Abstract
The asymmetric unit of the title compound, C20H24N2O2S, contains two independent molecules having very similar geometries. The main N-(6-methoxy-1,3-benzothiazol-2-yl)acetamide moiety adopts an almost planar structure (r.m.s. deviations of 0.091 and 0.051 Å for the two independent molecules). The adamantyl substituent occupies the gauche position relative to the C—N bond of the acetamide moiety [the corresponding N–C–C–C dihedral angles are −100.3 (3) and −96.5 (3)° for the two independent molecules]. In the crystal, the two independent molecules form a dimer via a pair of N—H⋯N hydrogen bonds. The dimers are further linked by C—H⋯O hydrogen bonds and attractive S⋯S [3.622 (2) Å] interactions into ribbons along [100].
Related literature
For properties of benzothiazoles as building blocks in organic synthesis, see: Gupta & Rawat (2010 ▶); Facchinetti et al. (2012 ▶); Sareen et al. (2012 ▶); Radatz et al. (2013 ▶). For syntheses and properties of 2-substituted benzothiazoles, see: Hussein et al. (2012 ▶); Ugale et al. (2012 ▶); Yoo et al. (2012 ▶); Zhu et al. (2012 ▶); Bhardwaj et al. (2013 ▶); Patel et al. (2013 ▶).
Experimental
Crystal data
C20H24N2O2S
M r = 356.48
Triclinic,

a = 11.0114 (13) Å
b = 13.6647 (18) Å
c = 13.9230 (18) Å
α = 61.554 (3)°
β = 80.252 (3)°
γ = 89.782 (4)°
V = 1808.3 (4) Å3
Z = 4
Mo Kα radiation
μ = 0.20 mm−1
T = 100 K
0.20 × 0.15 × 0.10 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2003 ▶) T min = 0.962, T max = 0.981
17897 measured reflections
7124 independent reflections
4424 reflections with I > 2σ(I)
R int = 0.071
Refinement
R[F 2 > 2σ(F 2)] = 0.049
wR(F 2) = 0.115
S = 0.91
7124 reflections
459 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.40 e Å−3
Δρmin = −0.37 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2001 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813023313/rk2412sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813023313/rk2412Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813023313/rk2412Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2N⋯N3 | 0.87 (3) | 2.13 (3) | 2.995 (3) | 169 (2) |
| N4—H4N⋯N1 | 0.78 (3) | 2.30 (3) | 3.077 (3) | 174 (2) |
| C6—H6⋯O3i | 0.95 | 2.58 | 3.452 (3) | 153 |
| C26—H26⋯O1ii | 0.95 | 2.45 | 3.392 (3) | 174 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The authors are grateful to the Ministry of Education and Science of the Russian Federation (State program No. 3.1168.2011).
supplementary crystallographic information
1. Comment
Benzothiazoles are important and versatile building blocks in organic synthesis, in particular, for production of various biologically active compounds in medicinal and industrial fields (Gupta & Rawat, 2010; Facchinetti et al., 2012; Sareen et al., 2012; Radatz et al., 2013). Notably, among all benzothiazole derivatives, 2-substituted benzothiazoles are of special interest due to their multiple applications as medicinal agents, agrochemicals, materials for chemical sensors etc. (Hussein et al., 2012; Ugale et al., 2012; Yoo et al., 2012; Zhu et al., 2012; Bhardwaj et al., 2013; Patel et al., 2013).
In this work, a 2-(1-adamantyl)-N-(6-methoxy-1,3-benzothiazol-2-yl)acetamide, C20H24N2O2S, (I) was prepared by the reaction of 1-(1-adamantylacetyl)-1H-imidazole with 6-methoxy-1,3-benzothiazol-2-amine (Fig. 1) and its structure was unambiguously established by the X-ray diffraction study.
Compound I crystallizes in the triclinic P1 space group with two crystallographically independent molecules forming an H-bonded dimer by the two classical intermolecular N–H···N hydrogen bonds (Table 1, Fig. 2). The geometries of these two independent molecules are very similar. The main N-(6-oxy-1,3-benzothiazol-2-yl)acetamide fragment adopts almost planar structure determined by the long chain of conjugated bonds. The adamantyl substituent occupies the gauche position in relative to the C–N bond of the acetamide moiety (the corresponding N–C–C–C dihedral angles are -100.3 (3)° and -96.5 (3)° for the two independent molecules, respectively).
In the crystal, the H-bonded dimers of I are linked by the intermolecular C6–H6···O3i and C26–H26···O1ii non-classical hydrogen bonds (Table 1) as well as attractive S1···S2i (3.622 (2)Å) interactions into ribbons toward [100] (Fig. 3). Symmetry codes: (i) x+1, y, z; (ii) x-1, y, z.
2. Experimental
A mixture 1-(1-adamantylacetyl)-1H-imidazole (1.06 g, 4.3 mmol) and 6-methoxy-1,3-benzothiazol-2-amine (0.9 g, 4.9 mmol) in CHCl3 (50 ml) were refluxed for 6 h. The precipitate was filtered, and then reaction mixture was concentrated in vacuo. The residue crystallized from 80% EtOH. Yield is 22%. The single crystals of the product I was obtained by slow crystallization from EtOH. M.p. = 485-486 K. IR (KBr), ν/cm-1: 3178, 2903, 2848, 1668, 1604, 1472, 1267, 1062, 827. 1H NMR (500 MHz, DMSO-d6, 304 K): δ = 1.52-1.49 (m, 6H), 1.66-1.63 (m, 6H), 1.96-1.93 (m, 3H), 2.63-2.61(m, 2H), 3.76 (s, 3H), 7.03-7.01 (m, 1H), 7.35-7.34 (m, 1H), 7.73 (dd, 1H, J = 8.87). Anal. Calcd for C20H24N2O2S: C, 67.38; H, 6.79. Found: C,67.32; H, 6.82.
3. Refinement
The hydrogen atoms of the amino groups were localized in the difference Fourier map and included in the refinement with fixed positional and isotropic displacement parameters - Uiso(H) = 1.2Ueq(N). The other hydrogen atoms were placed in the calculated positions with C–H = 0.95Å (for aryl H), 0.98Å (for methyl H), 0.99Å (for methylene H), 1.00Å (for methine H) and refined in the riding model with fixed isotropic displacement parameters: Uiso(H) = 1.5Ueq(C) for the CH3 groups and 1.2Ueq(C) for the other CH groups.
Figures
Fig. 1.
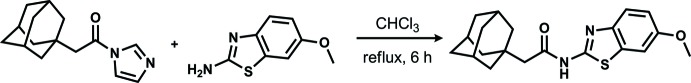
The reaction of 1-(1-adamantylacetyl)-1H-imidazole with 6-methoxy-1,3-benzothiazol-2-amine.
Fig. 2.
Molecular structure of I. The two crystallographically independent molecules forming the H-bonded dimer are shown. Displacement ellipsoids are presented at the 40% probability level. H atoms are depicted as small spheres of arbitrary radius. The dashed lines indicate the intermolecular N–H···N hydrogen bonds.
Fig. 3.
A portion of the crystal structure of I demonstrating the H-bonded ribbons toward [100]. The hydrogen atoms participating in the formation of hydrogen bonds are shown only. The intermolecular N–H···N and C–H···O hydrogen bonds as well as attractive S···S interactions are depicted by dashed lines.
Crystal data
| C20H24N2O2S | Z = 4 |
| Mr = 356.48 | F(000) = 760 |
| Triclinic, P1 | Dx = 1.309 Mg m−3 |
| Hall symbol: -P 1 | Melting point = 485–486 K |
| a = 11.0114 (13) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 13.6647 (18) Å | Cell parameters from 1305 reflections |
| c = 13.9230 (18) Å | θ = 2.8–24.8° |
| α = 61.554 (3)° | µ = 0.20 mm−1 |
| β = 80.252 (3)° | T = 100 K |
| γ = 89.782 (4)° | Prism, colourless |
| V = 1808.3 (4) Å3 | 0.20 × 0.15 × 0.10 mm |
Data collection
| Bruker APEXII CCD diffractometer | 7124 independent reflections |
| Radiation source: fine-focus sealed tube | 4424 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.071 |
| φ and ω scans | θmax = 26.0°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Bruker, 2003) | h = −13→13 |
| Tmin = 0.962, Tmax = 0.981 | k = −16→16 |
| 17897 measured reflections | l = −17→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.115 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.91 | w = 1/[σ2(Fo2) + (0.0515P)2] where P = (Fo2 + 2Fc2)/3 |
| 7124 reflections | (Δ/σ)max < 0.001 |
| 459 parameters | Δρmax = 0.40 e Å−3 |
| 0 restraints | Δρmin = −0.37 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.64296 (6) | 0.03880 (6) | 0.19027 (6) | 0.02258 (17) | |
| O1 | 0.59221 (16) | −0.16836 (15) | 0.23783 (15) | 0.0273 (4) | |
| O2 | 0.80076 (16) | 0.44718 (15) | 0.07773 (17) | 0.0336 (5) | |
| N1 | 0.47389 (18) | 0.14101 (17) | 0.07440 (17) | 0.0209 (5) | |
| N2 | 0.4527 (2) | −0.05386 (18) | 0.14838 (18) | 0.0211 (5) | |
| H2N | 0.382 (2) | −0.052 (2) | 0.128 (2) | 0.025* | |
| C1 | 0.5129 (2) | 0.0436 (2) | 0.1310 (2) | 0.0201 (6) | |
| C2 | 0.5505 (2) | 0.2228 (2) | 0.0741 (2) | 0.0211 (6) | |
| C3 | 0.5429 (2) | 0.3383 (2) | 0.0151 (2) | 0.0254 (6) | |
| H3 | 0.4801 | 0.3674 | −0.0276 | 0.030* | |
| C4 | 0.6279 (2) | 0.4093 (2) | 0.0197 (2) | 0.0291 (7) | |
| H4 | 0.6226 | 0.4877 | −0.0197 | 0.035* | |
| C5 | 0.7219 (2) | 0.3676 (2) | 0.0817 (2) | 0.0254 (6) | |
| C6 | 0.7324 (2) | 0.2535 (2) | 0.1401 (2) | 0.0234 (6) | |
| H6 | 0.7962 | 0.2246 | 0.1817 | 0.028* | |
| C7 | 0.6449 (2) | 0.1830 (2) | 0.1350 (2) | 0.0213 (6) | |
| C8 | 0.4930 (2) | −0.1576 (2) | 0.2068 (2) | 0.0205 (6) | |
| C9 | 0.4037 (2) | −0.2551 (2) | 0.2358 (2) | 0.0235 (6) | |
| H9A | 0.4499 | −0.3122 | 0.2243 | 0.028* | |
| H9B | 0.3437 | −0.2295 | 0.1851 | 0.028* | |
| C10 | 0.3327 (2) | −0.3087 (2) | 0.3571 (2) | 0.0219 (6) | |
| C11 | 0.2361 (2) | −0.3993 (2) | 0.3743 (2) | 0.0257 (6) | |
| H11A | 0.1786 | −0.3650 | 0.3219 | 0.031* | |
| H11B | 0.2781 | −0.4555 | 0.3583 | 0.031* | |
| C12 | 0.1631 (2) | −0.4565 (2) | 0.4939 (2) | 0.0303 (7) | |
| H12 | 0.1003 | −0.5148 | 0.5035 | 0.036* | |
| C13 | 0.2510 (3) | −0.5111 (2) | 0.5755 (2) | 0.0320 (7) | |
| H13A | 0.2927 | −0.5690 | 0.5622 | 0.038* | |
| H13B | 0.2036 | −0.5475 | 0.6529 | 0.038* | |
| C14 | 0.3477 (2) | −0.4215 (2) | 0.5595 (2) | 0.0295 (7) | |
| H14 | 0.4058 | −0.4571 | 0.6122 | 0.035* | |
| C15 | 0.2822 (3) | −0.3343 (3) | 0.5832 (3) | 0.0392 (8) | |
| H15A | 0.3441 | −0.2768 | 0.5741 | 0.047* | |
| H15B | 0.2358 | −0.3705 | 0.6608 | 0.047* | |
| C16 | 0.1931 (3) | −0.2794 (2) | 0.5030 (2) | 0.0347 (7) | |
| H16 | 0.1501 | −0.2226 | 0.5190 | 0.042* | |
| C17 | 0.2657 (2) | −0.2223 (2) | 0.3826 (2) | 0.0278 (6) | |
| H17A | 0.3270 | −0.1635 | 0.3719 | 0.033* | |
| H17B | 0.2082 | −0.1866 | 0.3306 | 0.033* | |
| C18 | 0.0973 (2) | −0.3689 (2) | 0.5174 (3) | 0.0347 (7) | |
| H18A | 0.0396 | −0.3336 | 0.4655 | 0.042* | |
| H18B | 0.0487 | −0.4053 | 0.5943 | 0.042* | |
| C19 | 0.4204 (2) | −0.3648 (2) | 0.4400 (2) | 0.0246 (6) | |
| H19A | 0.4636 | −0.4210 | 0.4247 | 0.029* | |
| H19B | 0.4836 | −0.3079 | 0.4303 | 0.029* | |
| C20 | 0.9080 (2) | 0.4100 (2) | 0.1270 (2) | 0.0304 (7) | |
| H20A | 0.9542 | 0.4734 | 0.1230 | 0.046* | |
| H20B | 0.9610 | 0.3784 | 0.0866 | 0.046* | |
| H20C | 0.8822 | 0.3527 | 0.2050 | 0.046* | |
| S2 | −0.03030 (6) | −0.00170 (5) | 0.16571 (5) | 0.02023 (16) | |
| O3 | 0.01738 (16) | 0.16742 (15) | 0.20276 (15) | 0.0245 (4) | |
| O4 | −0.15109 (16) | −0.34505 (15) | 0.11643 (15) | 0.0272 (4) | |
| N3 | 0.19426 (18) | −0.04250 (17) | 0.10519 (16) | 0.0190 (5) | |
| N4 | 0.18972 (19) | 0.11000 (18) | 0.13802 (18) | 0.0205 (5) | |
| H4N | 0.262 (2) | 0.122 (2) | 0.118 (2) | 0.025* | |
| C21 | 0.1303 (2) | 0.0230 (2) | 0.1335 (2) | 0.0173 (5) | |
| C22 | 0.1119 (2) | −0.1226 (2) | 0.1075 (2) | 0.0191 (6) | |
| C23 | 0.1451 (2) | −0.2080 (2) | 0.0829 (2) | 0.0230 (6) | |
| H23 | 0.2297 | −0.2164 | 0.0625 | 0.028* | |
| C24 | 0.0539 (2) | −0.2798 (2) | 0.0887 (2) | 0.0225 (6) | |
| H24 | 0.0763 | −0.3388 | 0.0733 | 0.027* | |
| C25 | −0.0717 (2) | −0.2682 (2) | 0.1169 (2) | 0.0234 (6) | |
| C26 | −0.1078 (2) | −0.1851 (2) | 0.1433 (2) | 0.0210 (6) | |
| H26 | −0.1926 | −0.1775 | 0.1641 | 0.025* | |
| C27 | −0.0139 (2) | −0.1133 (2) | 0.1380 (2) | 0.0187 (5) | |
| C28 | 0.1302 (2) | 0.1771 (2) | 0.1762 (2) | 0.0203 (6) | |
| C29 | 0.2130 (2) | 0.2567 (2) | 0.1874 (2) | 0.0225 (6) | |
| H29A | 0.1748 | 0.3282 | 0.1650 | 0.027* | |
| H29B | 0.2932 | 0.2721 | 0.1360 | 0.027* | |
| C30 | 0.2369 (2) | 0.2112 (2) | 0.3078 (2) | 0.0214 (6) | |
| C31 | 0.3265 (2) | 0.2979 (2) | 0.3073 (2) | 0.0269 (6) | |
| H31A | 0.4045 | 0.3099 | 0.2544 | 0.032* | |
| H31B | 0.2892 | 0.3701 | 0.2825 | 0.032* | |
| C32 | 0.3540 (3) | 0.2569 (3) | 0.4246 (2) | 0.0321 (7) | |
| H32 | 0.4122 | 0.3137 | 0.4233 | 0.039* | |
| C33 | 0.2330 (3) | 0.2398 (3) | 0.5059 (2) | 0.0352 (7) | |
| H33A | 0.2503 | 0.2138 | 0.5815 | 0.042* | |
| H33B | 0.1951 | 0.3115 | 0.4826 | 0.042* | |
| C34 | 0.1442 (3) | 0.1534 (2) | 0.5075 (2) | 0.0317 (7) | |
| H34 | 0.0653 | 0.1424 | 0.5605 | 0.038* | |
| C35 | 0.2028 (3) | 0.0416 (2) | 0.5449 (2) | 0.0343 (7) | |
| H35A | 0.2189 | 0.0135 | 0.6211 | 0.041* | |
| H35B | 0.1453 | −0.0145 | 0.5458 | 0.041* | |
| C36 | 0.3239 (3) | 0.0588 (2) | 0.4647 (2) | 0.0298 (7) | |
| H36 | 0.3627 | −0.0135 | 0.4893 | 0.036* | |
| C37 | 0.2961 (2) | 0.0997 (2) | 0.3474 (2) | 0.0248 (6) | |
| H37A | 0.3739 | 0.1095 | 0.2949 | 0.030* | |
| H37B | 0.2392 | 0.0430 | 0.3486 | 0.030* | |
| C38 | 0.4127 (2) | 0.1466 (2) | 0.4617 (2) | 0.0312 (7) | |
| H38A | 0.4319 | 0.1205 | 0.5366 | 0.037* | |
| H38B | 0.4910 | 0.1575 | 0.4095 | 0.037* | |
| C39 | 0.1164 (2) | 0.1944 (2) | 0.3902 (2) | 0.0265 (6) | |
| H39A | 0.0577 | 0.1388 | 0.3916 | 0.032* | |
| H39B | 0.0776 | 0.2658 | 0.3659 | 0.032* | |
| C40 | −0.2808 (2) | −0.3360 (2) | 0.1403 (2) | 0.0265 (6) | |
| H40A | −0.3270 | −0.3987 | 0.1428 | 0.040* | |
| H40B | −0.3021 | −0.2655 | 0.0819 | 0.040* | |
| H40C | −0.3023 | −0.3377 | 0.2123 | 0.040* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0177 (4) | 0.0212 (4) | 0.0290 (4) | 0.0032 (3) | −0.0085 (3) | −0.0108 (3) |
| O1 | 0.0208 (11) | 0.0255 (11) | 0.0340 (11) | 0.0054 (8) | −0.0092 (8) | −0.0119 (9) |
| O2 | 0.0232 (11) | 0.0238 (11) | 0.0607 (14) | 0.0043 (8) | −0.0137 (10) | −0.0241 (11) |
| N1 | 0.0162 (12) | 0.0213 (12) | 0.0255 (12) | 0.0040 (9) | −0.0062 (9) | −0.0108 (10) |
| N2 | 0.0159 (12) | 0.0215 (12) | 0.0275 (13) | 0.0032 (9) | −0.0069 (10) | −0.0123 (11) |
| C1 | 0.0142 (13) | 0.0246 (15) | 0.0218 (14) | 0.0024 (11) | −0.0028 (10) | −0.0117 (12) |
| C2 | 0.0166 (14) | 0.0218 (14) | 0.0252 (15) | 0.0002 (11) | 0.0006 (11) | −0.0131 (12) |
| C3 | 0.0175 (14) | 0.0235 (15) | 0.0343 (16) | 0.0065 (11) | −0.0065 (12) | −0.0128 (13) |
| C4 | 0.0239 (15) | 0.0207 (15) | 0.0415 (18) | 0.0052 (12) | −0.0044 (13) | −0.0145 (14) |
| C5 | 0.0189 (15) | 0.0258 (16) | 0.0366 (17) | 0.0008 (11) | −0.0022 (12) | −0.0202 (14) |
| C6 | 0.0187 (14) | 0.0245 (15) | 0.0287 (15) | 0.0029 (11) | −0.0044 (11) | −0.0142 (13) |
| C7 | 0.0166 (14) | 0.0222 (14) | 0.0253 (15) | 0.0035 (11) | −0.0016 (11) | −0.0123 (12) |
| C8 | 0.0205 (15) | 0.0215 (14) | 0.0231 (14) | 0.0056 (11) | −0.0062 (11) | −0.0129 (12) |
| C9 | 0.0231 (15) | 0.0223 (15) | 0.0323 (16) | 0.0058 (11) | −0.0095 (12) | −0.0177 (13) |
| C10 | 0.0185 (14) | 0.0204 (14) | 0.0288 (15) | 0.0031 (11) | −0.0057 (11) | −0.0130 (12) |
| C11 | 0.0226 (15) | 0.0213 (15) | 0.0371 (17) | 0.0031 (11) | −0.0122 (12) | −0.0152 (13) |
| C12 | 0.0190 (15) | 0.0248 (16) | 0.0458 (18) | −0.0010 (11) | −0.0070 (13) | −0.0158 (14) |
| C13 | 0.0255 (16) | 0.0283 (17) | 0.0342 (17) | 0.0001 (12) | −0.0035 (13) | −0.0094 (14) |
| C14 | 0.0241 (16) | 0.0334 (17) | 0.0263 (16) | 0.0020 (12) | −0.0088 (12) | −0.0094 (14) |
| C15 | 0.0351 (19) | 0.053 (2) | 0.0327 (17) | −0.0077 (15) | 0.0019 (14) | −0.0264 (16) |
| C16 | 0.0288 (17) | 0.0324 (17) | 0.0457 (19) | 0.0024 (13) | 0.0027 (14) | −0.0241 (16) |
| C17 | 0.0210 (15) | 0.0239 (15) | 0.0426 (18) | 0.0030 (12) | −0.0054 (12) | −0.0196 (14) |
| C18 | 0.0191 (16) | 0.0361 (18) | 0.0429 (19) | 0.0003 (13) | 0.0001 (13) | −0.0163 (15) |
| C19 | 0.0207 (15) | 0.0254 (15) | 0.0297 (16) | 0.0035 (11) | −0.0104 (12) | −0.0132 (13) |
| C20 | 0.0215 (16) | 0.0326 (17) | 0.0452 (18) | 0.0015 (12) | −0.0085 (13) | −0.0244 (15) |
| S2 | 0.0164 (4) | 0.0229 (4) | 0.0257 (4) | 0.0029 (3) | −0.0058 (3) | −0.0146 (3) |
| O3 | 0.0173 (10) | 0.0285 (11) | 0.0343 (11) | 0.0055 (8) | −0.0073 (8) | −0.0196 (9) |
| O4 | 0.0214 (11) | 0.0266 (11) | 0.0414 (12) | −0.0012 (8) | −0.0064 (8) | −0.0224 (10) |
| N3 | 0.0173 (12) | 0.0215 (12) | 0.0200 (12) | 0.0045 (9) | −0.0068 (9) | −0.0105 (10) |
| N4 | 0.0137 (11) | 0.0224 (12) | 0.0279 (13) | 0.0015 (10) | −0.0055 (10) | −0.0138 (11) |
| C21 | 0.0190 (14) | 0.0183 (14) | 0.0178 (13) | 0.0048 (10) | −0.0089 (10) | −0.0095 (11) |
| C22 | 0.0196 (14) | 0.0189 (14) | 0.0184 (13) | 0.0017 (11) | −0.0077 (11) | −0.0073 (11) |
| C23 | 0.0174 (14) | 0.0311 (16) | 0.0245 (15) | 0.0078 (11) | −0.0066 (11) | −0.0158 (13) |
| C24 | 0.0254 (15) | 0.0228 (14) | 0.0268 (15) | 0.0046 (11) | −0.0065 (11) | −0.0174 (13) |
| C25 | 0.0221 (15) | 0.0226 (15) | 0.0245 (15) | −0.0008 (11) | −0.0067 (11) | −0.0100 (12) |
| C26 | 0.0143 (14) | 0.0234 (15) | 0.0231 (14) | 0.0033 (11) | −0.0031 (11) | −0.0097 (12) |
| C27 | 0.0210 (14) | 0.0172 (13) | 0.0206 (14) | 0.0032 (10) | −0.0074 (11) | −0.0103 (11) |
| C28 | 0.0203 (15) | 0.0190 (14) | 0.0217 (14) | 0.0057 (11) | −0.0055 (11) | −0.0094 (12) |
| C29 | 0.0215 (15) | 0.0184 (14) | 0.0286 (15) | 0.0025 (11) | −0.0070 (11) | −0.0114 (12) |
| C30 | 0.0156 (14) | 0.0206 (14) | 0.0306 (15) | 0.0036 (10) | −0.0078 (11) | −0.0133 (12) |
| C31 | 0.0230 (15) | 0.0275 (16) | 0.0367 (17) | 0.0024 (12) | −0.0107 (12) | −0.0192 (14) |
| C32 | 0.0266 (17) | 0.0415 (18) | 0.0403 (18) | 0.0022 (13) | −0.0118 (13) | −0.0276 (16) |
| C33 | 0.0354 (18) | 0.0459 (19) | 0.0395 (18) | 0.0121 (14) | −0.0155 (14) | −0.0300 (16) |
| C34 | 0.0227 (16) | 0.0449 (19) | 0.0338 (17) | 0.0048 (13) | −0.0020 (12) | −0.0250 (15) |
| C35 | 0.0372 (19) | 0.0398 (19) | 0.0252 (16) | −0.0032 (14) | −0.0090 (13) | −0.0142 (14) |
| C36 | 0.0305 (17) | 0.0292 (16) | 0.0323 (16) | 0.0104 (13) | −0.0127 (13) | −0.0149 (14) |
| C37 | 0.0223 (15) | 0.0266 (15) | 0.0301 (16) | 0.0054 (12) | −0.0066 (12) | −0.0167 (13) |
| C38 | 0.0213 (16) | 0.0486 (19) | 0.0311 (16) | 0.0079 (13) | −0.0122 (12) | −0.0227 (15) |
| C39 | 0.0197 (15) | 0.0333 (16) | 0.0334 (16) | 0.0050 (12) | −0.0070 (12) | −0.0211 (14) |
| C40 | 0.0205 (15) | 0.0250 (15) | 0.0344 (16) | −0.0008 (11) | −0.0077 (12) | −0.0139 (13) |
Geometric parameters (Å, º)
| S1—C7 | 1.739 (3) | S2—C21 | 1.742 (2) |
| S1—C1 | 1.756 (2) | S2—C27 | 1.743 (3) |
| O1—C8 | 1.225 (3) | O3—C28 | 1.223 (3) |
| O2—C5 | 1.368 (3) | O4—C25 | 1.372 (3) |
| O2—C20 | 1.436 (3) | O4—C40 | 1.429 (3) |
| N1—C1 | 1.299 (3) | N3—C21 | 1.300 (3) |
| N1—C2 | 1.401 (3) | N3—C22 | 1.409 (3) |
| N2—C8 | 1.374 (3) | N4—C28 | 1.376 (3) |
| N2—C1 | 1.385 (3) | N4—C21 | 1.391 (3) |
| N2—H2N | 0.87 (3) | N4—H4N | 0.78 (3) |
| C2—C7 | 1.397 (3) | C22—C23 | 1.396 (3) |
| C2—C3 | 1.399 (3) | C22—C27 | 1.403 (3) |
| C3—C4 | 1.381 (4) | C23—C24 | 1.372 (3) |
| C3—H3 | 0.9500 | C23—H23 | 0.9500 |
| C4—C5 | 1.404 (4) | C24—C25 | 1.401 (3) |
| C4—H4 | 0.9500 | C24—H24 | 0.9500 |
| C5—C6 | 1.389 (4) | C25—C26 | 1.389 (4) |
| C6—C7 | 1.398 (3) | C26—C27 | 1.397 (3) |
| C6—H6 | 0.9500 | C26—H26 | 0.9500 |
| C8—C9 | 1.506 (3) | C28—C29 | 1.502 (3) |
| C9—C10 | 1.542 (4) | C29—C30 | 1.556 (3) |
| C9—H9A | 0.9900 | C29—H29A | 0.9900 |
| C9—H9B | 0.9900 | C29—H29B | 0.9900 |
| C10—C11 | 1.537 (3) | C30—C39 | 1.538 (3) |
| C10—C17 | 1.539 (3) | C30—C37 | 1.539 (3) |
| C10—C19 | 1.543 (3) | C30—C31 | 1.540 (3) |
| C11—C12 | 1.530 (4) | C31—C32 | 1.539 (4) |
| C11—H11A | 0.9900 | C31—H31A | 0.9900 |
| C11—H11B | 0.9900 | C31—H31B | 0.9900 |
| C12—C13 | 1.532 (4) | C32—C38 | 1.523 (4) |
| C12—C18 | 1.534 (4) | C32—C33 | 1.533 (4) |
| C12—H12 | 1.0000 | C32—H32 | 1.0000 |
| C13—C14 | 1.534 (4) | C33—C34 | 1.526 (4) |
| C13—H13A | 0.9900 | C33—H33A | 0.9900 |
| C13—H13B | 0.9900 | C33—H33B | 0.9900 |
| C14—C15 | 1.528 (4) | C34—C35 | 1.540 (4) |
| C14—C19 | 1.528 (4) | C34—C39 | 1.540 (4) |
| C14—H14 | 1.0000 | C34—H34 | 1.0000 |
| C15—C16 | 1.529 (4) | C35—C36 | 1.526 (4) |
| C15—H15A | 0.9900 | C35—H35A | 0.9900 |
| C15—H15B | 0.9900 | C35—H35B | 0.9900 |
| C16—C18 | 1.534 (4) | C36—C38 | 1.532 (4) |
| C16—C17 | 1.538 (4) | C36—C37 | 1.540 (4) |
| C16—H16 | 1.0000 | C36—H36 | 1.0000 |
| C17—H17A | 0.9900 | C37—H37A | 0.9900 |
| C17—H17B | 0.9900 | C37—H37B | 0.9900 |
| C18—H18A | 0.9900 | C38—H38A | 0.9900 |
| C18—H18B | 0.9900 | C38—H38B | 0.9900 |
| C19—H19A | 0.9900 | C39—H39A | 0.9900 |
| C19—H19B | 0.9900 | C39—H39B | 0.9900 |
| C20—H20A | 0.9800 | C40—H40A | 0.9800 |
| C20—H20B | 0.9800 | C40—H40B | 0.9800 |
| C20—H20C | 0.9800 | C40—H40C | 0.9800 |
| C7—S1—C1 | 87.98 (12) | C21—S2—C27 | 88.08 (12) |
| C5—O2—C20 | 117.2 (2) | C25—O4—C40 | 117.6 (2) |
| C1—N1—C2 | 109.1 (2) | C21—N3—C22 | 108.6 (2) |
| C8—N2—C1 | 123.1 (2) | C28—N4—C21 | 124.1 (2) |
| C8—N2—H2N | 116.6 (17) | C28—N4—H4N | 117 (2) |
| C1—N2—H2N | 119.9 (17) | C21—N4—H4N | 119 (2) |
| N1—C1—N2 | 122.0 (2) | N3—C21—N4 | 120.3 (2) |
| N1—C1—S1 | 117.39 (19) | N3—C21—S2 | 118.23 (19) |
| N2—C1—S1 | 120.53 (19) | N4—C21—S2 | 121.46 (19) |
| C7—C2—C3 | 118.8 (2) | C23—C22—C27 | 118.9 (2) |
| C7—C2—N1 | 115.4 (2) | C23—C22—N3 | 125.9 (2) |
| C3—C2—N1 | 125.7 (2) | C27—C22—N3 | 115.3 (2) |
| C4—C3—C2 | 119.2 (2) | C24—C23—C22 | 119.1 (2) |
| C4—C3—H3 | 120.4 | C24—C23—H23 | 120.4 |
| C2—C3—H3 | 120.4 | C22—C23—H23 | 120.4 |
| C3—C4—C5 | 121.2 (3) | C23—C24—C25 | 121.6 (2) |
| C3—C4—H4 | 119.4 | C23—C24—H24 | 119.2 |
| C5—C4—H4 | 119.4 | C25—C24—H24 | 119.2 |
| O2—C5—C6 | 124.1 (2) | O4—C25—C26 | 124.9 (2) |
| O2—C5—C4 | 115.1 (2) | O4—C25—C24 | 114.3 (2) |
| C6—C5—C4 | 120.8 (2) | C26—C25—C24 | 120.8 (2) |
| C5—C6—C7 | 117.1 (2) | C25—C26—C27 | 117.0 (2) |
| C5—C6—H6 | 121.4 | C25—C26—H26 | 121.5 |
| C7—C6—H6 | 121.4 | C27—C26—H26 | 121.5 |
| C2—C7—C6 | 122.9 (2) | C26—C27—C22 | 122.7 (2) |
| C2—C7—S1 | 110.02 (19) | C26—C27—S2 | 127.5 (2) |
| C6—C7—S1 | 126.9 (2) | C22—C27—S2 | 109.81 (18) |
| O1—C8—N2 | 121.3 (2) | O3—C28—N4 | 120.8 (2) |
| O1—C8—C9 | 123.1 (2) | O3—C28—C29 | 123.6 (2) |
| N2—C8—C9 | 115.5 (2) | N4—C28—C29 | 115.6 (2) |
| C8—C9—C10 | 112.4 (2) | C28—C29—C30 | 113.0 (2) |
| C8—C9—H9A | 109.1 | C28—C29—H29A | 109.0 |
| C10—C9—H9A | 109.1 | C30—C29—H29A | 109.0 |
| C8—C9—H9B | 109.1 | C28—C29—H29B | 109.0 |
| C10—C9—H9B | 109.1 | C30—C29—H29B | 109.0 |
| H9A—C9—H9B | 107.9 | H29A—C29—H29B | 107.8 |
| C11—C10—C17 | 108.7 (2) | C39—C30—C37 | 108.9 (2) |
| C11—C10—C9 | 108.1 (2) | C39—C30—C31 | 108.6 (2) |
| C17—C10—C9 | 111.7 (2) | C37—C30—C31 | 108.8 (2) |
| C11—C10—C19 | 108.2 (2) | C39—C30—C29 | 111.3 (2) |
| C17—C10—C19 | 108.9 (2) | C37—C30—C29 | 111.3 (2) |
| C9—C10—C19 | 111.2 (2) | C31—C30—C29 | 107.9 (2) |
| C12—C11—C10 | 110.6 (2) | C32—C31—C30 | 110.2 (2) |
| C12—C11—H11A | 109.5 | C32—C31—H31A | 109.6 |
| C10—C11—H11A | 109.5 | C30—C31—H31A | 109.6 |
| C12—C11—H11B | 109.5 | C32—C31—H31B | 109.6 |
| C10—C11—H11B | 109.5 | C30—C31—H31B | 109.6 |
| H11A—C11—H11B | 108.1 | H31A—C31—H31B | 108.1 |
| C11—C12—C13 | 110.0 (2) | C38—C32—C33 | 109.5 (2) |
| C11—C12—C18 | 109.1 (2) | C38—C32—C31 | 109.3 (2) |
| C13—C12—C18 | 109.1 (2) | C33—C32—C31 | 109.4 (2) |
| C11—C12—H12 | 109.6 | C38—C32—H32 | 109.5 |
| C13—C12—H12 | 109.6 | C33—C32—H32 | 109.5 |
| C18—C12—H12 | 109.6 | C31—C32—H32 | 109.5 |
| C12—C13—C14 | 109.3 (2) | C34—C33—C32 | 109.4 (2) |
| C12—C13—H13A | 109.8 | C34—C33—H33A | 109.8 |
| C14—C13—H13A | 109.8 | C32—C33—H33A | 109.8 |
| C12—C13—H13B | 109.8 | C34—C33—H33B | 109.8 |
| C14—C13—H13B | 109.8 | C32—C33—H33B | 109.8 |
| H13A—C13—H13B | 108.3 | H33A—C33—H33B | 108.2 |
| C15—C14—C19 | 109.4 (2) | C33—C34—C35 | 109.7 (2) |
| C15—C14—C13 | 109.2 (2) | C33—C34—C39 | 109.8 (2) |
| C19—C14—C13 | 109.8 (2) | C35—C34—C39 | 109.2 (2) |
| C15—C14—H14 | 109.5 | C33—C34—H34 | 109.4 |
| C19—C14—H14 | 109.5 | C35—C34—H34 | 109.4 |
| C13—C14—H14 | 109.5 | C39—C34—H34 | 109.4 |
| C14—C15—C16 | 109.9 (2) | C36—C35—C34 | 109.4 (2) |
| C14—C15—H15A | 109.7 | C36—C35—H35A | 109.8 |
| C16—C15—H15A | 109.7 | C34—C35—H35A | 109.8 |
| C14—C15—H15B | 109.7 | C36—C35—H35B | 109.8 |
| C16—C15—H15B | 109.7 | C34—C35—H35B | 109.8 |
| H15A—C15—H15B | 108.2 | H35A—C35—H35B | 108.3 |
| C15—C16—C18 | 109.5 (2) | C35—C36—C38 | 110.1 (2) |
| C15—C16—C17 | 109.5 (2) | C35—C36—C37 | 108.9 (2) |
| C18—C16—C17 | 108.8 (2) | C38—C36—C37 | 109.1 (2) |
| C15—C16—H16 | 109.7 | C35—C36—H36 | 109.6 |
| C18—C16—H16 | 109.7 | C38—C36—H36 | 109.6 |
| C17—C16—H16 | 109.7 | C37—C36—H36 | 109.6 |
| C16—C17—C10 | 110.2 (2) | C30—C37—C36 | 110.3 (2) |
| C16—C17—H17A | 109.6 | C30—C37—H37A | 109.6 |
| C10—C17—H17A | 109.6 | C36—C37—H37A | 109.6 |
| C16—C17—H17B | 109.6 | C30—C37—H37B | 109.6 |
| C10—C17—H17B | 109.6 | C36—C37—H37B | 109.6 |
| H17A—C17—H17B | 108.1 | H37A—C37—H37B | 108.1 |
| C16—C18—C12 | 109.8 (2) | C32—C38—C36 | 109.9 (2) |
| C16—C18—H18A | 109.7 | C32—C38—H38A | 109.7 |
| C12—C18—H18A | 109.7 | C36—C38—H38A | 109.7 |
| C16—C18—H18B | 109.7 | C32—C38—H38B | 109.7 |
| C12—C18—H18B | 109.7 | C36—C38—H38B | 109.7 |
| H18A—C18—H18B | 108.2 | H38A—C38—H38B | 108.2 |
| C14—C19—C10 | 110.4 (2) | C30—C39—C34 | 109.9 (2) |
| C14—C19—H19A | 109.6 | C30—C39—H39A | 109.7 |
| C10—C19—H19A | 109.6 | C34—C39—H39A | 109.7 |
| C14—C19—H19B | 109.6 | C30—C39—H39B | 109.7 |
| C10—C19—H19B | 109.6 | C34—C39—H39B | 109.7 |
| H19A—C19—H19B | 108.1 | H39A—C39—H39B | 108.2 |
| O2—C20—H20A | 109.5 | O4—C40—H40A | 109.5 |
| O2—C20—H20B | 109.5 | O4—C40—H40B | 109.5 |
| H20A—C20—H20B | 109.5 | H40A—C40—H40B | 109.5 |
| O2—C20—H20C | 109.5 | O4—C40—H40C | 109.5 |
| H20A—C20—H20C | 109.5 | H40A—C40—H40C | 109.5 |
| H20B—C20—H20C | 109.5 | H40B—C40—H40C | 109.5 |
| C2—N1—C1—N2 | 178.0 (2) | C22—N3—C21—N4 | 179.9 (2) |
| C2—N1—C1—S1 | 0.4 (3) | C22—N3—C21—S2 | 0.2 (3) |
| C8—N2—C1—N1 | 179.2 (2) | C28—N4—C21—N3 | −174.2 (2) |
| C8—N2—C1—S1 | −3.2 (3) | C28—N4—C21—S2 | 5.6 (3) |
| C7—S1—C1—N1 | 0.8 (2) | C27—S2—C21—N3 | −0.4 (2) |
| C7—S1—C1—N2 | −176.8 (2) | C27—S2—C21—N4 | 179.9 (2) |
| C1—N1—C2—C7 | −1.9 (3) | C21—N3—C22—C23 | −179.6 (2) |
| C1—N1—C2—C3 | 175.1 (2) | C21—N3—C22—C27 | 0.2 (3) |
| C7—C2—C3—C4 | −0.7 (4) | C27—C22—C23—C24 | 0.4 (4) |
| N1—C2—C3—C4 | −177.7 (2) | N3—C22—C23—C24 | −179.9 (2) |
| C2—C3—C4—C5 | 0.5 (4) | C22—C23—C24—C25 | 1.1 (4) |
| C20—O2—C5—C6 | 7.2 (4) | C40—O4—C25—C26 | 2.8 (4) |
| C20—O2—C5—C4 | −171.8 (2) | C40—O4—C25—C24 | −177.7 (2) |
| C3—C4—C5—O2 | 179.1 (2) | C23—C24—C25—O4 | 178.5 (2) |
| C3—C4—C5—C6 | 0.2 (4) | C23—C24—C25—C26 | −2.0 (4) |
| O2—C5—C6—C7 | −179.4 (2) | O4—C25—C26—C27 | −179.2 (2) |
| C4—C5—C6—C7 | −0.6 (4) | C24—C25—C26—C27 | 1.3 (4) |
| C3—C2—C7—C6 | 0.3 (4) | C25—C26—C27—C22 | 0.1 (4) |
| N1—C2—C7—C6 | 177.6 (2) | C25—C26—C27—S2 | 179.76 (19) |
| C3—C2—C7—S1 | −174.76 (19) | C23—C22—C27—C26 | −1.0 (4) |
| N1—C2—C7—S1 | 2.5 (3) | N3—C22—C27—C26 | 179.3 (2) |
| C5—C6—C7—C2 | 0.4 (4) | C23—C22—C27—S2 | 179.33 (19) |
| C5—C6—C7—S1 | 174.5 (2) | N3—C22—C27—S2 | −0.4 (3) |
| C1—S1—C7—C2 | −1.78 (19) | C21—S2—C27—C26 | −179.3 (2) |
| C1—S1—C7—C6 | −176.6 (2) | C21—S2—C27—C22 | 0.41 (19) |
| C1—N2—C8—O1 | −7.3 (4) | C21—N4—C28—O3 | −4.5 (4) |
| C1—N2—C8—C9 | 168.9 (2) | C21—N4—C28—C29 | 172.7 (2) |
| O1—C8—C9—C10 | 75.8 (3) | O3—C28—C29—C30 | 80.6 (3) |
| N2—C8—C9—C10 | −100.3 (3) | N4—C28—C29—C30 | −96.5 (3) |
| C8—C9—C10—C11 | 174.6 (2) | C28—C29—C30—C39 | −63.1 (3) |
| C8—C9—C10—C17 | 55.1 (3) | C28—C29—C30—C37 | 58.5 (3) |
| C8—C9—C10—C19 | −66.8 (3) | C28—C29—C30—C31 | 177.8 (2) |
| C17—C10—C11—C12 | −59.1 (3) | C39—C30—C31—C32 | 59.5 (3) |
| C9—C10—C11—C12 | 179.5 (2) | C37—C30—C31—C32 | −58.9 (3) |
| C19—C10—C11—C12 | 59.0 (3) | C29—C30—C31—C32 | −179.7 (2) |
| C10—C11—C12—C13 | −59.8 (3) | C30—C31—C32—C38 | 60.0 (3) |
| C10—C11—C12—C18 | 59.7 (3) | C30—C31—C32—C33 | −60.0 (3) |
| C11—C12—C13—C14 | 59.0 (3) | C38—C32—C33—C34 | −60.1 (3) |
| C18—C12—C13—C14 | −60.6 (3) | C31—C32—C33—C34 | 59.7 (3) |
| C12—C13—C14—C15 | 60.8 (3) | C32—C33—C34—C35 | 60.0 (3) |
| C12—C13—C14—C19 | −59.2 (3) | C32—C33—C34—C39 | −60.0 (3) |
| C19—C14—C15—C16 | 60.0 (3) | C33—C34—C35—C36 | −59.4 (3) |
| C13—C14—C15—C16 | −60.2 (3) | C39—C34—C35—C36 | 61.0 (3) |
| C14—C15—C16—C18 | 59.4 (3) | C34—C35—C36—C38 | 58.7 (3) |
| C14—C15—C16—C17 | −59.8 (3) | C34—C35—C36—C37 | −60.9 (3) |
| C15—C16—C17—C10 | 59.4 (3) | C39—C30—C37—C36 | −59.3 (3) |
| C18—C16—C17—C10 | −60.2 (3) | C31—C30—C37—C36 | 58.9 (3) |
| C11—C10—C17—C16 | 59.2 (3) | C29—C30—C37—C36 | 177.7 (2) |
| C9—C10—C17—C16 | 178.4 (2) | C35—C36—C37—C30 | 60.5 (3) |
| C19—C10—C17—C16 | −58.4 (3) | C38—C36—C37—C30 | −59.7 (3) |
| C15—C16—C18—C12 | −59.2 (3) | C33—C32—C38—C36 | 59.6 (3) |
| C17—C16—C18—C12 | 60.4 (3) | C31—C32—C38—C36 | −60.3 (3) |
| C11—C12—C18—C16 | −60.2 (3) | C35—C36—C38—C32 | −59.3 (3) |
| C13—C12—C18—C16 | 59.9 (3) | C37—C36—C38—C32 | 60.1 (3) |
| C15—C14—C19—C10 | −59.7 (3) | C37—C30—C39—C34 | 59.0 (3) |
| C13—C14—C19—C10 | 60.1 (3) | C31—C30—C39—C34 | −59.3 (3) |
| C11—C10—C19—C14 | −59.2 (3) | C29—C30—C39—C34 | −178.0 (2) |
| C17—C10—C19—C14 | 58.8 (3) | C33—C34—C39—C30 | 60.2 (3) |
| C9—C10—C19—C14 | −177.7 (2) | C35—C34—C39—C30 | −60.1 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2N···N3 | 0.87 (3) | 2.13 (3) | 2.995 (3) | 169 (2) |
| N4—H4N···N1 | 0.78 (3) | 2.30 (3) | 3.077 (3) | 174 (2) |
| C6—H6···O3i | 0.95 | 2.58 | 3.452 (3) | 153 |
| C26—H26···O1ii | 0.95 | 2.45 | 3.392 (3) | 174 |
Symmetry codes: (i) x+1, y, z; (ii) x−1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: RK2412).
References
- Bhardwaj, V. K., Saluja, P., Hundal, G., Hundal, M. S., Singh, N. & Jang, D. O. (2013). Tetrahedron, 69, 1606–1610.
- Bruker (2001). SAINT . Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2003). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2005). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Facchinetti, V., da Reis, R., Gomes, C. R. B. & Vasconcelos, T. R. A. (2012). Mini-Rev. Org. Chem. 9, 44–53. [DOI] [PubMed]
- Gupta, A. & Rawat, S. (2010). J. Curr. Pharm. Res. 3, 13–23.
- Hussein, B. H. M., Azab, H. A., El-Azab, M. F. & El-Falouji, A. I. (2012). Eur. J. Med. Chem. 51, 99–109. [DOI] [PubMed]
- Patel, N. B., Khan, I. H., Pannecouque, C. & De Clercq, E. (2013). Med. Chem. Res. 22, 1320–1329.
- Radatz, C. S., Alves, D. & Schneider, P. H. (2013). Tetrahedron, 69, 1316–1321.
- Sareen, S., Shinde, D., Khatri, V. & Sareen, V. (2012). Heterocycl. Lett. 2, 361–377.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Ugale, V. G., Patel, H. M., Wadodkar, S. G., Bari, S. B., Shirkhedkar, A. A. & Surana, S. J. (2012). Eur. J. Med. Chem. 53, 107–113. [DOI] [PubMed]
- Yoo, E., Hayat, F., Rhim, H. & Choo, H. P. (2012). Bioorg. Med. Chem. 20, 2707–2712. [DOI] [PubMed]
- Zhu, X. Y., Etukala, J. R., Eyunni, S. V. K., Setola, V., Roth, B. L. & Ablordeppey, S. Y. (2012). Eur. J. Med. Chem. 53, 124–132. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813023313/rk2412sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813023313/rk2412Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813023313/rk2412Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




