Abstract
Head and neck squamous cell carcinoma (HNSCC) is characterized by significant genomic instability that could lead to clonal diversity. Intratumor clonal heterogeneity has been proposed as a major attribute underlying tumor evolution, progression, and resistance to chemotherapy and radiation. Understanding genetic heterogeneity could lead to treatments specific to resistant and metastatic tumor cells. To characterize the degree of intratumor genetic heterogeneity within a single tumor, we performed whole-genome sequencing on three separate regions of an human papillomavirus (HPV)-positive oropharyngeal squamous cell carcinoma and two separate regions from one corresponding cervical lymph node metastasis. This approach achieved coverage of approximately 97.9% of the genome across all samples. In total, 5701 somatic point mutations (SPMs) and 4347 small somatic insertions and deletions (indels)were detected in at least one sample. Ninety-two percent of SPMs and 77% of indels were validated in a second set of samples adjacent to the discovery set. All five tumor samples shared 41% of SPMs, 57% of the 1805 genes with SPMs, and 34 of 55 cancer genes. The distribution of SPMs allowed phylogenetic reconstruction of this tumor's evolutionary pathway and showed that the metastatic samples arose as a late event. The degree of intratumor heterogeneity showed that a single biopsy may not represent the entire mutational landscape of HNSCC tumors. This approach may be used to further characterize intratumor heterogeneity in more patients, and their sample-to-sample variations could reveal the evolutionary process of cancer cells, facilitate our understanding of tumorigenesis, and enable the development of novel targeted therapies.
Introduction
For patients with head and neck squamous cell carcinoma (HNSCC), a major challenge that contributes to treatment failures is the emergence of chemotherapy and radiation resistance after an initial response. A major factor contributing to such treatment resistance is intratumor genomic heterogeneity [1]. Heterogeneous cancer cell populations with different genetic mutations are typically retained within tumors [1,2]. Recent studies using whole-exome sequencing of multiple tumor samples from patients with pancreatic cancer and renal cell carcinoma have demonstrated significant intratumor heterogeneity and clonal evolution, with metastatic potential possibly existing only in a small proportion of tumor cells [3,4]. These studies also suggest that metastatic clones may develop late in the course of tumor progression. Thus, understanding the genomic heterogeneity of tumors and the clonal events that lead to metastasis is paramount to eradicate all tumor clones and successfully treat HNSCC.
Previous studies have identified a large number of genetic mutations found in single samples from patients with HNSCC [5,6]. The recent adoption of next-generation sequencing technologies has greatly expanded our ability to identify the full extent of genomic instability and heterogeneity; however, an analysis of whole-genome intratumor heterogeneity within HNSCC tumors, particularly at the individual nucleotide level, has not been performed. Here, we performed one such study to demonstrate the feasibility of obtaining multiple intratumor samples and the application of next-generation sequencing technologies to determine the presence of intratumor heterogeneity in HNSCC. By obtaining physically separated samples from a primary tumor and corresponding metastatic lymph node, we detected widespread intratumor heterogeneity, which we used to reconstruct the evolutionary path of tumor clones.
Materials and Methods
Sample Collection
Research involving collection of tissue from human subjects was approved by the institutional review board at the Fred Hutchinson Cancer Research Center, and informed consent was obtained from the patient before participation. All tumor tissue specimens were collected from a 71-year-old male patient with stage IV, T2N2a, human papillomavirus (HPV)-positive oropharyngeal squamous cell carcinoma (OSCC) at the time of diagnostic direct laryngoscopy and cervical lymphadenectomy. The tumor measured 2.6 x 2.3 x 3.5 cm, whereas the metastatic lymph node measured 3.3 x 2.3 x 2.9 cm. Three separate cup biopsies (∼4-mm punch), denoted as T1, T2, and T3, were obtained from separate sections approximately 1 cm apart from each other. In a similar fashion, two spatially separated samples were obtained from one metastatic lymph node, denoted as M1 and M2. The tissue specimens were snap frozen in the operating theater and stored in liquid nitrogen until use. The HPV status was ascertained by testing one of the samples for presence of E6 sequence by polymerase chain reaction (PCR) through a well-established method published by Sotlar et al. [7]. In addition, as part of standard of care, a tumor sample was sent for diagnosis and p16 immunohistochemistry (IHC) by the Department of Pathology at the University of Washington (Seattle, WA); uniformly positive p16 staining was confirmed.
DNA Purification
Tissue specimens were embedded in Tissue-Tek OCT compound (Sakura Finetek USA, Torrance, CA), sectioned, and prepared as previously described [8]. One section from each sample was stained with hematoxylin and eosin, and OSCC tumor cells were outlined by our study pathologist. Adjacent sections were then macrodissected under microscopic visualization to enrich for >80% tumor epithelial cells. DNA was extracted from the purified tumor cells using a DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) following the manufacturer's instructions. Quantity and quality of the purified DNA were assessed with an ND-1000 spectrophotometer (Thermo Fisher Scientific, Rockford, IL). A DNA sample was also extracted from the patient's peripheral blood (denoted as “N”) using an ArchivePure DNA Blood Kit (5 PRIME Inc, Gaithersburg, MD) following the manufacturer's instructions. DNA was additionally isolated in the same fashion from adjacent sections of each tissue specimen as a validation batch. Batch “a” DNA served as the discovery set of samples, which included T1a, T2a, T3a, M1a, M2a, and blood DNA Na. Batch “b” DNA served as validation samples to confirm the discoveries in batch “a,” which included T1b, T2b, T3b, M1b, M2b, and blood DNA Nb (Na and Nb were from the same DNA prep). All samples yielded sufficient DNA for sequencing with an OD260/280 ratio between 1.8 and 2.0.
Whole-Genome DNA Sequencing
Whole-genome sequencing (WGS) was conducted by Illumina (San Diego, CA) using HiSeq 2000 technology. After sequencing batch “b” samples, we chose a second round of sequencing, using the adjacent samples named batch “b” to assess if somatic mutations were largely consistent. Batch “a” and batch “b” samples were sequenced 6 months apart. More than 1 billion paired-end reads of 100 bp each in length were generated for each DNA sample. Batch “a” was sequenced in June 2011. Batch “b” samples were sequenced in January 2012, after Illumina made improvements in sequencing technology and calling algorithms (Illumina, personal communication). After aligning and assembling the short-read sequences to human genome reference sequence National Center for Biotechnology Information (NCBI, Bethesda, MD) Build 37/hg19, Illumina provided initial variant callings using CASAVA 1.8 for batch “a” samples and CASAVA version 1.9 for batch “b” samples, with all computational parameters per recommendation. The primary difference between these two versions of CASAVA pertains to detection of large structural mutations, such as copy number variations or structure variations, but not detection of single nucleotide polymorphisms (SNPs) or small insertions/deletions (indels). To mitigate this issue and for analytic consistency between the two batches of samples, we primarily focused on small mutations (SNPs/indels) for this study. Using either HPV16 or HPV18 genome sequence as a reference, we used Burrows-Wheeler Aligner software (Wellcome Trust Sanger Institute, Cambridge, United Kingdom) [9] to align the short-read sequences of our WGS samples (both tumor and blood) to the HPV genome and look for integration sites.
Somatic Mutation Detection
Using germline DNA samples from blood as references (sequenced in both batches and denoted as Na and Nb), we identified somatic point mutations (SPMs) and somatic indels in each tumor specimen. To minimize false-positive discovery rate, we applied a set of stringent and relatively conservative criteria on the basis of quality scores of individual reads, read depth, and variant calling confidence. (For details, see Supplemental Methods section.) In addition, we excluded mutations that have been identified in public databases, including the NCBI single nucleotide polymorphism database (dbSNP) and the 1000 Genomes Project (The EMBL-European Bioinformatics Institute, Cambridge, United Kingdom), on the basis of the assumption that the mutations in these databases are less likely to represent somatic, cancer-causing mutations because they were mostly discovered from germline DNA samples.
To test for reproducibility, we used batch “b” samples to validate the somatic mutations detected in batch “a” samples. By study design, each sample in batch “b” corresponded to the next physically adjacent sample in batch “a.” Thereby, we evaluated the validation rate as the percentage of somatic mutations detected in each (or any) batch “a” sample that were also detected in the corresponding (or any) batch “b” sample.
A Venn diagram plot showing shared and unique genes with identified SPMs among all batch “a” samples was made using an R code from https://stat.ethz.ch/pipermail/bioconductor/2007-October/019703.html.
Annotating Mutations
We annotated somatic mutations on the basis of RefSeq gene hg19 using ANNOVAR (University of Pennsylvania, Philadelphia, PA) [10]. Firstly, somatic mutations were annotated against 488 cancer-associated genes that have mutations causally implicated in cancer (http://www.sanger.ac.uk/genetics/CGP/Census) on the basis of whether their locations were in the gene-coding, intronic, downstream, upstream, or intergenic regions. The cancer-associated genes implicated in our patient with OSCC were examined for their functions using Ingenuity Pathway Analysis (version 16542223; Ingenuity Systems Inc, Redwood City, CA) and heterogeneity among tumor samples. Secondly, possible functions such as stop-gain, synonymous/nonsynonymous were annotated whenever possible, particularly for those mutations in genecoding regions.
Cancer Genome Evolution
To explore the evolutionary history of this tumor, we focused on the variations within 1-kb flanking coding regions of genes containing somatic mutations [11]. For each pair of tumor samples, we calculated their distance as the number of nonshared mutational genes. On the basis of this distance, a coalescent tree was then constructed using the neighbor-joining method [12] in MATLAB version 2011b (The MathWorks, Inc, Natick, MA). On the basis of the set of somatic mutations, we estimated the chronological time for the evolution of the sampled tumor cell populations on the basis of previous estimations of mutation frequency as described by Yachida et al. [4]. Major phases of tumor evolution were estimated as described in Supplemental Methods section. Briefly, we set the time from tumor initialization to the parental clone as the time to accumulate the number of somatic mutations shared in all 10 tumor samples. The average time from the parental clone to subclones was calculated as the time to accumulate the average number of mutations in each primary tumor but not in the parental clone. The average time to develop metastasis was calculated as the time to accumulate the average number of mutations in each metastasis but neither in the primary tumor nor in the parental clone. To evaluate the significance of a coalescent tree, we performed “bootstrap sampling,” in which a randomly selected subset (80%) of genes containing at least one SPM in one tumor sample were used to plot the coalescent tree. We also performed “jackknife sampling” to evaluate the stability of the coalescent tree. Specifically, we left one sample out at a time and performed a coalescent analysis on the remaining samples. Then, we evaluated whether the coalescent tree from each jackknife sample was consistent to the complete tree.
Results
Sequencing Characteristics
Sequencing statistics for both batches of DNA samples were summarized in Table 1. The sequence coverage of the six batch “a” samples was comparable, with a mean depth of 44.7X (range = 41.7–48.0) and coverage of 97.9%. Using the cutoff of quality score Q ≥ 20, approximately 3.6 million SNPs and approximately 600 thousand indels were detected (Table 1). The coverage in batch “b” samples was similar to that of batch “a” samples, but mean depth was lower at 36.8X (range = 34.9–39.3). On average, the number of SNPs and indels detected in batch “b” samples was 32,860 (0.9%) and 34,246 (5%), lower than that in the corresponding samples in batch “a.” In addition, the concordance between the genotypes of SNPs detected by sequencing versus Illumina Quad SNP array was slightly lower for samples in batch “b” (99.3%) compared to that in batch “a” (99.9%) samples. Other sequencing characteristics were comparable between all tumor samples both within or between the batches. For example, the ratio of transition and transversion type of mutations was consistent at 2.06, and greater than 93% of SNPs detected in each tumor was reported in dbSNP version 131. Interestingly, despite detection of HPV E6 sequences by PCR and uniformly positive p16 staining by IHC, we did not observe alignment of short-read sequences from our WGS data (either from tumor or blood) with sequences from HPV 16 or 18 reference genomes.
Table 1.
Sequencing Statistics.
| Batch a | T1a | T2a | T3a | M1a | M2a | Na |
| At Known Sites | ||||||
| Total bases mapped | 120.8 | 135.6 | 130.1 | 137.2 | 123.3 | 119.1 |
| Mean depth | 42.3 | 47.5 | 45.5 | 48.0 | 43.1 | 41.7 |
| Coverage (%) | 97.93 | 97.93 | 97.9 | 97.91 | 97.89 | 97.96 |
| No. of SNPs Detected (Q ≥ 20) | ||||||
| SNPs | 3,690,282 | 3,680,844 | 3,678,250 | 3,679,869 | 3,648,729 | 3,691,527 |
| Transition/transversion | 2.06 | 2.06 | 2.06 | 2.06 | 2.06 | 2.06 |
| Heterogeneous/homogeneous | 1.5 | 1.48 | 1.48 | 1.48 | 1.46 | 1.49 |
| Concordance with Quad SNPs (%) | 99.89 | 99.85 | 99.83 | 99.74 | 99.63 | 99.89 |
| In dbSNP (%) | 93.8 | 93.7 | 93.8 | 94 | 93.9 | 93.9 |
| No. of Indels Detected (Q ≥ 20, length ≤ 300) | ||||||
| Insertions | 311,033 | 309,245 | 305,466 | 318,674 | 293,919 | 315,539 |
| Deletions | 329,908 | 329,198 | 327,838 | 338,497 | 316,097 | 331,755 |
| Breakpoints | 26,636 | 26,536 | 25,823 | 28,802 | 24,867 | 27,365 |
| Batch b | T1b | T2b | T3b | M1b | M2b | Nb |
| At Known Sites | ||||||
| Total bases mapped | 110.3 | 112.5 | 100.7 | 102.3 | 106.3 | 99.8 |
| Mean depth | 38.6 | 39.3 | 35.2 | 35.8 | 37.2 | 34.9 |
| Coverage (%) | 97.9 | 97.95 | 97.84 | 97.92 | 97.92 | 97.85 |
| No. of SNPs Detected (Q ≥ 20) | ||||||
| SNPs | 3,655,604 | 3,659,173 | 3,658,838 | 3,616,641 | 3,621,011 | 3,661,077 |
| Transition/transversion | 2.06 | 2.05 | 2.06 | 2.06 | 2.06 | 2.06 |
| Heterogeneous/homogeneous | 1.48 | 1.47 | 1.48 | 1.46 | 1.45 | 1.48 |
| Concordance with Quad SNPs (%) | 99.28 | 99.27 | 99.28 | 99.27 | 99.25 | 99.26 |
| In dbSNP (%) | 93.8 | 93.7 | 93.8 | 94 | 93.9 | 93.9 |
| No. of Indels Detected (Q ≥ 20, length ≤ 300) | ||||||
| Insertions | 295,118 | 306,338 | 293,880 | 280,595 | 285,086 | 295,240 |
| Deletions | 313,896 | 321,806 | 311,007 | 301,312 | 304,709 | 312,708 |
| Breakpoints | 9,605 | 9,860 | 9,073 | 8,447 | 9,062 | 9,224 |
Somatic Point Mutations
In total, 5701 SPMs were detected in at least one of the five primary tumor or metastatic samples in the discovery set (batch “a”). Of these, 5236 were confirmed in at least one tumor/metastatic sample in the validation set (batch “b”) for an overall validation rate of 92%. The number of SPMs in a single sample varied from 3228 to 4043, around 85% of which could be confirmed in the corresponding physically adjacent sample in the validation set (Table 2). Furthermore, the genomic distribution of SPMs for samples from the two batches were largely concordant, as shown by comparing the number of SPMs in sliding windows of size 500 kb (Figure W1A). Among the 10 samples in both batches, we detected a total of 6440 SPMs.
Table 2.
Validation Results of Somatic Mutations.
| SPMs | Somatic Indels | |||
| No. Validated (Total) | Validation Rate* | No. Validated (Total) | Validation Rate* | |
| T1 | 2747 (3228) | 0.85 | 2248 (3085) | 0.73 |
| T2 | 3028 (3696) | 0.82 | 2356 (3279) | 0.72 |
| T3 | 3402 (4043) | 0.84 | 2256 (3202) | 0.7 |
| M1 | 3532 (4102) | 0.86 | 2166 (3254) | 0.67 |
| M2 | 3476 (3932) | 0.88 | 2219 (3028) | 0.73 |
| Overall† | 5236 (5701) | 0.92‡ | 3368 (4347) | 0.77‡ |
Validation rate for T1 to M2 shows the proportion of variants in a batch “a” sample that were validated in the corresponding batch “b” sample.
Overall shows the number of variants validated (identified) in at least one of the five tumor samples (T1, T2, T3, M1, or M2).
Overall validation rate shows the proportion of variants identified in at least one tumor sample in batch “a” that were validated in at least one tumor sample in batch “b.”
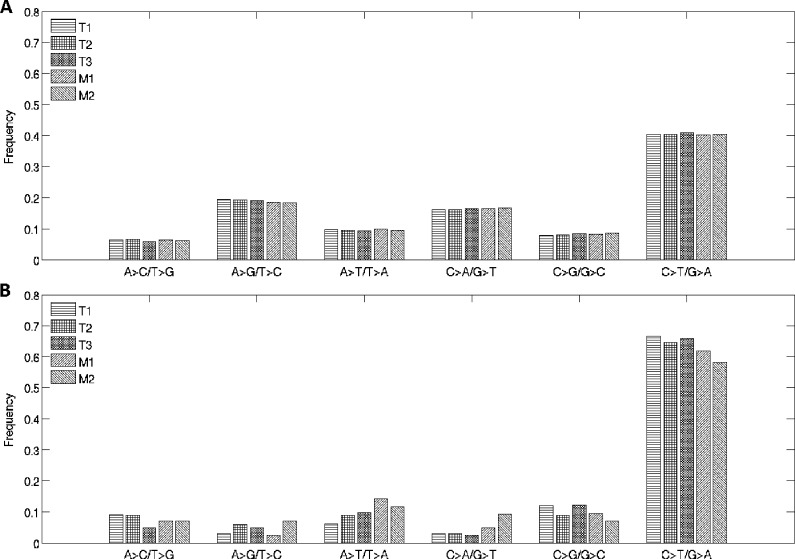
Examining the distributions of SPMs, we found that the two transition types of mutations C > T/G > A and A > G/T > C were most frequent at 40.5% and 18.9%, respectively, over the entire genome (Figure 1A). However, the C > T/G > A mutations were much more enriched in coding regions, with a frequency of 63.5%, whereas A > G/T > C mutations were at 4.63% (Figure 1B). This distribution was reminiscent of the SPM pattern observed in malignant melanoma [13]. The distribution pattern was largely consistent across the five samples in batch “a,” with no apparent differences between tumor and metastatic samples.
Figure 1.
Distribution of transition and transversion types of SPMs over the entire genome (A) and in the protein-coding region of the genome (B).
Of the SPMs detected in the discovery set, 1153 were only in the primary tumors, and 792 were only in the metastatic samples. Moreover, the primary tumors were more heterogeneous than the metastatic ones, with 51.7% and 76.7% shared SPMs, respectively. Overall, 41% SPMs were shared among all five tumors (primary or metastatic).
Small Somatic Indels
In this study, somatic indels were determined as insertions or deletions up to 300 bp in length. Following a set of stringent criteria (see Materials and Methods section), we identified a total of 4347 somatic indels in at least one of the five tumor samples in the discovery set (batch “a”). Of these, 3368 were confirmed in at least one tumor/metastatic sample in the validation set (batch “b”) for an overall validation rate of 77% (Table 2). The average number of somatic indels in a single sample was 3170. The genomic distributions of somatic indels were largely concordant, similar to the SPMs (Figure W1 B). Among the 10 samples in both batches, there were a total of 4638 somatic indels detected.
Similar to SPMs, the number of somatic indels specific to primary tumors was greater than the number specific to metastatic samples (613 and 329, respectively). The primary tumors shared 59% of somatic indels, whereas the metastatic samples shared 68%. Overall, 46% somatic indels were shared among all five tumors (primary or metastatic).
Annotations of Mutations
As shown in Table 3, of the entire 6440 SPMs in the two batches of samples, only 61 (<1%) SPMs resided in the coding region. Among these, 41 were nonsynonymous; 16 were synonymous; three SPMs could not be annotated due to errors in the gene structure definition in the database; and one represented a stop-gain SPM in gene poly(A) polymerase beta (testis specific) (PAPOLB) at chr7:4900509, which leads to the creation of stop codon at the site. Of SPMs in the noncoding region, 17 SPMs were in the exonic region of noncoding RNA, and 1 SPM was located in an intron 1 bp from a splicing junction (Table W1). The majority (68.7%) of SPMs were in intergenic regions, which were more than 1 kb away from the closest genes. For somatic indels, 13 (0.28%) indels were in the coding region, 5 were in the exonic region of noncoding RNA, and 2 in introns were overlapping with a splicing site.
Table 3.
Annotations of Somatic Mutations.
| Total | Subcategories | SPMs | Somatic Indels | |||
| 6440 | % | 4638 | % | |||
| Intragenic | 2017 | 31.32 | 1550 | 33.42 | ||
| Coding | 61 | 0.95 | 13 | 0.28 | ||
| Nonsynonymous | 41 | |||||
| Synonymous | 16 | |||||
| Stop-gain | 1 | |||||
| Unknown | 3 | |||||
| Noncoding, transcribed | 247 | 3.84 | 164 | 3.54 | ||
| ncRNA (exonic) | 17 | 5 | ||||
| ncRNA (intronic) | 188 | 125 | ||||
| UTR3 | 35 | 31 | ||||
| UTR5 | 7 | 3 | ||||
| Intronic | 1632 | 25.34 | 1313 | 28.31 | ||
| Splicing | 1 | 2 | ||||
| Other | 1631 | 1311 | ||||
| Flanking | 77 | 1.20 | 60 | 1.29 | ||
| Downstream (1 kb) | 38 | 26 | ||||
| Upstream (1 kb) | 39 | 34 | ||||
| Intergenic | 4423 | 68.68 | 3088 | 66.58 | ||
ncRNA indicates noncoding RNA; UTR3/UTR5, untranslated region 3/5.
Intragenic Mutations
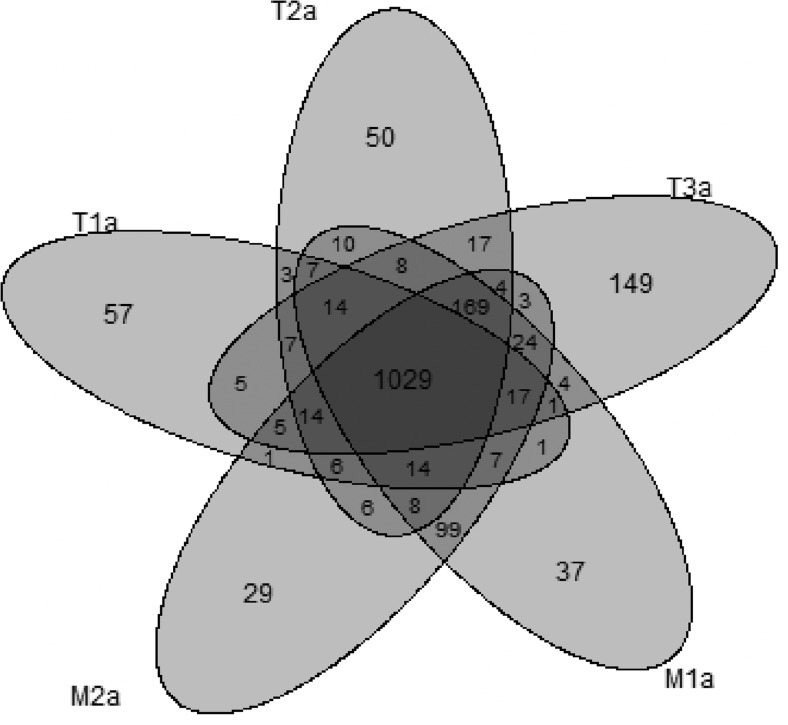
Besides the mutations in coding regions, many mutations in intronic or gene flanking regions may potentially affect gene expression or regulation and thus may have functional consequences [14,15]. To characterize such likely functional somatic mutations, we grouped the somatic mutations by genes. There are a total of 27,925 genes of nonidentical genomic positions in RefSeq hg19 [as assembled by ANNOVAR, June 2012 [10]]. Within 1 kb downstream or upstream of each gene, we found SPMs and somatic indels in 2729 genes among 10 tumor samples. Given that detected SPMs had much higher validation rate than somatic indels, we compared the heterogeneity of somatic mutations among tumors on the basis of genes containing SPMs. In particular, for the five tumor samples in the discovery set, there were 1805 genes with SPMs. Among these genes, 1029 were shared among all five samples, whereas T1a, T2a, T3a, M1a, and M2a contained 57, 50, 149, 37, and 29 unique genes with SPMs that were not shared by any other tumors (see Venn diagram in Figure 2). Among genes with SPMs that were shared between several but not all tumors, the largest group was the 169 genes shared by all four tumors except for T1a, followed by the 99 genes shared only between the two metastatic samples. This likely indicated a closer evolutionary relationship among these samples.
Figure 2.
Venn diagram plot shows number of shared and unique genes with identified SPMs among batch “a” samples.
Among all functional genes, there are 488 genes classified as cancer-associated genes by the Cancer Genome Project (Wellcome Trust Sanger Institute). Of these cancer genes, there are 428 unique geneIDs on autosomes. We identified their locations using their Entrez ID in Ensembl genome database (The EMBL-European Bioinformatics Institute) or NCBI Gene (NCBI). In total, we found 162 somatic mutations (SPMs or somatic indels) located within 1 kb of 71 cancer genes. Of these, 55 cancer genes contained somatic mutations that were validated by the corresponding sample in batch “b” (Table W2; for a list of contained mutations, see Table W3). Similar to the entire set of functional genes, the majority (34 of 55) of cancer genes were shared by all the five samples in batch “a.” This probably indicates the possible common root of all these tumors. Two genes previously implicated in squamous cell carcinomas, specifically notch (Drosophila) homolog 2 (NOTCH2) and epidermal growth factor receptor (EGFR) [16–18], both contained intronic mutations in all tumor samples. However, mutations in five cancer genes [K(lysine) acetyltransferase 6A (KAT6A), PR domain containing 16 (PRDM16), mastermind-like 2 (Drosophila) (MAML2), zinc finger and BTB domain containing 16 (ZBTB16), and B-cell CLL/lymphoma 10 (BCL10)] had mutations only in three primary tumor samples (T3, T2, T2, T1, and T1, respectively), and four cancer genes [c-abl oncogene 2, nonreceptor tyrosine kinase (ABL2), pre-B-cell leukemia homeobox 1 (PBX1), SET domain containing 2 (SETD2), and tripartite motif containing 24 (TRIM24)] had mutations only in the two metastatic samples. Ingenuity Pathway Analysis (version 16542223; Ingenuity Systems; http://www.ingenuity.com) was applied to these genes to determine pathways potentially affected by these mutations (Table 4). Genes with mutations in primary tumor samples only were significantly associated with Notch signaling, transcriptional regulation in embryonic stem cells, high mobility group-B1 (HMGB1) signaling, and multiple pathways involving signaling in lymphocytes. Those genes with mutations in the lymph node samples only were significantly associated with transcriptional regulation in embryonic stem cells, PDGF signaling, Ras homolog gene family, member A signaling, and retinoic acid receptor activation.
Table 4.
Cancer Genes with Mutations in Primary Tumor and Lymph Node Samples.
| Pathway | P Value | Genes |
| Genes Mutated in Primary Tumor Samples Only | ||
| Notch signaling | .011 | MAML2 |
| Transcriptional regulatory network in embryonic stem cells | .012 | MYST3 |
| Regulation of IL-2 expression in T lymphocytes | .024 | BCL10 |
| HMGB1 signaling | .028 | MYST3 |
| T cell receptor signaling | .029 | BCL10 |
| Cluster of differentiation 28 signaling in T helper cells | .034 | BCL10 |
| Protein kinase C θ signaling in T lymphocytes | .034 | BCL10 |
| Phosphoinositide 3-kinase signaling in B lymphocytes | .038 | BCL10 |
| B cell receptor signaling | .047 | BCL10 |
| Nuclear factor kappa-light-chain-enhancer of activated B cells signaling | .049 | BCL10 |
| Genes Mutated in Lymph Node Samples Only | ||
| Transcriptional regulatory network in embryonic stem cells | .009 | TRIM24 |
| PDGF signaling | .018 | ABL2 |
| Ras homolog gene family, member A signaling | .028 | ABL2 |
| Retinoic acid receptor activation | .041 | TRIM24 |
Cancer Genome Evolution
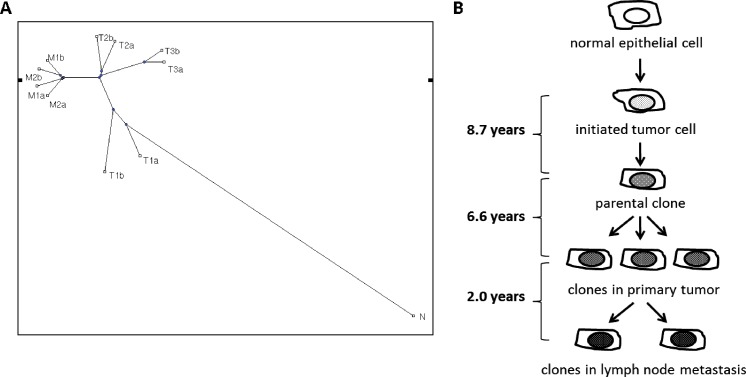
Following the clonal expansion hypothesis, we expected that multiple clones were present within a single tumor but in different physical locations of the primary and metastatic tumors. Dissecting heterogeneities across their sequences allowed us to capture the evolutionary relationship among these 10 tumor samples. A coalescent tree (Figure 3) was built for the 10 tumor samples together with the sample from peripheral blood (“N”) on the basis of their shared cancer-associated genes containing SPMs. It appeared that tumor samples physically close to each other were more homogeneous and were clustered to the same branch. The primary tumors at the first site (T1a and T1b) were closer to the founder clone, whereas those at the second and the third sites contained additional mutations and occurred in more distant branches. All four metastatic samples were clustered in the same branch and appeared to have evolved from the same parental clone. Bootstrap and jackknife sampling were done to evaluate the significance of the coalescent tree, and the structure was found to be consistent throughout (data not shown).
Figure 3.
(A) Coalescent tree is based on number of shared genes containing SPMs. (B) Timeline of tumor progression based on estimated mutation rate indicates significant passage of time from the development of the initial cancerous cell to the development of the parental clone of all tumor samples obtained.
We estimated that the tumor in this patient took approximately 8.7 years to develop an initial tumor clone from one or more single somatic mutations. From this initial clone, it took approximately another 6.6 years for the single clone to evolve into the six primary tumor cell populations that have been observed. It took another 2 years for one of the clones to establish four tumor cell populations present in the metastatic lymph node.
Discussion
By sequencing whole genomes on multiple cancerous cells extracted from different portions of the primary tumor and also from two different metastatic sites on a single patient with OSCC, we were able to characterize the extent of small somatic mutations (SPMs and small indels) on the genome-wide scale, examine the tumor heterogeneity among different tumor sites, and also investigate the pattern of tumor evolution from initiation to metastasis. This is the first such study in HNSCC to examine intratumor heterogeneity by performing whole-genome sequencing in multiple tumor samples from a single patient. Although one previous study performed whole-genome sequencing on an HPV-negative oropharyngeal tumor and an HPV-negative hypopharyngeal tumor [[5]], the current study is the first to perform whole-genome sequencing on an HPV-positive HNSCC. This study provides a model that can be applied to the investigation of additional HNSCC tumors to determine shared and unique patterns of mutation and clonal evolutionary breakpoints.
Overall, this HPV+ tumor had 6440 SPMs and 4638 small indels based on the discovery set with an overall validation rate of 92% and 77%, respectively. Thus, despite the potentially higher detection sensitivity by the revised chemistry of the Illumina sequencer, the higher sequence coverage still seems to increase the sensitivity of mutation detection. Our validation samples were sequenced at approximately 37x coverage compared to approximately 45x coverage in the discovery set. Around 0.9% fewer SNPs and 5% fewer small indels were detected in the validation set. The much lower detection rate of indels in the validation set could be due to the difficulty of aligning reads with indels, which also partially explained the lower validation rate for small indels. Although indels may have significant impacts on cellular phenotype, we restricted further analysis to the SPMs due to their higher validation rates.
The greater frequency of mutations found in this study compared with those found by Stransky et al. and Agrawal et al. [5,6] in HPV+ tumors is likely due to the fact that we report mutations on the whole genome as opposed to focusing on the exome as was done in these two studies. In fact, the majority of the mutations we found were in noncoding regions, which would not have been detected by exome sequencing alone. In accordance with these two studies, only 61 SPMs were located within exons of protein-coding genes. Several tumor suppressors and oncogenes have been previously implicated in HNSCC including tumor protein p53 (TP53), cyclin-dependent kinase inhibitor 2A (CDKN2A), TP63, phosphatase and tensin homolog (PTEN), NOTCH1, NOTCH2, NOTCH3, F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase (FBXW7), phosphatidylinositol-4, 5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA), cyclin D1 (CCND1), HRAS, and EGFR [5,6,18]. In our study, none of these genes had exonic mutations in any tumor sample. Two of these genes, NOTCH2 and EGFR, both contained intronic mutations that were present in all tumor samples we assessed. Our exclusion of known SNPs identified in the 1000 Genomes Project and dbSNP databases potentially excluded SPMs that coincided with known SNPs. As suggested in previous studies [5,6], HPV-positive tumors, such as the one we examined, may be expected to have fewer mutations compared to other tumors, particularly those associated with heavy exposure to tobacco and alcohol. The major risk factors for HNSCC include tobacco use, alcohol abuse, and infection with high-risk HPV strains [18]. Tobacco and alcohol are thought to contribute to tumor pathogenicity by inducing DNA mutations [19]. Although high-risk HPV strains likely cause tumor pathogenicity through functional inactivation of p53, genomic instability may still occur during clonal expansion of tumor cells and result in intratumor heterogeneity. A more widespread analysis is needed to clarify which mutations in tumor suppressors and oncogenes are involved in HPV-positive and HPV-negative tumors and which may be important in the development and testing of therapeutic agents targeting the pathways associated with these genes. It is likely that targeted pathways may be different depending on HPV status.
Interestingly, despite detection of HPV E6 sequences by PCR and uniformly positive p16 staining by IHC, we did not observe HPV integration in our data by WGS, nor could we map short-read sequences to the HPV reference genome. The following reasons for this could be multifactorial: insufficient depth of sequencing, algorithm/software difficulty of aligning short reads at insertion sites, HPV sequence variations from reference, and other factors. In the future, more sensitive sequencing tools might allow detection of HPV genome integration and localization of insertion site(s).
In earlier studies of HNSCC somatic mutations, a common study design is to compare genetic profiles between a single tumor sample and a single normal sample, e.g., adjacent normal tissue or blood from the same patient [5,6,20,21]. Although these studies have provided important insight into cancer genomic instability, they cannot easily provide insight into tumor heterogeneity without making unsubstantiated assumptions [22]. If single tumor biopsies are used to identify molecular biomarkers for guiding therapy without fully documenting intratumor heterogeneity and identifying essential somatic mutations, one may be misled, hence, subject patients to unnecessary risk or inadequate treatment. In the current feasibility study, we demonstrated that, although a slight majority of genes with SPMs were shared by all five tumor samples, only 41% SPMs and 46% of somatic indels were shared by all.
The degree of intratumor heterogeneity detected in this tumor permitted us to estimate the branching process of clonal expansion and to approximate the timeline of tumor development by phylogenetic construction. It was shown that the physically adjacent tumor samples (in batch “a” and batch “b”) were placed in the same branch of the phylogenetic tree, as might be expected from the clonal expansion process. This branching type of evolution has been observed in several other cancer types as well [1]. It is a commonly held belief that the majority of solid tumors originate from a single cell that develops the potential for unregulated growth and, eventually, invasive and metastatic potential. Our results here support a branching evolutionary process that can be traced backward to a common progenitor clone, and they suggest a significant amount of time during which it may be possible to detect tumors at early stages. By estimating the sequence of genetic mutations that occur during clonal expansion, this approach may help to identify driver mutations contributing to early tumor growth, which can then be targeted with selective therapies.
Currently, the single greatest prognostic factor in HNSCC is the presence or absence of cervical lymph node metastases, which are associated with approximately 50% reduced survival [23]. By establishing genetic mutations unique to metastatic clones, targeted therapies may be developed with improved efficacy against disseminated disease. Our results suggest that the two metastatic samples had a high degree of similarity. The estimated timeline of tumor development placed the metastatic samples at a later time than the development of the primary tumor samples. The two metastatic samples shared a total of 99 intragenic mutations that were not present in any primary tumor sample, and the metastatic samples then had an additional 29 and 37 unique mutations, respectively. Those mutations that are shared by both lymph node samples but not present in the primary tumor may confer metastatic ability. Four proposed cancer genes had intronic mutations that were present in the metastatic samples only. One of these, ABL2, has recently been shown to function downstream of the EGFR and contribute to tumor cell invasion and blood vessel intravasation [24]. A recent study showed an inverse relationship between expression of TRIM24, a protein thought to interact between chromatin and several nuclear receptors, in HNSCC tumors and patient survival and that knockdown of TRIM24 in HNSCC cell lines inhibited cell growth [25]. It is unknown whether the intronic mutations we identified in the metastatic samples of our patient have any effect on protein expression or function, however.
In this study, we studied the tumor heterogeneity and tumor clonal expansion in OSCC on the basis of SPMs and small indels. Larger mutations such as loss of heterozygosity, copy number variation, and translocations were not exploited. It is expected that larger mutations may exhibit larger phenotypic effects. However, a systematic examination of such mutations remains to be done. Additionally, the majority of mutations, including those within cancer genes, were observed within introns or intergenic segments. The impact of mutations within these regions on gene expression has become increasingly accepted. Future studies to determine how intratumor heterogeneity affects gene expression will help to elucidate biologically relevant genomic alterations. Our data support the presence of significant intratumor heterogeneity, and larger studies with more patients and larger number of tumor samples are warranted to further determine the degree of intratumor heterogeneity in HNSCC tumors. Larger studies will be valuable to identify any common mutations and any common clonal branching points.
Supplemental Methods
Somatic Mutation Detection
To reduce false-positive discoveries of somatic mutations, we applied a set of stringent and relatively conservative criteria on the basis of quality scores of individual reads, read depth, and variant calling confidence. Firstly, mutation sites were filtered to be those sites of high confidence in the tumor specimen. For SNPs in each tumor DNA, we required the phred-based quality Q(SNP) [1] to be at least 40, at least 20 reads coving the site (read depth), unique alternative allele, and at least 20% of alternative alleles among all reads. For indels, we required Q(indels) ≥ 40, read depth ≥ 40, at least 10 reads of indel calls, and at least 10 reads of reference allele calls for heterozygous indel sites. Secondly, the mutation sites were further filtered to be those of low mutation possibility in the blood samples. In both Na and Nb, for SNPs, we required that Q(SNP) = 0, a read depth of at least 20, and at least 80% of reference alleles; whereas for indels, we required Q(indels) < 3, a read depth of at least 20, and no indel calls. Thirdly, using ANNOVAR [2], we excluded the mutations that have been identified in public databases, including dbSNP (version 132) and the 1000 Genomes Project [3] (Phase 1, March 2012 release). Lastly, to minimize the technical differences (such as random read sampling or tumor tissue dissections) among these tumor specimens of the same individual, we re-examined the possibilities of mutations in each tumor DNA, at the somatic mutation sites that have been identified with high confidence in at least one tumor DNA. We rescued a number of such mutations using relaxed conditional quality score criteria as Q(SNP|polysite) ≥ 20 or Q(indel|polysite) ≥ 20, instead of Q(SNP) or Q(indel) score. In the end, a set of high-confidence somatic mutations was obtained for each tumor sample.
Estimation of Chronological Time of Tumor Evolution
Following exactly as [4], the accumulation of N somatic mutations was modeled as a Poisson process, where the number of cell divisions Ci to obtain the ith mutation follows an exponential distribution with rate r. If assuming a mutation rate per base pair per cell generation is 5 x 10-10 [4] and the length of human genome is approximately 3 x 109 bp, then r, the number of mutations per cell generation, would be 1.5. Assuming each new somatic mutation occurs independently, the total number of cell generations to accumulate N mutations would be the summation of all Ci, which have a Gamma distribution with mean N/r and
. Thus,
.
Given that the average cell doubling time (cell generation Tgen) in oral cancer is not currently known, we used the estimation of 2–3 days in pancreatic cancer [4] and estimated the time to accumulate N somatic mutations as
The tumor evolution was divided into the following three phases: P1, from tumor initialization to the parental clone; P2, from the parental clone to subclones; and P3, to develop metastasis after subclones. The evolutional time for each phase was estimated using the above formula with N1, N2, and N3 as the corresponding number of somatic mutations. We estimated N1 as the number of somatic mutations shared in all 10 tumor samples. Then, N2 was calculated as the average number of mutations in each primary tumor but not in the parental clone. Similarly, N3 was calculated as the average number of mutations in each metastasis but neither in the primary tumor nor in the parental clone.
Supplementary Material
Footnotes
This work was supported in part by grants CA119225, MH084621, R01MH084621-03S2, R01CA124574-01, and 5T32DC000018-29 from National Institutes of Health, Early Physician-Scientist Career Development Award from the Howard Hughes Medical Institute, grant RSG TBG-123653 from the American Cancer Society, and center funds from the Department of Otolaryngology-Head and Neck Surgery, University of Washington and VA Puget Sound Health Care System, Seattle, WA.
This article refers to supplementary materials, which are designated by Tables W1 to W3 and Figure W1 and are available online at www.neoplasia.com.
References
- 1.Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Research. 2012;72:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sotlar K, Diemer D, Dethleffs A, Hack Y, Stubner A, Vollmer N, Menton S, Menton M, Dietz K, Wallwiener D, et al. Detection and typing of human papillomavirus by e6 nested multiplex PCR. J Clin Microbiol. 2004;42:3176–3184. doi: 10.1128/JCM.42.7.3176-3184.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu C, Houck JR, Fan W, Wang P, Chen Y, Upton M, Futran ND, Schwartz SM, Zhao LP, Chen C, et al. Simultaneous isolation of DNA and RNA from the same cell population obtained by laser capture microdissection for genome and transcriptome profiling. J Mol Diagn. 2008;10:129–134. doi: 10.2353/jmoldx.2008.070131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenwood TA, Kelsoe JR. Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics. 2003;82:511–520. doi: 10.1016/s0888-7543(03)00142-3. [DOI] [PubMed] [Google Scholar]
- 15.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Diseaseassociated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16:129–141. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- 16.Egloff AM, Grandis JR. Molecular pathways: context-dependent approaches to Notch targeting as cancer therapy. Clin Cancer Res. 2012;18:5188–5195. doi: 10.1158/1078-0432.CCR-11-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smilek P, Neuwirthova J, Jarkovsky J, Dusek L, Rottenberg J, Kostrica R, Srovnal J, Hajduch M, Drabek J, Klozar J. Epidermal growth factor receptor (EGFR) expression and mutations in the EGFR signaling pathway in correlation with anti-EGFR therapy in head and neck squamous cell carcinomas. Neoplasma. 2012;59:508–515. doi: 10.4149/neo_2012_065. [DOI] [PubMed] [Google Scholar]
- 18.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 19.Sabitha K, Reddy MV, Jamil K. Smoking related risk involved in individuals carrying genetic variants of CYP1A1 gene in head and neck cancer. Cancer Epidemiol. 2010;34:587–592. doi: 10.1016/j.canep.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Challen C, Brown H, Cai C, Betts G, Paterson I, Sloan P, West C, Birch-Machin M, Robinson M. Mitochondrial DNA mutations in head and neck cancer are infrequent and lack prognostic utility. Br J Cancer. 2011;104:1319–1324. doi: 10.1038/bjc.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedrero JM, Carracedo DG, Pinto CM, Zapatero AH, Rodrigo JP, Nieto CS, Gonzalez MV. Frequent genetic and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head and neck squamous cell carcinoma. Int J Cancer. 2005;114:242–248. doi: 10.1002/ijc.20711. [DOI] [PubMed] [Google Scholar]
- 22.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49:211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson JT, Barnes EL, Myers EN, Schramm VL, Jr, Borochovitz D, Sigler BA. The extracapsular spread of tumors in cervical node metastasis. Arch Otolaryngol. 1981;107:725–729. doi: 10.1001/archotol.1981.00790480001001. [DOI] [PubMed] [Google Scholar]
- 24.Gil-Henn H, Patsialou A, Wang Y, Warren MS, Condeelis JS, Koleske AJ. Arg/Abl2 promotes invasion and attenuates proliferation of breast cancer in vivo. Oncogene. 2013;32:2622–2630. doi: 10.1038/onc.2012.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Z, Cao W, Li J, Song X, Mao L, Chen W. TRIM24 overexpression is common in locally advanced head and neck squamous cell carcinoma and correlates with aggressive malignant phenotypes. PLoS One. 2013;8:e63887. doi: 10.1371/journal.pone.0063887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- 1.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 2.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.1000 Genomes Project Consortium, author. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.