Abstract
In the title structure, C8H6BrNS, the molecules are planar with the exception of the methyl H atoms. In the crystal, molecules are linked by intermolecular C—H⋯N interactions to form ribbons parallel to the b axis. Groups of ribbons are arranged in a herringbone pattern to form a layered structure parallel to the ab plane.
Related literature
For related structures and their applications, see: Perner et al. (2003 ▶); Kose (2004 ▶); Chandra et al. (2006 ▶); Zhao et al. (2009 ▶); Pu et al. (2010 ▶); Dinçalp et al. (2011 ▶).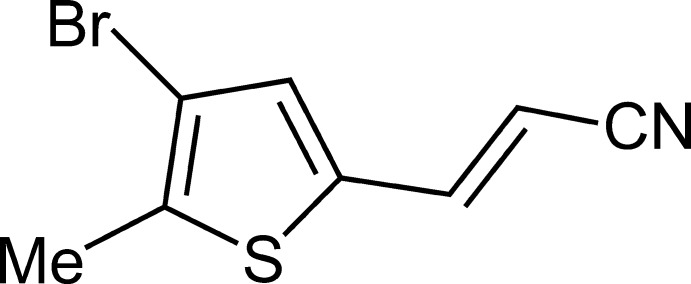
Experimental
Crystal data
C8H6BrNS
M r = 228.11
Orthorhombic,

a = 6.1347 (5) Å
b = 7.1124 (3) Å
c = 19.8245 (13) Å
V = 864.99 (10) Å3
Z = 4
Mo Kα radiation
μ = 4.92 mm−1
T = 150 K
0.40 × 0.30 × 0.10 mm
Data collection
Nonius KappaCCD diffractometer
Absorption correction: empirical (using intensity measurements) (DENZO/SCALEPACK; Otwinowski & Minor, 1997 ▶) T min = 0.243, T max = 0.639
3294 measured reflections
1910 independent reflections
1769 reflections with I > 2σ(I)
R int = 0.060
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.120
S = 1.05
1910 reflections
102 parameters
H-atom parameters constrained
Δρmax = 0.74 e Å−3
Δρmin = −1.12 e Å−3
Absolute structure: Flack (1983 ▶), 699 Friedel pairs
Absolute structure parameter: 0.03 (2)
Data collection: COLLECT (Nonius, 2000 ▶); cell refinement: DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: DENZO and SCALEPACK; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶) and CHEMDRAW Ultra (Cambridge Soft, 2001 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536813019752/hg5330sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813019752/hg5330Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813019752/hg5330Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C3—H3⋯N1i | 0.93 | 2.59 | 3.501 (8) | 166 |
Symmetry code: (i) 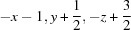 .
.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for its funding for this research through Research Group Project No. RGP-VPP-239.
supplementary crystallographic information
1. Comment
During the research focused on new synthetic routes towards novel substituted thiophene derivatives, we have synthesized the title compound (I), which was isolated in high yield. Thiophene derivatives are interesting compounds (Zhao et al., 2009). They can be used in a wide range of applications such as enzyme inhibitors (Perner et al., 2003), photochromic materials (Kose, 2004; Pu et al., 2010), bioprobes (Chandra et al., 2006) and dyes (Dinçalp et al., 2011).
In the structure, the molecules of (E)-3-(4-bromo-5-methylthiophen-2-yl)-acrylonitrile (I) are planar, except for H atoms of the methyl group (Fig. 1). The molecules are linked by C—H···N interactions (Table 1) to form corrugated ribbons. The ribbons run parallel to the b axis and, within a ribbon, the orientation of consecutive molecules alternates to the left and right (Fig. 2). Groups of ribbons are arranged in a herringbone pattern to form a layered structure with layers parallel to the ab plane (Fig. 3).
2. Experimental
Synthesis ofE-3-(4-bromo-5-methylthiophen-2-yl)acrylonitrile (I)
Diethyl (cyanomethyl)phosphonate (0.94 g, 5.3 mmol) was added to sodium hydride (6.25 mmol) suspended in dry THF (50 ml) under inert atmosphere. The mixture was stirred for 1 h, 3-bromo-2-methylthiophene-5-carboxaldehyde (1.00 g, 4.90 mmol) was added and stirring was continued overnight. Saturated aqueous ammonium chloride solution (25 ml) was added and the mixture was extracted with diethyl ether (4 × 50 ml). The organic phase was washed with saturated aqueous sodium hydrogen carbonate solution (50 ml) and brine (25 ml) and dried over anhydrous magnesium sulfate. The solvent was removed under reduced pressure and the crude product was separated by column chromatography (silica gel, Et2O:hexane in 1:1 by volume) to give a mixture of E- and Z-isomers of 3-(4-bromo-5-methylthiophen-2-yl)acrylonitrile in 4:1 ratio. m.p. 80–81°C. 1H NMR (400 MHz, CDCl3, δ, p.p.m.): 7.23 (d, J = 16.3 Hz, 0.8H), 7.20 (d, J = 11.7 Hz, 0.2H), 6.98 (s, 1H), 5.46 (d, J = 16.3 Hz, 0.8H), 5.15 (d, J = 11.7 Hz, 0.2H), 2.37 (s, 0.6H), 2.35 (s, 2.4H). 13C NMR (100 MHz, CDCl3, δ, p.p.m.): 141.6 (d), 140.0 (d), 138.9 (s), 135.2 (s), 134.8 (d), 133.5 (d), 117.8 (s), 110.9 (s), 94.5 (d), 91.2 (d), 15.4 (q). EI–MS (m/z, %): 229 ([M81Br]+, 80), 227 ([M79Br]+, 78), 148 (100), 121 (10). HRMS (EI): Calculated for C8H6BrNS [M79Br]+ 226.9404; found: 226.9402. FT–IR (νmax, cm-1): 2211. Recrystallization from diethyl ether gave colorless crystals of the E-isomer (I).
3. Refinement
H atoms were positioned geometrically and refined using a riding model. For sp2 H atoms, Uiso(H) is constrained to 1.2 times the Ueq for the atoms they are bonded to and the C—H distance is 0.93 Å. For the methyl group, Uiso(H) is 1.5 times the Ueq for C atom they are bonded to and the C—H distance is 0.96 Å, with free rotation about the C—C bond.
Figures
Fig. 1.

A molecule of I showing atom labels and 50% probability displacement ellipsoids for non-H atoms.
Fig. 2.

A segment of the crystal structure showing C—H···N interactions as dashed lines.
Fig. 3.
A segment of the crystal structure of I showing the herringbone arrangement to form layers of ribbons.
Crystal data
| C8H6BrNS | F(000) = 448 |
| Mr = 228.11 | Dx = 1.752 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 1769 reflections |
| a = 6.1347 (5) Å | θ = 3.0–28.4° |
| b = 7.1124 (3) Å | µ = 4.92 mm−1 |
| c = 19.8245 (13) Å | T = 150 K |
| V = 864.99 (10) Å3 | Plate, yellow |
| Z = 4 | 0.40 × 0.30 × 0.10 mm |
Data collection
| Nonius KappaCCD diffractometer | 1910 independent reflections |
| Radiation source: fine-focus sealed tube | 1769 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.060 |
| CCD scans | θmax = 27.4°, θmin = 3.0° |
| Absorption correction: empirical (using intensity measurements) (DENZO/SCALEPACK; Otwinowski & Minor, 1997) | h = −4→7 |
| Tmin = 0.243, Tmax = 0.639 | k = −9→7 |
| 3294 measured reflections | l = −25→20 |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.048 | w = 1/[σ2(Fo2) + (0.0412P)2 + 2.3854P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.120 | (Δ/σ)max < 0.001 |
| S = 1.05 | Δρmax = 0.74 e Å−3 |
| 1910 reflections | Δρmin = −1.12 e Å−3 |
| 102 parameters | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.030 (3) |
| Primary atom site location: structure-invariant direct methods | Absolute structure: Flack (1983), 699 Friedel pairs |
| Secondary atom site location: difference Fourier map | Absolute structure parameter: 0.03 (2) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | −0.4336 (10) | −0.1420 (8) | 0.7590 (3) | 0.0265 (11) | |
| C2 | −0.2429 (10) | −0.1841 (9) | 0.7200 (3) | 0.0264 (12) | |
| H2 | −0.1959 | −0.3079 | 0.7159 | 0.032* | |
| C3 | −0.1322 (9) | −0.0462 (8) | 0.6895 (3) | 0.0258 (12) | |
| H3 | −0.1824 | 0.0764 | 0.6946 | 0.031* | |
| C4 | 0.0617 (10) | −0.0763 (7) | 0.6489 (3) | 0.0235 (11) | |
| C5 | 0.1860 (10) | 0.0569 (8) | 0.6186 (3) | 0.0222 (11) | |
| H5 | 0.1553 | 0.1849 | 0.6203 | 0.027* | |
| C6 | 0.3668 (10) | −0.0199 (8) | 0.5843 (3) | 0.0241 (12) | |
| C7 | 0.3838 (8) | −0.2120 (7) | 0.5887 (2) | 0.0191 (11) | |
| C8 | 0.5528 (11) | −0.3400 (7) | 0.5583 (3) | 0.0264 (12) | |
| H8A | 0.5059 | −0.3791 | 0.5142 | 0.040* | |
| H8B | 0.5713 | −0.4485 | 0.5865 | 0.040* | |
| H8C | 0.6888 | −0.2741 | 0.5546 | 0.040* | |
| N1 | −0.5883 (9) | −0.1203 (8) | 0.7912 (3) | 0.0331 (11) | |
| S1 | 0.1701 (3) | −0.2987 (2) | 0.63524 (7) | 0.0246 (3) | |
| Br1 | 0.57000 (10) | 0.12692 (8) | 0.53635 (3) | 0.0319 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.022 (3) | 0.028 (3) | 0.030 (3) | −0.001 (3) | 0.000 (2) | 0.000 (2) |
| C2 | 0.024 (3) | 0.027 (3) | 0.029 (3) | 0.003 (2) | 0.001 (2) | −0.005 (2) |
| C3 | 0.023 (3) | 0.025 (3) | 0.029 (3) | 0.006 (2) | −0.002 (2) | −0.003 (2) |
| C4 | 0.016 (2) | 0.025 (3) | 0.030 (3) | 0.003 (2) | −0.002 (2) | −0.001 (2) |
| C5 | 0.023 (3) | 0.018 (2) | 0.026 (3) | 0.004 (2) | −0.006 (2) | −0.006 (2) |
| C6 | 0.025 (3) | 0.024 (3) | 0.023 (3) | −0.001 (2) | −0.001 (2) | −0.004 (2) |
| C7 | 0.021 (3) | 0.019 (2) | 0.017 (2) | 0.002 (2) | −0.0057 (19) | 0.0017 (19) |
| C8 | 0.032 (3) | 0.019 (3) | 0.028 (3) | 0.002 (2) | 0.002 (2) | 0.003 (2) |
| N1 | 0.031 (3) | 0.031 (2) | 0.038 (3) | −0.008 (3) | 0.002 (2) | −0.004 (2) |
| S1 | 0.0240 (7) | 0.0207 (6) | 0.0291 (7) | 0.0007 (6) | 0.0021 (6) | −0.0004 (5) |
| Br1 | 0.0332 (3) | 0.0259 (3) | 0.0368 (3) | −0.0021 (3) | 0.0065 (3) | 0.0040 (2) |
Geometric parameters (Å, º)
| C1—N1 | 1.154 (8) | C5—H5 | 0.9300 |
| C1—C2 | 1.434 (8) | C6—C7 | 1.373 (7) |
| C2—C3 | 1.338 (8) | C6—Br1 | 1.884 (6) |
| C2—H2 | 0.9300 | C7—C8 | 1.506 (8) |
| C3—C4 | 1.452 (8) | C7—S1 | 1.717 (5) |
| C3—H3 | 0.9300 | C8—H8A | 0.9600 |
| C4—C5 | 1.356 (8) | C8—H8B | 0.9600 |
| C4—S1 | 1.737 (5) | C8—H8C | 0.9600 |
| C5—C6 | 1.412 (8) | ||
| N1—C1—C2 | 175.6 (7) | C7—C6—C5 | 114.5 (5) |
| C3—C2—C1 | 120.3 (5) | C7—C6—Br1 | 122.3 (4) |
| C3—C2—H2 | 119.8 | C5—C6—Br1 | 123.3 (4) |
| C1—C2—H2 | 119.8 | C6—C7—C8 | 128.9 (5) |
| C2—C3—C4 | 123.9 (5) | C6—C7—S1 | 109.5 (4) |
| C2—C3—H3 | 118.0 | C8—C7—S1 | 121.6 (4) |
| C4—C3—H3 | 118.0 | C7—C8—H8A | 109.5 |
| C5—C4—C3 | 127.1 (5) | C7—C8—H8B | 109.5 |
| C5—C4—S1 | 110.6 (4) | H8A—C8—H8B | 109.5 |
| C3—C4—S1 | 122.3 (4) | C7—C8—H8C | 109.5 |
| C4—C5—C6 | 112.6 (5) | H8A—C8—H8C | 109.5 |
| C4—C5—H5 | 123.7 | H8B—C8—H8C | 109.5 |
| C6—C5—H5 | 123.7 | C7—S1—C4 | 92.8 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C3—H3···N1i | 0.93 | 2.59 | 3.501 (8) | 166 |
Symmetry code: (i) −x−1, y+1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG5330).
References
- Cambridge Soft (2001). CHEMDRAW Ultra Cambridge Soft Corporation, Cambridge, Massachusetts, USA.
- Chandra, R., Kung, M.-P. & Kung, H. F. (2006). Bioorg. Med. Chem. Lett. 16, 1350–1352. [DOI] [PubMed]
- Dinçalp, H., Aşkar, Z., Zafer, C. & İçli, S. (2011). Dyes Pigm. 91, 182–191.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Kose, M. (2004). J. Photochem. Photobiol. A, 165, 97–102.
- Nonius (2000). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Perner, R. J., Gu, Y.-G., Lee, C.-H., Bayburt, E. K., McKie, J., Alexander, K. M., Kohlhaas, K. L., Wismer, C. T., Mikusa, J., Jarvis, M. F., Kowaluk, E. A. & Bhagwat, S. S. (2003). J. Med. Chem. 46, 5249–5257. [DOI] [PubMed]
- Pu, S., Liu, W. & Liu, G. (2010). Dyes Pigm. 87, 1–9.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Zhao, J., Huang, L., Cheng, K. & Zhang, Y. (2009). Tetrahedron Lett. 50, 2758–2761.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536813019752/hg5330sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813019752/hg5330Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813019752/hg5330Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



