Abstract
In the title compound, C24H18N2O5S, the S atom has a distorted tetrahedral configuration, with bond angles varying from 105.11 (7) to 119.98 (8)°. As a result of the electron-withdrawing character of the phenylsulfonyl group, the N—Csp 2 bond lengths [1.414 (2) and 1.413 (2) Å] are slightly longer than the reported value of 1.355 (14) Å for N atoms with a planar configuration. The indole moiety is essentially planar, with a maximum deviation of 0.0177 (14) Å for the N atom. The phenyl ring of the sulfonyl substituent makes a dihedral angle of 85.70 (7)° with the mean plane of the indole moiety. The molecular structure features intramolecular C—H⋯O hydrogen bonds, which generate S(6) and S(12) ring motifs. In the crystal, adjacent molecules are linked via C—H⋯O hydrogen bonds, forming infinite C(7) chains running along the a-axis direction. The crystal packing also features C—H⋯π interactions, which form a three-dimensional structure.
Related literature
For the biological activity of Indole derivatives, see: Rodriguez et al. (1985 ▶); Chai et al. (2006 ▶); Olgen & Coban (2003 ▶). For related crystal structures, see: Karthikeyan et al. (2011 ▶, 2012 ▶). For related bond distances and bond-angle geometries and distortions, see: Allen (1981 ▶); Allen et al. (1987 ▶). For graph-set notation, see: Bernstein et al. (1995 ▶). For the Thorpe–Ingold effect, see: Bassindale (1984 ▶).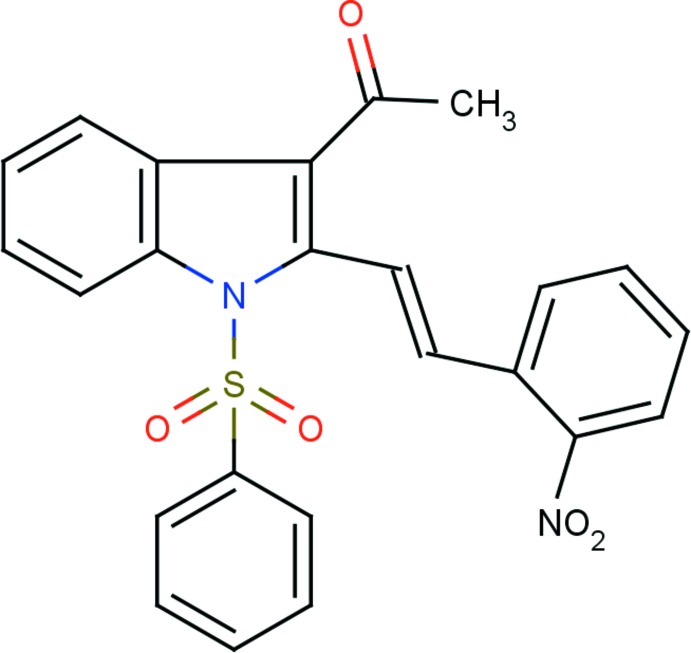
Experimental
Crystal data
C24H18N2O5S
M r = 446.46
Monoclinic,
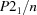
a = 8.2409 (8) Å
b = 16.1702 (15) Å
c = 15.3700 (15) Å
β = 95.775 (5)°
V = 2037.8 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.20 mm−1
T = 296 K
0.28 × 0.25 × 0.23 mm
Data collection
Bruker SMART APEXII CCD diffractometer
18837 measured reflections
4960 independent reflections
3893 reflections with I > 2σ(I)
R int = 0.021
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.118
S = 1.02
4960 reflections
290 parameters
H-atom parameters constrained
Δρmax = 0.42 e Å−3
Δρmin = −0.38 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶) and Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813022241/su2631sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813022241/su2631Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813022241/su2631Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1, Cg2 and Cg3 are the centroids of the C17–C22, C11–C16 and C1–C6 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2⋯O1 | 0.93 | 2.35 | 2.935 (2) | 121 |
| C18—H18⋯O3 | 0.93 | 2.46 | 3.108 (2) | 127 |
| C20—H20⋯O2i | 0.93 | 2.68 | 3.319 (2) | 126 |
| C5—H5⋯Cg1ii | 0.93 | 2.89 | 3.720 (2) | 149 |
| C22—H22⋯Cg2iii | 0.93 | 2.73 | 3.4618 (18) | 137 |
| C24—H24B⋯Cg3iv | 0.96 | 2.90 | 3.601 (3) | 131 |
Symmetry codes: (i) 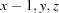 ; (ii)
; (ii) 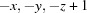 ; (iii)
; (iii) 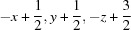 ; (iv)
; (iv) 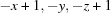 .
.
Acknowledgments
The authors thank Mr T. Srinivasan and Dr D. Velmurugan, The Head, CAS in Crystallography and Biophysics, University of Madras, Chennai, India, for the data collection.
supplementary crystallographic information
1. Comment
The indole ring system is present in a number of natural products, many of which are found to possess pharmacological properties like anti-microbial, anti-inflammatory and anti-implantation activities (Rodriguez et al., 1985). Indole derivatives are also known to exhibit anti-oxidant activities (Olgen & Coban, 2003) and antihepatitis B virus activities (Chai et al., 2006).
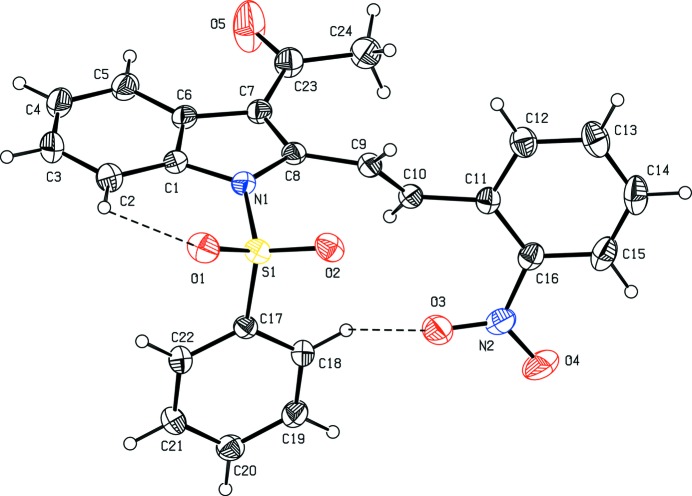
In the title compound, Fig. 1, the indole ring is essentially planar with a maximum deviation of 0.0177 (14) Å for atom N1. The indole moiety is perpendicular to the phenylsulfonyl ring with a dihedral angle 85.70 (7)°. The nitro-group is inclined at an angle of 7.59 (9)° to the benzene ring to which it is attached. The nitro-phenyl ring makes a dihedral angle of 76.41 (7)° with the indole moiety mean plane. Short intramolecular C2—H2···O1 and C18—H18···O3 hyrogen bonds result in S(6) and S(12) ring motifs (Table 1 and Fig. 1). The molecular dimensions in the title compound are in excellent agreement with those reported for a closely related compound (Karthikeyan et al., 2012).
The expansion of the ispo angles at C1, C3 and C4 [122.34 (16), 121.59 (18) and 121.13 (18)°, respectively] and contraction of the apical angles at C2, C5 and C6 [117.12 (19), 118.54 (18) and 119.28 (16)°, respectively] are caused by the fusion of the smaller pyrrole ring to the six-membered benzene ring and the strain is taken up by the angular distortion rather than by bond-length distortions (Allen, 1981). As a result of the electron withdrawing character of the phenylsulfonyl group, the N—Csp2 bond lengths, viz, N1—C1 [1.414 (2) Å] and N1—C8 [1.413 (2) Å], are longer than the mean value of 1.355 (14) Å reported for N atoms with planar configurations (Allen et al., 1987).
Atom S1 has a distorted tetrahedral configuration. The widening of the angle O2═S1═O1 [119.98 (8)°] and the narrowing of the angle N1—S1—C17 [105.11 (7)°] from the ideal tetrahedral value are attributed to the Thorpe-Ingold effect (Bassindale, 1984). The widening of the angles may be due to the repulsive interactions between the two short S═O bonds, similar to what is observed in a related structure (Karthikeyan et al., 2011).
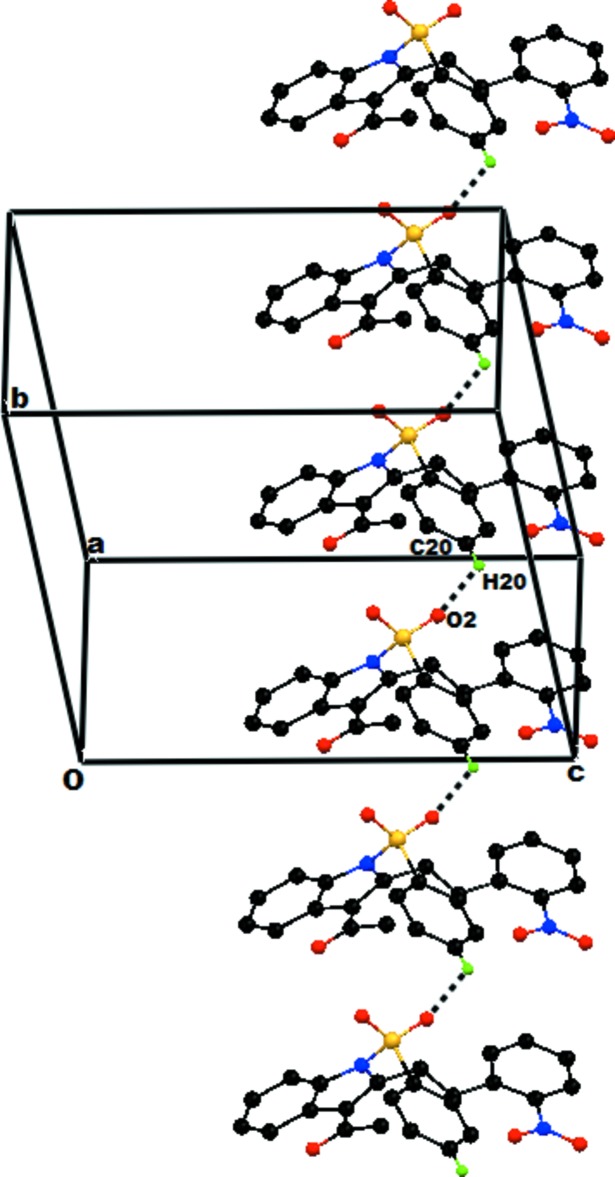
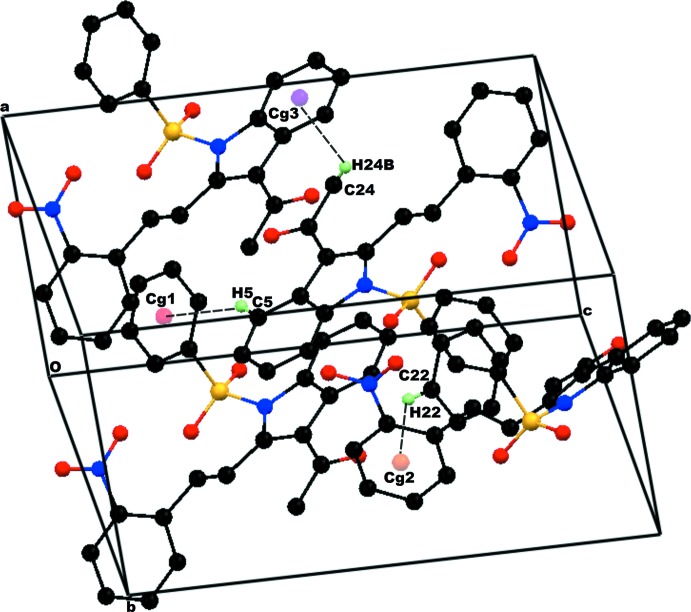
The crystal packing is stabilized by C—H···O hydrogen bonds (Fig. 2 and Table 1) resulting in C(7) infinite chains of molecules running along the a axis direction (Bernstein et al., 1995). The crystal packing is further stabilized by C—H···π interactions forming a three-dimensional structure (Table 1 and Fig. 3).
2. Experimental
A solution of the ylide, 1-[1-(phenylsulfonyl)-2-[(triphenyl-$15-phosphanylidene)methyl]-1H-indol-3-yl]ethan-1-one (1 g, 1.745 mmol), and 2-nitrobenzaldehyde (0.29 g, 1.919 mmol) in dry DCM (20 ml) was refluxed for 8 h under N2. Removal of solvent in vacuo followed by trituration of the crude product with cold methanol (5 ml) afforded the title compound as a yellow solid [Yield: 0.71 g (92%); M.p. 427-429 K]. Block-like yellow crystals were obtained by slow evaporation of a solution in CHCl3.
3. Refinement
The hydrogen atoms were localized from the difference electron density maps and were refined as riding atoms: C—H = 0.93 and 0.96 Å for CH(aromatic) and methyl H atoms, respectively, with Uiso(H) = 1.5 Ueq(C-methyl) and = 1.2Ueq(C) for other H atoms. The rotation angles for the methyl groups were optimized by least squares.
Figures
Fig. 1.
The molecular structure of the title molecule, with the atom labelling. Displacement ellipsoids are drawn at 30% probability level. The intramolecular C-H···O hydrogen bonds are shown as dashed lines (see Table 1 for details).
Fig. 2.
The crystal packing of the title compound viewed along the a axis, showing the formation of infinite C(7) chains. The dashed lines indicate C—H···O hydrogen bonds (see Table 1 for details).
Fig. 3.
The crystal packing of the title compound, showing the intermolecular C—H···π interactions as dashed lines (centroid = red sphere; see Table 1 for details).
Crystal data
| C24H18N2O5S | F(000) = 928 |
| Mr = 446.46 | Dx = 1.455 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -p 2yn | Cell parameters from 4960 reflections |
| a = 8.2409 (8) Å | θ = 1.8–28.4° |
| b = 16.1702 (15) Å | µ = 0.20 mm−1 |
| c = 15.3700 (15) Å | T = 296 K |
| β = 95.775 (5)° | Block, yellow |
| V = 2037.8 (3) Å3 | 0.28 × 0.25 × 0.23 mm |
| Z = 4 |
Data collection
| Bruker SMART APEXII CCD diffractometer | 3893 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.021 |
| Graphite monochromator | θmax = 28.4°, θmin = 1.8° |
| ω and φ scans | h = −10→7 |
| 18837 measured reflections | k = −17→21 |
| 4960 independent reflections | l = −19→20 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.118 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.059P)2 + 0.5665P] where P = (Fo2 + 2Fc2)/3 |
| 4960 reflections | (Δ/σ)max = 0.001 |
| 290 parameters | Δρmax = 0.42 e Å−3 |
| 0 restraints | Δρmin = −0.38 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.21108 (19) | 0.10784 (11) | 0.52063 (10) | 0.0405 (4) | |
| C2 | 0.1313 (3) | 0.17176 (13) | 0.47358 (13) | 0.0570 (5) | |
| H2 | 0.1284 | 0.2250 | 0.4963 | 0.068* | |
| C3 | 0.0567 (2) | 0.15297 (15) | 0.39184 (13) | 0.0627 (5) | |
| H3 | 0.0019 | 0.1944 | 0.3587 | 0.075* | |
| C4 | 0.0611 (2) | 0.07382 (14) | 0.35767 (12) | 0.0579 (5) | |
| H4 | 0.0089 | 0.0631 | 0.3023 | 0.070* | |
| C5 | 0.1414 (2) | 0.01077 (13) | 0.40421 (11) | 0.0499 (4) | |
| H5 | 0.1450 | −0.0421 | 0.3806 | 0.060* | |
| C6 | 0.21746 (19) | 0.02793 (11) | 0.48764 (10) | 0.0395 (4) | |
| C7 | 0.31131 (18) | −0.02214 (10) | 0.55255 (10) | 0.0379 (3) | |
| C8 | 0.35638 (18) | 0.02700 (9) | 0.62314 (10) | 0.0356 (3) | |
| C9 | 0.45622 (19) | 0.00804 (10) | 0.70516 (10) | 0.0375 (3) | |
| H9 | 0.5562 | 0.0346 | 0.7165 | 0.045* | |
| C10 | 0.41139 (19) | −0.04492 (10) | 0.76375 (10) | 0.0381 (3) | |
| H10 | 0.3066 | −0.0668 | 0.7559 | 0.046* | |
| C11 | 0.5202 (2) | −0.07076 (9) | 0.84085 (10) | 0.0386 (3) | |
| C12 | 0.6855 (2) | −0.08209 (11) | 0.83104 (13) | 0.0500 (4) | |
| H12 | 0.7229 | −0.0696 | 0.7775 | 0.060* | |
| C13 | 0.7946 (2) | −0.11089 (13) | 0.89725 (15) | 0.0618 (5) | |
| H13 | 0.9040 | −0.1172 | 0.8885 | 0.074* | |
| C14 | 0.7415 (3) | −0.13056 (13) | 0.97719 (14) | 0.0652 (6) | |
| H14 | 0.8147 | −0.1514 | 1.0218 | 0.078* | |
| C15 | 0.5818 (3) | −0.11937 (12) | 0.99067 (12) | 0.0566 (5) | |
| H15 | 0.5464 | −0.1316 | 1.0448 | 0.068* | |
| C16 | 0.4726 (2) | −0.08979 (10) | 0.92347 (10) | 0.0422 (4) | |
| C17 | 0.10318 (18) | 0.16933 (9) | 0.72059 (10) | 0.0339 (3) | |
| C18 | 0.0997 (2) | 0.11710 (11) | 0.79151 (10) | 0.0426 (4) | |
| H18 | 0.1937 | 0.0897 | 0.8143 | 0.051* | |
| C19 | −0.0445 (2) | 0.10613 (12) | 0.82802 (11) | 0.0478 (4) | |
| H19 | −0.0483 | 0.0711 | 0.8757 | 0.057* | |
| C20 | −0.1833 (2) | 0.14712 (12) | 0.79391 (11) | 0.0460 (4) | |
| H20 | −0.2800 | 0.1404 | 0.8195 | 0.055* | |
| C21 | −0.1796 (2) | 0.19756 (12) | 0.72260 (12) | 0.0466 (4) | |
| H21 | −0.2743 | 0.2241 | 0.6994 | 0.056* | |
| C22 | −0.03571 (19) | 0.20929 (10) | 0.68482 (11) | 0.0415 (4) | |
| H22 | −0.0328 | 0.2434 | 0.6363 | 0.050* | |
| C23 | 0.3572 (2) | −0.10893 (11) | 0.53594 (12) | 0.0513 (4) | |
| C24 | 0.4614 (3) | −0.15953 (12) | 0.59949 (13) | 0.0617 (5) | |
| H24A | 0.3984 | −0.1781 | 0.6450 | 0.093* | |
| H24B | 0.5515 | −0.1268 | 0.6245 | 0.093* | |
| H24C | 0.5018 | −0.2065 | 0.5702 | 0.093* | |
| N1 | 0.30044 (16) | 0.10825 (8) | 0.60412 (8) | 0.0394 (3) | |
| N2 | 0.3042 (2) | −0.07812 (9) | 0.94350 (10) | 0.0501 (4) | |
| O1 | 0.28218 (16) | 0.25924 (7) | 0.62838 (9) | 0.0549 (3) | |
| O2 | 0.41828 (14) | 0.17130 (8) | 0.74404 (8) | 0.0498 (3) | |
| O3 | 0.20813 (17) | −0.04423 (10) | 0.89080 (9) | 0.0653 (4) | |
| O4 | 0.2671 (2) | −0.10333 (12) | 1.01319 (10) | 0.0862 (5) | |
| O5 | 0.3144 (3) | −0.13858 (12) | 0.46632 (13) | 0.1255 (10) | |
| S1 | 0.28946 (5) | 0.18452 (2) | 0.67705 (3) | 0.03893 (12) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0387 (8) | 0.0493 (9) | 0.0345 (8) | 0.0064 (7) | 0.0078 (6) | 0.0042 (7) |
| C2 | 0.0639 (12) | 0.0574 (11) | 0.0499 (10) | 0.0185 (9) | 0.0072 (9) | 0.0088 (9) |
| C3 | 0.0602 (12) | 0.0790 (14) | 0.0479 (11) | 0.0200 (11) | 0.0012 (9) | 0.0180 (10) |
| C4 | 0.0465 (10) | 0.0853 (15) | 0.0409 (10) | 0.0039 (10) | −0.0008 (8) | 0.0064 (9) |
| C5 | 0.0443 (9) | 0.0639 (11) | 0.0414 (9) | −0.0026 (8) | 0.0030 (7) | −0.0025 (8) |
| C6 | 0.0354 (8) | 0.0484 (9) | 0.0356 (8) | −0.0002 (7) | 0.0080 (6) | 0.0027 (7) |
| C7 | 0.0364 (8) | 0.0411 (8) | 0.0367 (8) | −0.0010 (6) | 0.0060 (6) | 0.0015 (6) |
| C8 | 0.0336 (7) | 0.0380 (8) | 0.0363 (8) | 0.0016 (6) | 0.0089 (6) | 0.0037 (6) |
| C9 | 0.0354 (8) | 0.0392 (8) | 0.0378 (8) | 0.0019 (6) | 0.0028 (6) | −0.0007 (6) |
| C10 | 0.0350 (8) | 0.0430 (9) | 0.0363 (8) | 0.0012 (6) | 0.0033 (6) | −0.0008 (7) |
| C11 | 0.0429 (9) | 0.0342 (8) | 0.0376 (8) | −0.0022 (6) | −0.0006 (6) | −0.0016 (6) |
| C12 | 0.0441 (9) | 0.0502 (10) | 0.0547 (10) | −0.0022 (8) | 0.0004 (8) | 0.0038 (8) |
| C13 | 0.0457 (10) | 0.0613 (12) | 0.0743 (14) | −0.0011 (9) | −0.0142 (9) | −0.0001 (10) |
| C14 | 0.0725 (14) | 0.0595 (12) | 0.0569 (12) | 0.0018 (10) | −0.0271 (10) | 0.0017 (10) |
| C15 | 0.0834 (15) | 0.0468 (10) | 0.0369 (9) | −0.0011 (9) | −0.0081 (9) | −0.0002 (8) |
| C16 | 0.0551 (10) | 0.0337 (8) | 0.0370 (8) | −0.0014 (7) | 0.0014 (7) | −0.0035 (6) |
| C17 | 0.0335 (7) | 0.0331 (7) | 0.0347 (7) | −0.0002 (6) | 0.0020 (6) | −0.0054 (6) |
| C18 | 0.0422 (9) | 0.0481 (9) | 0.0366 (8) | 0.0067 (7) | −0.0005 (7) | 0.0026 (7) |
| C19 | 0.0499 (10) | 0.0556 (10) | 0.0382 (9) | 0.0001 (8) | 0.0066 (7) | 0.0065 (8) |
| C20 | 0.0377 (9) | 0.0561 (10) | 0.0446 (9) | −0.0045 (7) | 0.0062 (7) | −0.0037 (8) |
| C21 | 0.0349 (8) | 0.0533 (10) | 0.0506 (10) | 0.0052 (7) | −0.0008 (7) | 0.0015 (8) |
| C22 | 0.0417 (9) | 0.0401 (8) | 0.0419 (9) | 0.0022 (7) | −0.0007 (7) | 0.0050 (7) |
| C23 | 0.0586 (11) | 0.0445 (10) | 0.0495 (10) | −0.0001 (8) | −0.0006 (8) | −0.0066 (8) |
| C24 | 0.0823 (15) | 0.0428 (10) | 0.0596 (12) | 0.0124 (10) | 0.0054 (10) | −0.0016 (9) |
| N1 | 0.0437 (7) | 0.0401 (7) | 0.0349 (7) | 0.0076 (6) | 0.0060 (5) | 0.0013 (5) |
| N2 | 0.0696 (10) | 0.0408 (8) | 0.0419 (8) | 0.0020 (7) | 0.0159 (7) | 0.0004 (6) |
| O1 | 0.0575 (8) | 0.0386 (6) | 0.0708 (8) | −0.0015 (5) | 0.0165 (6) | 0.0090 (6) |
| O2 | 0.0369 (6) | 0.0521 (7) | 0.0589 (8) | −0.0016 (5) | −0.0030 (5) | −0.0118 (6) |
| O3 | 0.0606 (9) | 0.0722 (10) | 0.0661 (9) | 0.0154 (7) | 0.0206 (7) | 0.0209 (7) |
| O4 | 0.1070 (13) | 0.1054 (13) | 0.0524 (9) | 0.0156 (11) | 0.0377 (9) | 0.0191 (9) |
| O5 | 0.191 (2) | 0.0768 (12) | 0.0917 (13) | 0.0510 (14) | −0.0689 (14) | −0.0432 (11) |
| S1 | 0.0365 (2) | 0.0349 (2) | 0.0456 (2) | −0.00095 (15) | 0.00548 (16) | −0.00227 (16) |
Geometric parameters (Å, º)
| C1—C2 | 1.389 (2) | C14—H14 | 0.9300 |
| C1—C6 | 1.391 (2) | C15—C16 | 1.385 (2) |
| C1—N1 | 1.414 (2) | C15—H15 | 0.9300 |
| C2—C3 | 1.376 (3) | C16—N2 | 1.464 (2) |
| C2—H2 | 0.9300 | C17—C22 | 1.380 (2) |
| C3—C4 | 1.385 (3) | C17—C18 | 1.382 (2) |
| C3—H3 | 0.9300 | C17—S1 | 1.7526 (15) |
| C4—C5 | 1.376 (3) | C18—C19 | 1.375 (2) |
| C4—H4 | 0.9300 | C18—H18 | 0.9300 |
| C5—C6 | 1.397 (2) | C19—C20 | 1.379 (2) |
| C5—H5 | 0.9300 | C19—H19 | 0.9300 |
| C6—C7 | 1.447 (2) | C20—C21 | 1.369 (3) |
| C7—C8 | 1.366 (2) | C20—H20 | 0.9300 |
| C7—C23 | 1.482 (2) | C21—C22 | 1.385 (2) |
| C8—N1 | 1.413 (2) | C21—H21 | 0.9300 |
| C8—C9 | 1.467 (2) | C22—H22 | 0.9300 |
| C9—C10 | 1.322 (2) | C23—O5 | 1.193 (2) |
| C9—H9 | 0.9300 | C23—C24 | 1.480 (3) |
| C10—C11 | 1.473 (2) | C24—H24A | 0.9600 |
| C10—H10 | 0.9300 | C24—H24B | 0.9600 |
| C11—C12 | 1.397 (2) | C24—H24C | 0.9600 |
| C11—C16 | 1.401 (2) | N1—S1 | 1.6752 (14) |
| C12—C13 | 1.370 (3) | N2—O3 | 1.2058 (19) |
| C12—H12 | 0.9300 | N2—O4 | 1.2135 (19) |
| C13—C14 | 1.382 (3) | O1—S1 | 1.4192 (13) |
| C13—H13 | 0.9300 | O2—S1 | 1.4187 (12) |
| C14—C15 | 1.365 (3) | ||
| C2—C1—C6 | 122.34 (16) | C16—C15—H15 | 120.1 |
| C2—C1—N1 | 130.19 (17) | C15—C16—C11 | 122.11 (17) |
| C6—C1—N1 | 107.45 (13) | C15—C16—N2 | 116.51 (16) |
| C3—C2—C1 | 117.12 (19) | C11—C16—N2 | 121.38 (15) |
| C3—C2—H2 | 121.4 | C22—C17—C18 | 121.22 (15) |
| C1—C2—H2 | 121.4 | C22—C17—S1 | 120.38 (12) |
| C2—C3—C4 | 121.59 (18) | C18—C17—S1 | 118.40 (12) |
| C2—C3—H3 | 119.2 | C19—C18—C17 | 119.23 (15) |
| C4—C3—H3 | 119.2 | C19—C18—H18 | 120.4 |
| C5—C4—C3 | 121.13 (18) | C17—C18—H18 | 120.4 |
| C5—C4—H4 | 119.4 | C18—C19—C20 | 120.04 (16) |
| C3—C4—H4 | 119.4 | C18—C19—H19 | 120.0 |
| C4—C5—C6 | 118.54 (18) | C20—C19—H19 | 120.0 |
| C4—C5—H5 | 120.7 | C21—C20—C19 | 120.40 (16) |
| C6—C5—H5 | 120.7 | C21—C20—H20 | 119.8 |
| C1—C6—C5 | 119.28 (16) | C19—C20—H20 | 119.8 |
| C1—C6—C7 | 107.80 (14) | C20—C21—C22 | 120.41 (16) |
| C5—C6—C7 | 132.91 (16) | C20—C21—H21 | 119.8 |
| C8—C7—C6 | 107.81 (14) | C22—C21—H21 | 119.8 |
| C8—C7—C23 | 129.34 (15) | C17—C22—C21 | 118.68 (15) |
| C6—C7—C23 | 122.62 (15) | C17—C22—H22 | 120.7 |
| C7—C8—N1 | 108.64 (13) | C21—C22—H22 | 120.7 |
| C7—C8—C9 | 130.18 (14) | O5—C23—C24 | 117.99 (18) |
| N1—C8—C9 | 121.02 (14) | O5—C23—C7 | 118.50 (18) |
| C10—C9—C8 | 123.36 (15) | C24—C23—C7 | 123.42 (16) |
| C10—C9—H9 | 118.3 | C23—C24—H24A | 109.5 |
| C8—C9—H9 | 118.3 | C23—C24—H24B | 109.5 |
| C9—C10—C11 | 122.86 (15) | H24A—C24—H24B | 109.5 |
| C9—C10—H10 | 118.6 | C23—C24—H24C | 109.5 |
| C11—C10—H10 | 118.6 | H24A—C24—H24C | 109.5 |
| C12—C11—C16 | 115.74 (15) | H24B—C24—H24C | 109.5 |
| C12—C11—C10 | 118.13 (15) | C8—N1—C1 | 108.22 (13) |
| C16—C11—C10 | 126.03 (15) | C8—N1—S1 | 125.77 (11) |
| C13—C12—C11 | 122.58 (19) | C1—N1—S1 | 123.54 (11) |
| C13—C12—H12 | 118.7 | O3—N2—O4 | 122.63 (17) |
| C11—C12—H12 | 118.7 | O3—N2—C16 | 119.31 (14) |
| C12—C13—C14 | 119.7 (2) | O4—N2—C16 | 118.06 (17) |
| C12—C13—H13 | 120.1 | O2—S1—O1 | 119.98 (8) |
| C14—C13—H13 | 120.1 | O2—S1—N1 | 106.73 (7) |
| C15—C14—C13 | 120.05 (18) | O1—S1—N1 | 106.04 (8) |
| C15—C14—H14 | 120.0 | O2—S1—C17 | 108.79 (7) |
| C13—C14—H14 | 120.0 | O1—S1—C17 | 109.17 (8) |
| C14—C15—C16 | 119.79 (19) | N1—S1—C17 | 105.11 (7) |
| C14—C15—H15 | 120.1 | ||
| C6—C1—C2—C3 | 0.3 (3) | S1—C17—C18—C19 | 178.28 (13) |
| N1—C1—C2—C3 | 178.49 (17) | C17—C18—C19—C20 | −0.1 (3) |
| C1—C2—C3—C4 | −0.2 (3) | C18—C19—C20—C21 | 1.3 (3) |
| C2—C3—C4—C5 | −0.3 (3) | C19—C20—C21—C22 | −1.2 (3) |
| C3—C4—C5—C6 | 0.7 (3) | C18—C17—C22—C21 | 1.4 (2) |
| C2—C1—C6—C5 | 0.1 (3) | S1—C17—C22—C21 | −178.17 (13) |
| N1—C1—C6—C5 | −178.46 (14) | C20—C21—C22—C17 | −0.1 (3) |
| C2—C1—C6—C7 | 179.14 (16) | C8—C7—C23—O5 | −174.0 (2) |
| N1—C1—C6—C7 | 0.59 (17) | C6—C7—C23—O5 | −0.3 (3) |
| C4—C5—C6—C1 | −0.6 (2) | C8—C7—C23—C24 | 2.5 (3) |
| C4—C5—C6—C7 | −179.37 (18) | C6—C7—C23—C24 | 176.22 (17) |
| C1—C6—C7—C8 | 1.22 (17) | C7—C8—N1—C1 | 2.95 (17) |
| C5—C6—C7—C8 | −179.90 (17) | C9—C8—N1—C1 | 178.81 (13) |
| C1—C6—C7—C23 | −173.71 (15) | C7—C8—N1—S1 | 165.52 (11) |
| C5—C6—C7—C23 | 5.2 (3) | C9—C8—N1—S1 | −18.6 (2) |
| C6—C7—C8—N1 | −2.55 (17) | C2—C1—N1—C8 | 179.46 (18) |
| C23—C7—C8—N1 | 171.93 (16) | C6—C1—N1—C8 | −2.14 (17) |
| C6—C7—C8—C9 | −177.91 (15) | C2—C1—N1—S1 | 16.4 (3) |
| C23—C7—C8—C9 | −3.4 (3) | C6—C1—N1—S1 | −165.19 (11) |
| C7—C8—C9—C10 | −65.7 (2) | C15—C16—N2—O3 | 172.02 (17) |
| N1—C8—C9—C10 | 119.47 (18) | C11—C16—N2—O3 | −7.1 (2) |
| C8—C9—C10—C11 | 172.71 (14) | C15—C16—N2—O4 | −8.0 (2) |
| C9—C10—C11—C12 | −37.6 (2) | C11—C16—N2—O4 | 172.85 (17) |
| C9—C10—C11—C16 | 146.09 (17) | C8—N1—S1—O2 | 31.41 (15) |
| C16—C11—C12—C13 | 0.8 (3) | C1—N1—S1—O2 | −168.55 (12) |
| C10—C11—C12—C13 | −175.90 (17) | C8—N1—S1—O1 | 160.41 (13) |
| C11—C12—C13—C14 | 0.6 (3) | C1—N1—S1—O1 | −39.55 (15) |
| C12—C13—C14—C15 | −1.6 (3) | C8—N1—S1—C17 | −84.02 (14) |
| C13—C14—C15—C16 | 1.3 (3) | C1—N1—S1—C17 | 76.02 (14) |
| C14—C15—C16—C11 | 0.1 (3) | C22—C17—S1—O2 | 152.91 (13) |
| C14—C15—C16—N2 | −178.97 (17) | C18—C17—S1—O2 | −26.65 (14) |
| C12—C11—C16—C15 | −1.1 (2) | C22—C17—S1—O1 | 20.30 (15) |
| C10—C11—C16—C15 | 175.24 (16) | C18—C17—S1—O1 | −159.27 (12) |
| C12—C11—C16—N2 | 177.94 (15) | C22—C17—S1—N1 | −93.09 (13) |
| C10—C11—C16—N2 | −5.7 (2) | C18—C17—S1—N1 | 87.35 (13) |
| C22—C17—C18—C19 | −1.3 (2) |
Hydrogen-bond geometry (Å, º)
Cg1, Cg2 and Cg3 are the centroids of the C17–C22, C11–C16 and C1–C6 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2···O1 | 0.93 | 2.35 | 2.935 (2) | 121 |
| C18—H18···O3 | 0.93 | 2.46 | 3.108 (2) | 127 |
| C20—H20···O2i | 0.93 | 2.68 | 3.319 (2) | 126 |
| C5—H5···Cg1ii | 0.93 | 2.89 | 3.720 (2) | 149 |
| C22—H22···Cg2iii | 0.93 | 2.73 | 3.4618 (18) | 137 |
| C24—H24B···Cg3iv | 0.96 | 2.90 | 3.601 (3) | 131 |
Symmetry codes: (i) x−1, y, z; (ii) −x, −y, −z+1; (iii) −x+1/2, y+1/2, −z+3/2; (iv) −x+1, −y, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2631).
References
- Allen, F. H. (1981). Acta Cryst. B37, 900–906.
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bassindale, A. (1984). The Third Dimension in Organic Chemistry, ch. 1, p. 11. New York: John Wiley and Sons.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2008). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chai, H., Zhao, C. & Gong, P. (2006). Bioorg. Med. Chem. 14, 911-917. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Karthikeyan, S., Sethusankar, K., Rajeswaran, G. G. & Mohanakrishnan, A. K. (2011). Acta Cryst. E67, o2245–o2246. [DOI] [PMC free article] [PubMed]
- Karthikeyan, S., Sethusankar, K., Rajeswaran, G. G. & Mohanakrishnan, A. K. (2012). Acta Cryst. E68, o9. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Olgen, S. & Coban, T. (2003). Biol. Pharm. Bull. 26, 736-738. [DOI] [PubMed]
- Rodriguez, J. G., Temprano, F., Esteban-Calderon, C., Martinez-Ripoll, M. & Garcia-Blanco, S. (1985). Tetrahedron, 41, 3813-3823.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813022241/su2631sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813022241/su2631Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813022241/su2631Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report