9500 |
| N1—H1N | 0.886 (10) | C23—C24 | 1.503 (3) |
| N2—C23 | 1.328 (3) | C24—H24A | 0.9800 |
| N2—C20 | 1.355 (3) | C24—H24B | 0.9800 |
| N3—C26 | 1.329 (3) | C24—H24C | 0.9800 |
| N3—C30 | 1.359 (3) | C25—C26 | 1.505 (3) |
| N4—C37 | 1.330 (3) | C25—H25A | 0.9800 |
| N4—C34 | 1.364 (3) | C25—H25B | 0.9800 |
| C1—C3i | 1.399 (3) | C25—H25C | 0.9800 |
| C1—C2 | 1.423 (2) | C26—C27 | 1.415 (3) |
| C1—C4 | 1.521 (2) | C27—C28 | 1.362 (4) |
| C2—C3 | 1.395 (2) | C27—H27 | 0.9500 |
| C2—C5 | 1.531 (3) | C28—C29 | 1.413 (4) |
| C3—C1i | 1.399 (3) | C28—H28 | 0.9500 |
| C3—H3 | 0.9500 | C29—C30 | 1.417 (3) |
| C6—C8ii | 1.396 (3) | C29—C31 | 1.433 (4) |
| C6—C7 | 1.402 (2) | C30—C34 | 1.445 (3) |
| C6—C9 | 1.507 (2) | C31—C32 | 1.349 (4) |
| C7—C8 | 1.395 (2) | C31—H31 | 0.9500 |
| C7—C10 | 1.503 (3) | C32—C33 | 1.442 (3) |
| C8—C6ii | 1.396 (3) | C32—H32 | 0.9500 |
| C8—H8 | 0.9500 | C33—C35 | 1.403 (3) |
| C11—C12 | 1.492 (3) | C33—C34 | 1.415 (3) |
| C11—H11A | 0.9800 | C35—C36 | 1.366 (3) |
| C11—H11B | 0.9800 | C35—H35 | 0.9500 |
| C11—H11C | 0.9800 | C36—C37 | 1.411 (3) |
| C12—C13 | 1.409 (3) | C36—H36 | 0.9500 |
| C13—C14 | 1.371 (3) | C37—C38 | 1.501 (3) |
| C13—H13 | 0.9500 | C38—H38A | 0.9800 |
| C14—C15 | 1.413 (3) | C38—H38B | 0.9800 |
| C14—H14 | 0.9500 | C38—H38C | 0.9800 |
| C5—O4—H1O | 112.6 (18) | C20—C19—C18 | 120.15 (17) |
| C9—O6—H2O | 109.7 (17) | N2—C20—C19 | 124.58 (18) |
| C10—O8—H3O | 103.9 (16) | N2—C20—C16 | 117.70 (18) |
| C12—N1—C16 | 123.66 (17) | C19—C20—C16 | 117.71 (18) |
| C12—N1—H1N | 120.4 (15) | C22—C21—C19 | 119.6 (2) |
| C16—N1—H1N | 115.9 (15) | C22—C21—H21 | 120.2 |
| C23—N2—C20 | 117.74 (18) | C19—C21—H21 | 120.2 |
| C26—N3—C30 | 118.95 (19) | C21—C22—C23 | 120.28 (19) |
| C37—N4—C34 | 119.62 (18) | C21—C22—H22 | 119.9 |
| C3i—C1—C2 | 117.94 (16) | C23—C22—H22 | 119.9 |
| C3i—C1—C4 | 114.11 (15) | N2—C23—C22 | 121.8 (2) |
| C2—C1—C4 | 127.95 (17) | N2—C23—C24 | 116.98 (19) |
| C3—C2—C1 | 117.58 (17) | C22—C23—C24 | 121.24 (19) |
| C3—C2—C5 | 114.00 (15) | C23—C24—H24A | 109.5 |
| C1—C2—C5 | 128.39 (16) | C23—C24—H24B | 109.5 |
| C2—C3—C1i | 124.48 (16) | H24A—C24—H24B | 109.5 |
| C2—C3—H3 | 117.8 | C23—C24—H24C | 109.5 |
| C1i—C3—H3 | 117.8 | H24A—C24—H24C | 109.5 |
| O2—C4—O1 | 122.35 (17) | H24B—C24—H24C | 109.5 |
| O2—C4—C1 | 117.46 (17) | C26—C25—H25A | 109.5 |
| O1—C4—C1 | 120.17 (16) | C26—C25—H25B | 109.5 |
| O3—C5—O4 | 121.32 (18) | H25A—C25—H25B | 109.5 |
| O3—C5—C2 | 119.44 (17) | C26—C25—H25C | 109.5 |
| O4—C5—C2 | 119.17 (16) | H25A—C25—H25C | 109.5 |
| C8ii—C6—C7 | 119.87 (16) | H25B—C25—H25C | 109.5 |
| C8ii—C6—C9 | 116.60 (15) | N3—C26—C27 | 121.9 (2) |
| C7—C6—C9 | 123.48 (16) | N3—C26—C25 | 116.8 (2) |
| C8—C7—C6 | 119.28 (17) | C27—C26—C25 | 121.4 (2) |
| C8—C7—C10 | 120.16 (15) | C28—C27—C26 | 119.5 (2) |
| C6—C7—C10 | 120.47 (16) | C28—C27—H27 | 120.3 |
| C7—C8—C6ii | 120.85 (16) | C26—C27—H27 | 120.3 |
| C7—C8—H8 | 119.6 | C27—C28—C29 | 120.2 (2) |
| C6ii—C8—H8 | 119.6 | C27—C28—H28 | 119.9 |
| O5—C9—O6 | 125.37 (17) | C29—C28—H28 | 119.9 |
| O5—C9—C6 | 122.40 (17) | C28—C29—C30 | 116.7 (2) |
| O6—C9—C6 | 112.04 (15) | C28—C29—C31 | 123.6 (2) |
| O7—C10—O8 | 125.23 (17) | C30—C29—C31 | 119.7 (2) |
| O7—C10—C7 | 120.91 (16) | N3—C30—C29 | 122.7 (2) |
| O8—C10—C7 | 113.84 (15) | N3—C30—C34 | 118.23 (18) |
| C12—C11—H11A | 109.5 | C29—C30—C34 | 119.0 (2) |
| C12—C11—H11B | 109.5 | C32—C31—C29 | 121.5 (2) |
| H11A—C11—H11B | 109.5 | C32—C31—H31 | 119.2 |
| C12—C11—H11C | 109.5 | C29—C31—H31 | 119.2 |
| H11A—C11—H11C | 109.5 | C31—C32—C33 | 120.4 (2) |
| H11B—C11—H11C | 109.5 | C31—C32—H32 | 119.8 |
| N1—C12—C13 | 118.27 (18) | C33—C32—H32 | 119.8 |
| N1—C12—C11 | 118.29 (18) | C35—C33—C34 | 118.0 (2) |
| C13—C12—C11 | 123.41 (18) | C35—C33—C32 | 122.3 (2) |
| C14—C13—C12 | 120.16 (18) | C34—C33—C32 | 119.8 (2) |
| C14—C13—H13 | 119.9 | N4—C34—C33 | 121.3 (2) |
| C12—C13—H13 | 119.9 | N4—C34—C30 | 119.23 (18) |
| C13—C14—C15 | 120.83 (18) | C33—C34—C30 | 119.49 (19) |
| C13—C14—H14 | 119.6 | C36—C35—C33 | 119.9 (2) |
| C15—C14—H14 | 119.6 | C36—C35—H35 | 120.0 |
| C16—C15—C14 | 117.30 (18) | C33—C35—H35 | 120.0 |
| C16—C15—C17 | 118.90 (18) | C35—C36—C37 | 119.3 (2) |
| C14—C15—C17 | 123.76 (19) | C35—C36—H36 | 120.4 |
| N1—C16—C15 | 119.75 (17) | C37—C36—H36 | 120.4 |
| N1—C16—C20 | 118.73 (18) | N4—C37—C36 | 121.9 (2) |
| C15—C16—C20 | 121.51 (18) | N4—C37—C38 | 117.36 (18) |
| C18—C17—C15 | 120.61 (19) | C36—C37—C38 | 120.71 (19) |
| C18—C17—H17 | 119.7 | C37—C38—H38A | 109.5 |
| C15—C17—H17 | 119.7 | C37—C38—H38B | 109.5 |
| C17—C18—C19 | 121.10 (19) | H38A—C38—H38B | 109.5 |
| C17—C18—H18 | 119.4 | C37—C38—H38C | 109.5 |
| C19—C18—H18 | 119.4 | H38A—C38—H38C | 109.5 |
| C21—C19—C20 | 115.99 (19) | H38B—C38—H38C | 109.5 |
| C21—C19—C18 | 123.85 (19) | ||
| C3i—C1—C2—C3 | −0.5 (3) | C18—C19—C20—N2 | 177.97 (17) |
| C4—C1—C2—C3 | −179.62 (17) | C21—C19—C20—C16 | −179.72 (16) |
| C3i—C1—C2—C5 | −178.40 (17) | C18—C19—C20—C16 | −0.6 (3) |
| C4—C1—C2—C5 | 2.4 (3) | N1—C16—C20—N2 | −0.6 (2) |
| C1—C2—C3—C1i | 0.5 (3) | C15—C16—C20—N2 | −179.19 (16) |
| C5—C2—C3—C1i | 178.73 (17) | N1—C16—C20—C19 | 178.12 (15) |
| C3i—C1—C4—O2 | −10.8 (2) | C15—C16—C20—C19 | −0.5 (3) |
| C2—C1—C4—O2 | 168.41 (18) | C20—C19—C21—C22 | 0.4 (3) |
| C3i—C1—C4—O1 | 167.52 (17) | C18—C19—C21—C22 | −178.69 (18) |
| C2—C1—C4—O1 | −13.3 (3) | C19—C21—C22—C23 | 1.0 (3) |
| C3—C2—C5—O3 | 15.2 (3) | C20—N2—C23—C22 | 1.1 (3) |
| C1—C2—C5—O3 | −166.75 (18) | C20—N2—C23—C24 | −178.25 (17) |
| C3—C2—C5—O4 | −161.82 (17) | C21—C22—C23—N2 | −1.8 (3) |
| C1—C2—C5—O4 | 16.2 (3) | C21—C22—C23—C24 | 177.49 (19) |
| C8ii—C6—C7—C8 | 0.0 (3) | C30—N3—C26—C27 | −1.1 (3) |
| C9—C6—C7—C8 | −177.05 (16) | C30—N3—C26—C25 | 179.39 (17) |
| C8ii—C6—C7—C10 | −176.53 (16) | N3—C26—C27—C28 | 2.7 (3) |
| C9—C6—C7—C10 | 6.4 (3) | C25—C26—C27—C28 | −177.8 (2) |
| C6—C7—C8—C6ii | 0.0 (3) | C26—C27—C28—C29 | −1.7 (3) |
| C10—C7—C8—C6ii | 176.54 (16) | C27—C28—C29—C30 | −0.8 (3) |
| C8ii—C6—C9—O5 | 64.4 (2) | C27—C28—C29—C31 | 178.0 (2) |
| C7—C6—C9—O5 | −118.4 (2) | C26—N3—C30—C29 | −1.5 (3) |
| C8ii—C6—C9—O6 | −110.89 (18) | C26—N3—C30—C34 | 179.41 (16) |
| C7—C6—C9—O6 | 66.3 (2) | C28—C29—C30—N3 | 2.5 (3) |
| C8—C7—C10—O7 | −158.38 (17) | C31—C29—C30—N3 | −176.38 (18) |
| C6—C7—C10—O7 | 18.2 (3) | C28—C29—C30—C34 | −178.51 (17) |
| C8—C7—C10—O8 | 20.0 (2) | C31—C29—C30—C34 | 2.7 (3) |
| C6—C7—C10—O8 | −163.44 (16) | C28—C29—C31—C32 | −180.0 (2) |
| C16—N1—C12—C13 | −2.0 (3) | C30—C29—C31—C32 | −1.2 (3) |
| C16—N1—C12—C11 | 176.50 (16) | C29—C31—C32—C33 | −0.6 (3) |
| N1—C12—C13—C14 | 1.8 (3) | C31—C32—C33—C35 | −179.5 (2) |
| C11—C12—C13—C14 | −176.64 (18) | C31—C32—C33—C34 | 1.0 (3) |
| C12—C13—C14—C15 | −0.4 (3) | C37—N4—C34—C33 | −1.0 (3) |
| C13—C14—C15—C16 | −0.8 (3) | C37—N4—C34—C30 | 177.42 (16) |
| C13—C14—C15—C17 | 176.99 (17) | C35—C33—C34—N4 | −0.6 (3) |
| C12—N1—C16—C15 | 0.8 (3) | C32—C33—C34—N4 | 178.94 (17) |
| C12—N1—C16—C20 | −177.88 (16) | C35—C33—C34—C30 | −179.09 (17) |
| C14—C15—C16—N1 | 0.7 (3) | C32—C33—C34—C30 | 0.5 (3) |
| C17—C15—C16—N1 | −177.25 (16) | N3—C30—C34—N4 | −1.7 (3) |
| C14—C15—C16—C20 | 179.30 (16) | C29—C30—C34—N4 | 179.23 (16) |
| C17—C15—C16—C20 | 1.4 (3) | N3—C30—C34—C33 | 176.79 (16) |
| C16—C15—C17—C18 | −1.1 (3) | C29—C30—C34—C33 | −2.3 (3) |
| C14—C15—C17—C18 | −178.89 (18) | C34—C33—C35—C36 | 1.3 (3) |
| C15—C17—C18—C19 | 0.0 (3) | C32—C33—C35—C36 | −178.27 (19) |
| C17—C18—C19—C21 | 179.92 (18) | C33—C35—C36—C37 | −0.4 (3) |
| C17—C18—C19—C20 | 0.9 (3) | C34—N4—C37—C36 | 2.0 (3) |
| C23—N2—C20—C19 | 0.4 (3) | C34—N4—C37—C38 | −175.67 (16) |
| C23—N2—C20—C16 | 179.00 (16) | C35—C36—C37—N4 | −1.4 (3) |
| C21—C19—C20—N2 | −1.1 (3) | C35—C36—C37—C38 | 176.27 (18) |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+2, −y, −z+1.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O4—H1o···O1 | 0.85 (2) | 1.55 (2) | 2.403 (2) | 176 (3) |
| O6—H2o···O2iii | 0.85 (1) | 1.74 (1) | 2.577 (2) | 168 (2) |
| O8—H3o···N4iv | 0.85 (2) | 1.79 (2) | 2.636 (2) | 173 (2) |
| N1—H1n···O3v | 0.89 (2) | 2.41 (2) | 3.257 (2) | 161 (2) |
| N1—H1n···O4v | 0.89 (2) | 2.35 (2) | 2.957 (2) | 126 (2) |
| C13—H13···O7vi | 0.95 | 2.28 | 3.225 (3) | 171 |
| C28—H28···O5vii | 0.95 | 2.40 | 3.320 (3) | 162 |
Symmetry codes: (iii) x+1/2, −y+1/2, z+1/2; (iv) −x+1, −y, −z+1; (v) −x+3/2, y+1/2, −z+1/2; (vi) x−1/2, −y+3/2, z−1/2; (vii) x−1/2, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU5731).
References
- Arman, H. D., Kaulgud, T. & Tiekink, E. R. T. (2010). Acta Cryst. E66, o2602. [DOI] [PMC free article] [PubMed]
- Arman, H. D. & Tiekink, E. R. T. (2013). Z. Kristallogr. Cryst. Mat 228, 289–294.
- Brandenburg, K. (2006). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Derikvand, Z. & Olmstead, M. M. (2011). Acta Cryst. E67, o87–o88. [DOI] [PMC free article] [PubMed]
- Higashi, T. (1995). ABSCOR Rigaku Corporation, Tokyo, Japan.
- Johnson, C. K. (1976). ORTEPII Report ORNL-5138. Oak Ridge National Laboratory, Tennessee, USA.
- Molecular Structure Corporation & Rigaku (2005). CrystalClear MSC, The Woodlands, Texas, USA, and Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813022691/xu5731sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813022691/xu5731Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813022691/xu5731Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report