Abstract
The Zfp57 gene encodes a KRAB (Krüppel-associated box) domain-containing C2H2 zinc finger transcription factor that is expressed in early development. Zfp57 protein recognizes methylated CpG dinucleotide within GCGGCA elements at multiple imprinting control regions. In the previously determined structure of the mouse Zfp57 DNA-binding domain in complex with DNA containing 5-methylcytosine (5mC), the side chains of Arg178 and Glu182 contact the methyl group via hydrophobic and van der Waals interactions. We examined the role of Glu182 in recognition of 5mC by mutagenesis. The majority of mutants examined lose selectivity of methylated (5mC) over unmodified (C) and oxidative derivatives, 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine (5caC), suggesting that the side chain of Glu182 (the size and the charge) is dispensable for methyl group recognition but negatively impacts the binding of unmodified cytosine as well as oxidized derivatives of 5mC to achieve 5mC selectivity. Substitution of Glu182 with its corresponding amide (E182Q) had no effect on methylated DNA binding but gained significant binding affinity for 5caC DNA, resulting in a binding affinity for 5caC DNA comparable to that of the wild-type protein for 5mC. We show structurally that the uncharged amide group of E182Q interacts favorably with the carboxylate group of 5caC. Furthermore, introducing a positively charged arginine at position 182 resulted in a mutant (E182R) having higher selectivity for the negatively charged 5caC.
Mammalian DNA cytosine modification is a dynamic process catalyzed by specific DNA methyltransferases that convert cytosine (C) to 5-methylcytosine (5mC).1,2 The 5mC may then be oxidized to 5-hydroxymethylcytosine (5hmC) by ten-eleven translocation (Tet) proteins.3,4 Tet proteins can further oxidize 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC).5,6 Thus, cytosine residues in mammalian DNA occur in at least five forms (C, 5mC, 5hmC, 5fC, and 5caC),7–15 and the ability to recognize and differentiate the modification status of DNA cytosine residues is essential for the control of gene expression in mammals.
The best-known mammalian DNA-binding domains that recognize methylated cytosine are the methyl-binding domains (MBDs), recognizing fully methylated CpG dinucleotides,16,17 and the “SET and RING finger-associated” (SRA) domains, binding to hemimethylated CpG sites generated transiently by DNA replication.18,19 Both SRA domain proteins UHRF1 and UHRF2 and MBD domain proteins MeCP2, MBD3, and MBD4 have been suggested to bind 5hmC;20–24 however, the 5hmC binding by these proteins is not selective, and in some studies, there is no difference between 5mC and 5hmC binding.20,22 We previously showed that the SRA domain of UHRF1 and the MBD domain of MeCP2 have a preference, by factors of 10 and 5, respectively, for binding the methylated DNA over 5hmC-containing oligonucleotides of identical sequence, whereas an only 2-fold preference was observed for MBD3 and MBD4.25 Furthermore, 5hmC levels in mouse brain at three different ages were inversely correlated with MeCP2 dosage, suggesting binding of MeCP2 to 5mC may protect the conversion of 5mC to 5hmC by Tet proteins.26 A high selectivity of 5hmC over 5mC and C is exemplified by a bacterial modification-dependent restriction endonuclease AbaSI with a 500:1 5hmC:5mC relative selectivity.27 In addition, the mammalian thymine DNA glycosylase excises the mismatched base as well as removing 5fC and 5caC, but not 5mC and 5hmC, when paired with a guanine.6,28,29
Zfp57 belongs to the family of Krüppel-associated box (KRAB) domain-containing C2H2 zinc finger (ZnF) transcription factors.30,31 KRAB ZnF transcription factors (KRAB-ZnFs) act mostly as chromatin-modulating transcription repressors,32 and the family has grown greatly during vertebrate evolution.33 Point mutations within the DNA-binding domain of Zfp57 have been found in patients with transient neonatal diabetes,30 and those mutations abolish DNA binding activity.34 Zfp57 recognizes the methylated CpG within a specific sequence GMGGCA (M = 5mC) (the sequence of the opposite strand, TGCCGC, was initially used35). Structural analysis of the complex between methylated DNA and the DNA-binding domain of mouse Zfp57, which has two ZnF motifs in tandem, revealed that the recognition of 5mCpG involves a 5mC-Arg-G triad.34,36 Biochemically, Zfp57 binds most strongly to methylated (5mC) DNA, with affinity decreasing in the following order: 5mC > 5hmC (or C) > 5fC ≫ 5caC (1:7–10:17:187).34
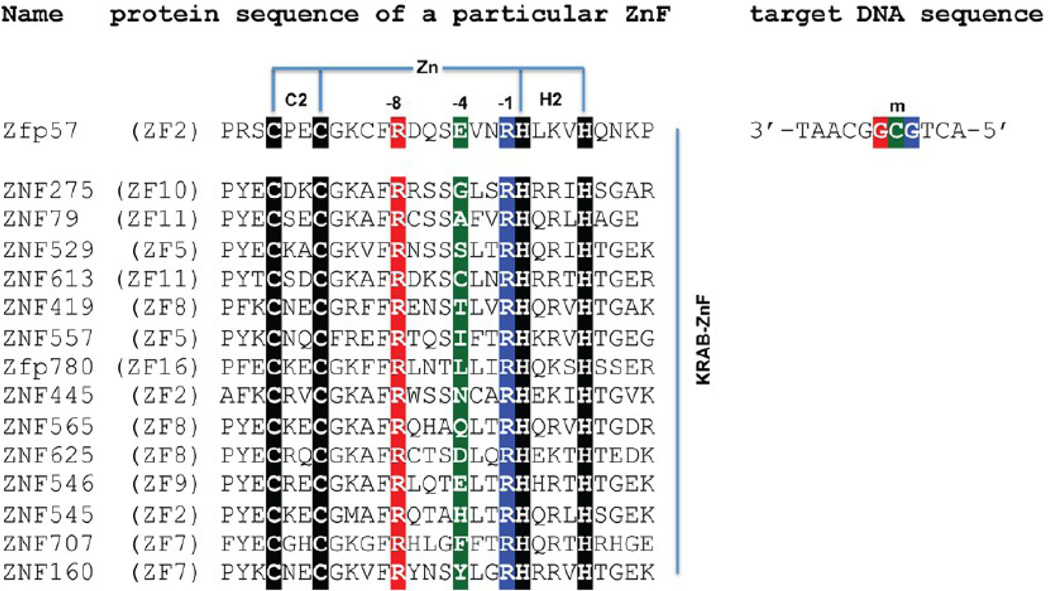
In classic C2H2 DNA-binding ZnF proteins, each finger is composed of two β-strands and one helix and generally recognizes three DNA base pairs.37,38 The DNA base contacts in the major groove are made by the side chains in the N-terminal portion of the helix together with the residue immediately preceding the helix. Because the first zinc-binding histidine (C2–4CX12H2–6H) is located almost always in the middle of the recognition helix, and the spacing between Cys2 and His2 is constant (12 residues), we use the amino acids at positions −1 to −8 (relative to the first zinc-binding histidine) in the following text to discuss the residues making base contacts. Most commonly, residues at position −1, −4, −7, or −8 (from the C- to-N-terminus) make contacts in the DNA major groove, and the identities of the amino acids determine the sequence specificity (from 5′ to 3′). In the second ZnF of Zfp57, Arg185 at position −1 (RH) makes direct base contact to the 5′ Gua, Glu182 at position −4 interacts with the central 5mC, and Arg178 at position −8 recognizes the 3′ Gua of methylated 5′-GCG-3′ (Figure 1).
Figure 1.
Sequence alignment of representatives of KRAB-C2H2 ZnF proteins with variation at position −4. We focus on the proteins that, like Zfp57, contain arginines at positions −1 and −8. The amino acids at positions −1, −4, and −8 and the primary DNA base recognition are highlighted with the same color code (red, green, and blue).
Using high-throughput sequencing to profile the mouse and human cortex from the fetus to young adult, Lister et al. revealed global epigenomic reconfiguration of 5hmC, the level of which is enriched in brain,7,8,39 during mammalian brain development.40 The genomic levels of 5fC and 5caC may also be similarly regulated, because both are derived from 5hmC oxidation. Although the global epigenomic profiles point to regulatory roles for 5mC and its oxidative derivatives (5hmC, 5fC, and 5caC) in the brain, the mechanisms by which these modification marks are read out to affect gene expression await elucidation. It will be essential to understand which DNA-binding factors are recruited to unmethylated, methylated, and hydroxymethylated DNA to mediate their regulatory functions.
Interestingly, KRAB-ZnF genes are also expressed at higher levels in embryonic cells and brain,33,41 so it seems entirely justifiable to ask the question of whether some of the KRAB-ZnF proteins might have gained the ability to bind the oxidative products of 5mC. Unfortunately, despite the large number of KRAB-ZnF proteins (in the hundreds), few of them have known biologic roles,42–48 and even fewer have known target DNA sequences. We searched the C2H2 zinc finger gene (SysZNF) database (http://lifecenter.sgst.cn/SysZNF/)49 for KRAB-ZnFs that, like Zfp57, contain arginines at positions −1 and −8. Among these, the amino acid at position −4 (corresponding to Glu182 of Zfp57) could be at least 14 different amino acids (Figure 1). Obviously, different amino acids at position −4 could recognize different DNA bases. Alternatively, we postulated that some of these amino acids could distinguish different modifications of cytosine at the C5 position in the DNA major groove, with 5mC carrying a hydrophobic methyl group, 5hmC carrying a hydroxyl oxygen with the potential to be a hydrogen bond donor, 5fC carrying a carbonyl oxygen with the potential to be a hydrogen bond acceptor, and 5caC carrying a carboxylate moiety with the potential to be both a hydrogen bond door and a hydrogen bond acceptor.
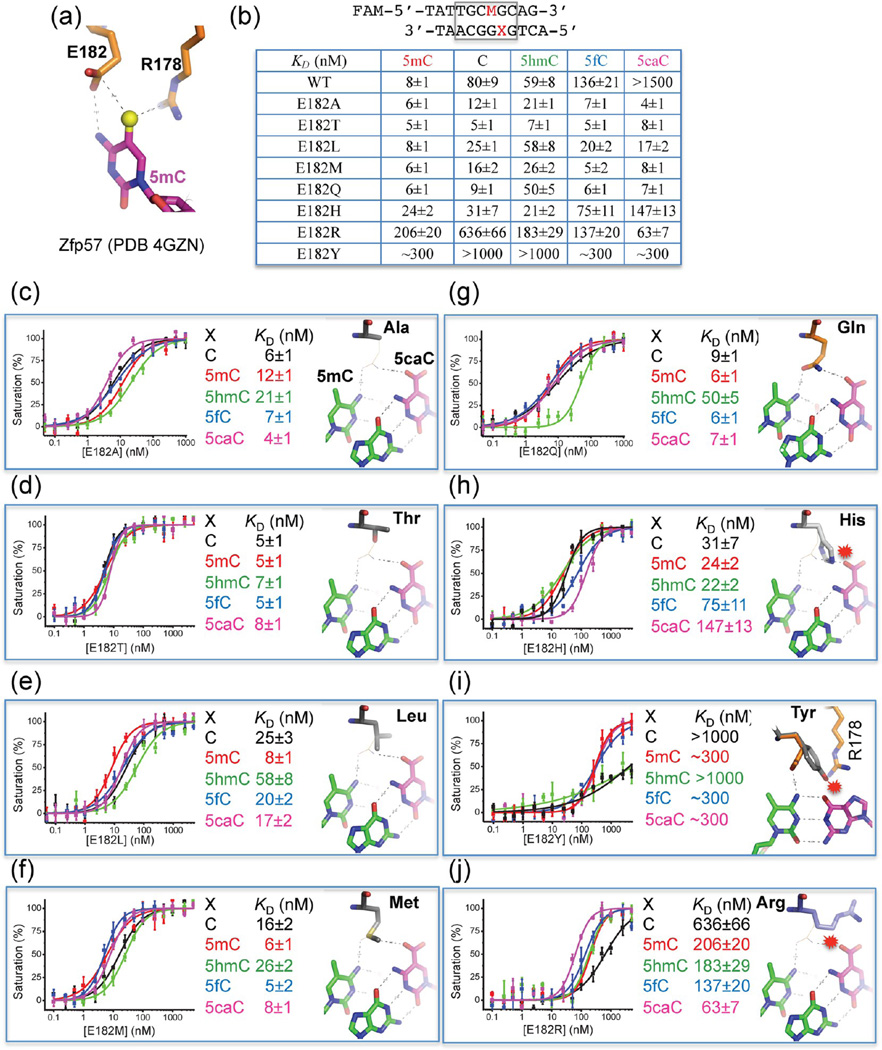
Here, using Zfp57 as a model system, we show that a single glutamate residue (Glu182 at position −4) in Zfp57 can critically discriminate against 5caC. Glu182 forms a van der Waals contact with the methyl group of 5mC, and one of its carboxylate oxygen atoms interacts with the exocyclic N4 amino group of the same 5mC base (Figure 2a).34 Substitution of Glu182 with its corresponding amide (E182Q) has no effect on 5mC DNA binding because the same interactions are maintained. Surprisingly, this E-to-Q mutant has a >200-fold higher binding affinity for 5caC DNA, resulting in a binding affinity for 5caC DNA comparable to that of the wild-type (WT) protein for 5mC. We show structurally that the uncharged amide group of E182Q interacts favorably with the carboxylate group of 5caC. This observation raises the possibility that some KRAB-ZnF proteins with other amino acids at the corresponding position might preferentially recognize the oxidative products of 5mC.
Figure 2.
Mutations of Glu182 in the binding site of 5mC recognition by Zfp57. (a) In Zfp57, in addition to Arg178, the side chain of Glu182 forms a van der Waals contact with the methyl group of 5mC and one of its carboxylate oxygen atoms interacts with the N4 atom of the same 5mC base. (b) Summary of DNA binding affinities of mutants for DNA substrates containing five different modification states. The sequences of FAM-labeled double-stranded oligonucleotides were shown. (c–j) Dissociation constants (KD) between Zfp57 mutants and double-stranded oligonucleotides containing a single CpG dinucleotide. For each mutant, a computational model was provided on the basis of the E182Q-5caC DNA structure. The mutated residue is colored gray, superimposed with the side chain of Gln182 as a thin line. The 5caC:G base pair is shown as 5caC colored magenta and the opposite Gua colored green. The 5mC, 5′ to the Gua, is also colored green. Base pairing hydrogen bonds are shown as dashed lines. The hydrogen bonds mediated by Gln182 are shown as dashed lines to the carboxylate moiety of 5caC and the exocyclic N4 amino group of 5mC. Minor clashes to the 5caC could occur for the E182H or E182R mutant (panels h and j). Serious clashes with protein side chains, either Arg178 (panel i) or Arg185 (not shown), could occur for the E182Y mutant, via the adoption of different side chain rotamer conformations.
EXPERIMENTAL PROCEDURES
Protein Purification
Proteins containing mouse Zfp57 residues 137–195 (pXC1127) and its mutants, E182A (pXC1150), E182T (pXC1242), E182L (pXC1151), E182M (pXC1240), E182Q (pXC1158), E182H (pXC1244), E182R (pXC1243), and E182Y (pXC1241), were prepared as previously described.34 For each mutant, recombinant GST-tagged protein was purified with Glutathione Sepharose 4B, and the GST tag was removed by incubating with PreScission protease overnight at 4 °C, resulting in an additional N-terminal Gly-Pro-Lys-Gly-Ser sequence. Protein was further purified to homogeneity by three columns of HiTrap-Q, HiTrap-SP, and Superdex-200 (16/60). The yields of the mutant proteins were lower than that of the WT protein: ~87% for E182M, 85% for E182Q, 80% for E182Y, 73% for E182A, 54% for E182T, 51% for E182R, 30% for E182L, and 23% for E182H. The difference in yield mostly reflected protein expression level, and all purified proteins appeared to be stable.
Fluorescence-Based DNA Binding Assay
The DNA binding assays were performed by measuring fluorescence polarization (mP), in 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5% (v/v) glycerol, and 1 mM tris(2-carboxyethyl)-phosphine (TCEP) at room temperature using a Synergy 4 Microplate Reader (BioTek). Fluorescently labeled DNA probe (1 nM) and various amounts of Zfp57 protein (WT or mutants) with a final volume of 50 µL were incubated in a 384-well plate for 0.5 h before measurements were taken. The sequences of 6-carboxyfluorescein (FAM)-labeled double-stranded oligonucleotides were FAM-5′-TATTGCMGCAG-3′ and 3′-TAACGGXGTCA-5′ (where M = 5mC and X = C, 5mC, 5hmC, 5fC, or 5caC). Curves were fit individually using Origin version 7.5 (OriginLab). KD values were calculated as [mP] = [maximal mP] × [C]/(KD + [C]) + [baseline mP], where [mP] is millipolarization and [C] is protein concentration. The averaged KD and its standard error are reported. We note that in some cases (E182T), the KD values are on the same order of magnitude as the probe concentration (1 nM, the minimum needed for the instrument to accurately measure mP), which could result in slight underestimation of binding constants.
Crystallography
The purified E182Q protein was incubated with annealed oligonucleotides at an equimolar ratio for 1 h on ice before crystallization. The mutant E182Q–DNA complexes, in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2.5% glycerol, and 0.5 mM TCEP, were crystallized at 16 °C with mother liquor that contained 35% 2-methyl-2,4-pentanediol (MPD), 15% polyethylene glycol 8000, 100 mM CaCl2, and 100 mM acetate (pH 4.2). The E182Q–DNA complexes crystallized in space group P21, containing two complexes per crystallographic asymmetric unit.
The crystals were flash-frozen by being plunged into liquid nitrogen. X-ray diffraction data were collected at 100 K at the SER-CAT 22-ID beamline of the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). HKL200050 was used for X-ray diffraction data processing. Because an appropriate choice of resolution cut off is difficult,51–53 we truncated the high-resolution data at 0.97 Å, with Rmerge not exceeding ~0.6–0.8. The structure was determined by the molecular replacement method using the WT Zfp57–DNA complex structure (Protein Data Bank entry 4GZN) as the initial searching model, using PHASER.54 PHENIX55 and COOT56 were used for structural refinement. The ReadySet module of the PHENIX suite was used to add hydrogen atoms.55 The Dali server57 was used for structural comparison.
RESULTS
DNA Binding Study of Glu182 Mutants of Zfp57
To see the effects on cytosine modification discrimination, we mutated the negatively charged Glu182 of mouse Zfp57 to residues with small (Ala and Thr), hydrophobic (Leu and Met), corresponding amide (Gln), polar (His), aromatic (Tyr), and positively charged side chains (Arg), in the context of residues 137–19534 (Figure 2b). The majority of mutants examined lose selectivity of methylated over unmodified and oxidative derivatives, not via a decrease in the affinity of 5mC but via an increase in the affinity for the others. Only ~3-fold variation of affinities was observed for E182A, suggesting that the side chain of Glu182 is dispensable for methyl group recognition (Figure 2c). E182T exhibited an almost equal high affinity for all five states of the DNA cytosine residue (Figure 2d). E182L, E182M, and E182Q maintain the 4–8-fold discrimination against 5hmC but have relatively similar affinities for the four other forms (Figure 2e–g). E157H has ~3-fold reduced affinity for methylated DNA but ~10-fold higher affinity for 5caC, in comparison to those of the WT protein, thereby diminishing the selectivity against 5caC (from ~200-fold for the WT to ~6-fold) (Figure 2h). Unexpectedly, E182Y has a binding affinity for methylated DNA reduced by a factor of approximately 38 in comparison to that of WT protein (Figure 2i). On the other hand, introducing a positively charged arginine at position 182 (E182R) resulted in a protein that was selective for 5caC over all four other forms of cytosine, gaining an approximately 10-fold selectivity against unmodified cytosine and a 2–3-fold selectivity against 5mC, 5hmC, and 5fC (Figure 2j).
Atomic Structure of the Zfp57 E182Q Mutant in Complex with 5-Carboxylcytosine
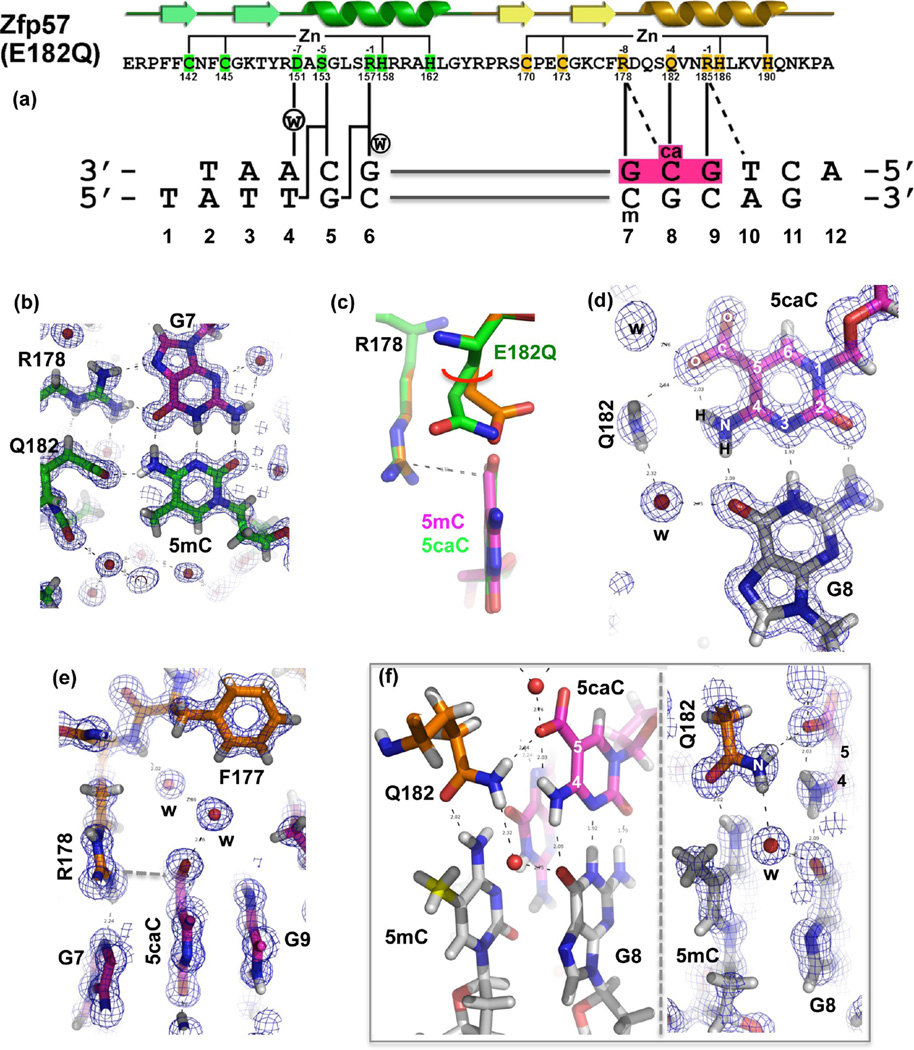
We decided to further analyze the E182Q mutant structurally because the E-to-Q mutation represents the smallest change from the WT (an amide group in place of the carboxylate), while the mutant (like E182A and E182T) can gain >200-fold binding affinity for 5caC without a change in the affinity of 5mC. We crystallized the E182Q mutant protein in complex with 5caC-containing DNA and determined the structure to a resolution of 0.97 Å (Table 1 and Figure 3). The DNA used for cocrystallization was a 10 bp oligonucleotide containing the recognition element, with a 5′-overhanging thymine on the T strand and a 5′-overhanging adenine on the A strand (Figure 3a). The oligonucleotide has a 5mC on the T strand and a 5caC on the A strand for the following reasons. (i) Zfp57 recognizes the 5mC residues of the two DNA strands asymmetrically: the 5mC of the A strand is contacted by the protein (Arg178 and Glu182), while the 5mC of the T strand is surrounded by a layer of water molecules34 (Figure 3b). (ii) Tet-mediated 5mC oxidation (most probably) generates asymmetric hydroxymethylation at CpG sequences,9,11 with one strand hydroxymethylated (5hmC). Because 5caC is derived from further oxidations of 5hmC,5,6 we replaced only 5mC of the A strand with 5caC for structural analysis (Figure 3a).
Table 1.
X-ray Data Collection and Refinement Statistics
| Data Collectiona | |
| space group | P21 |
| cell dimensions | |
| a, b, c (Å) | 36.388, 96.093, 36.408 |
| α, γ (deg) | 90 |
| β (deg) | 113.145 |
| beamline | APS 22-ID (SERCAT) |
| detector–crystal distance (mm) | 125 |
| wavelength (Å) | 0.8 |
| exposure time (s/image) | 1 |
| rotation degree (deg/image) | 1 |
| total no. of images | 140 |
| resolution (Å) | 33.5–0.97 (1.00–0.97) |
| Rmerge (%) | 0.070 (0.648) |
| 〈I〉/σ(I) | 11.7 (1.4) |
| completeness (%) | 95.7 (92.7) |
| redundancy | 2.6 (2.2) |
| no. of observed reflections | 331787 |
| no. of unique reflections | 129957 (12599) |
| Refinement | |
| Rwork/Rfree (%) | 13.01/14.26 |
| no. of atoms | |
| protein | 1177 (2005 with hydrogens) |
| DNA | 1076 (1524 with hydrogens) |
| water | 501 |
| others | 54 (122 with hydrogens) (4 Zn2+, 2 Ca2+, 4 acetate molecules, 4 MPD molecules) |
| overall B factor (Å2) | 13.7 |
| B factor for protein (Å2) | 11.4 |
| B factor for DNA (Å2) | 13.2 |
| B factor for water (Å2) | 22.2 |
| B factor for others (Å2) | 21.6 |
| root-mean-square deviation | |
| bond lengths (Å) | 0.009 |
| bond angles (deg) | 1.815 |
Data for the highest-resolution shell are given in parentheses.
Figure 3.
Atomic structure of Zfp57 E182Q in complex with 5caC DNA. (a) The secondary structure elements (arrows for β-strands and ribbons for α-helices) are shown above the protein sequence. The amino acid positions highlighted are responsible for Zn ligand binding (C2H2) and DNA base-specific interactions (−1 to −8 relative to the first Zn-binding His). (b) Details of E182Q base-specific interactions for the 5mC:G base pair. Electron densities (2Fo − Fc) contoured at 1σ above the mean are shown. (c) Structural comparison of Zfp57 WT (orange) and the E182Q mutant (green) interacting with 5mC and 5caC, respectively. (d) Details of E182Q base-specific interactions for the 5caC:G base pair. (e) Arg178 forms an ion pair with the carboxylate moiety of 5caC. (f) The side chain of Gln182 links 5caC (via the amide group) of the top strand to 5mC (via the carbonyl oxygen) of the bottom strand. The left panel illustrates interactions of Gln182 with the CpG duplex, and the right panel shows a superimposition with the electron density.
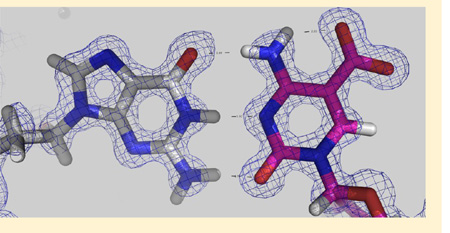
The atomic resolution (0.97 Å) of our crystallographic data allowed us to include hydrogen atoms in the final stage of the structural refinement, resulting in a structure with an overall crystallographic thermal B factor of 13.7 Å2. Sequential refinement and model building followed the order of adding water molecules (Rwork/Rfree = 0.2189/0.2203), the anisotropic B factor refinement (0.1523/0.1605), and the alternative conformations (0.1301/0.1426). The atomic-resolution electron density allowed us to include a large number of water molecules (~500) in addition to positioning every atom of the DNA nucleotides and protein residues. The mutant complex structure resembles closely that of the WT Zfp57–DNA complex, with an overall root-mean-square deviation of 0.1 Å. Among the side chains involved in DNA base-specific interactions, E182Q undergoes the largest conformational change upon binding of 5caC (as compared to 5mC) DNA. Superimposing the two structures (mutant and WT) reveals that E182Q moves from the 5mC-interacting conformation to the 5caC-interacting conformation via an ~25° rotation of both side chain torsion angles χ1 and χ2 (Figure 3c).
Recognition of 5caC
The 5caC is involved in a network of intra- and intermolecular interactions. The polar groups of the 5caC base along the Watson–Crick edge are all hydrogen bonded with the guanine of the opposite strand (Figure 3d). An intramolecular hydrogen bond is formed between the exocyclic N4 amino group and one of the carboxylate oxygen atoms at C5 of 5caC (Figure 3d). The same carboxylate oxygen atom forms a hydrogen bond with the amide group of Gln182 (Figure 3d). In addition, an ionic interaction is formed between the negatively charged carboxylate group and the positively charged guanidino group of R178, which in turn makes bifurcated hydrogen bonding interactions with the 3′ guanine of the same strand (Figure 3e). The two carboxylate oxygen atoms of 5caC are further stabilized by water-mediated interactions (Figure 3e). Furthermore, the side chain of Q182 links 5caC to 5mC of the bottom strand (Figure 3f).
Taken together, we suggest that the side chain of Glu182 of Zfp57 (the size and the charge) negatively impacts the binding of unmodified cytosine as well as oxidative derivatives of 5mC, particularly the negatively charged 5caC. Indeed, introducing a positively charged arginine at position 182 (E182R) resulted in a protein that was selective for 5caC over all four other forms of cytosine, gaining approximately 10-fold selectivity against unmodified cytosine and 2–3-fold selectivity against 5mC, 5hmC, and 5fC (Figure 2j). This reversal of selectivity is achieved via significantly increasing the affinity for 5caC (~24-fold) as well as decreasing the affinities for other cytosine forms, particularly 5mC (~26-fold), in comparison to that of the WT protein.
Computational Modeling of the Glu182 Mutants
To understand the lack of differences in the measured binding affinity of Glu182 mutants, particularly those with smaller side chains, we further performed computational modeling of the mutants based on the structures of the WT–5mC DNA complex34 as well as the E182Q–5caC DNA complex. Because the two structures are highly similar except for the differences mentioned above (Figure 3c), we focus our comparisons on that of the E182Q–5caC DNA complex. Substituting an alanine in the place of Glu182 (E182A) resulted in a void space between the Ala Cβ atom and the carboxylate group of 5caC (separated by ~4.5 Å) (Figure 2c). The space could be easily filled with water molecules, as the carboxylate group is already involved in water-mediated interactions (Figure 3e). The hydroxyl oxygen atom of the E182T mutant is approximately 3.6 Å from the 5caC carboxylate group (Figure 2d); a water molecule between them could form a water-mediated hydrogen bond. Replacement of Glu182 with either Leu or Met, each of which is similar in size to E/Q, could form a van der Waals contact or unconventional C–H⋯O (Leu) or S–H⋯O (Met) type of hydrogen bond to one of the carboxylate oxygen atoms (Figure 2e,f). It seems that it is the size of the amino acid, either similar to or smaller than that of E/Q, that is required to maintain the binding affinity, rather than the ability to form direct interaction with 5mC or 5caC, as the five mutants (A, T, L, M, and Q) have little effect on binding either 5mC or 5caC (Figure 2c–g). Interestingly, the E182T mutant seems to selectively bind 5hmC stronger than E182A and E182M by ~3–4-fold and E182Q and E182L by ~7–8-fold.
On the other hand, the replacement by His or Tyr had variable effects. The less flexible imidazole ring would crash into the DNA base, resulting in reduced binding affinity (Figure 2h). The bulkier aromatic ring of tyrosine would collide with either 3′ Gua-interacting Arg178 (Figure 2i) or 5′ Gua-interacting Arg185 via an alternative rotamer conformation (not shown). A methyl–π interaction has recently been observed in ZNF217 with the aromatic ring of a tyrosine at the corresponding −4 position contacting a thymine methyl group in a TpG dinucleotide.58 However, in ZNF217, the residue corresponding to Arg185 at position −1 is an isoleucine, which does not directly interact with DNA and leaves space to accommodate the tyrosine at position −4. Finally, the larger but more flexible side chain of arginine (E182R) could be modeled to occupy the space vacated by the carboxylate-interacting water molecules (Figure 2j), allowing a more favorable interaction to form between the positively charged guanidino group and the negatively charged carboxylate.
DISCUSSION
Using Zfp57 as an example, we showed that the identity of the amino acid at position 182 (corresponding to position −4 of the second ZnF of Zfp57) might be important in differential binding of 5mC and its oxidative derivatives in the sequence context of GCG. The WT protein (Glu182) binds with decreasing affinity in the following order: 5mC > 5hmC > 5fC ≫ 5caC. The negatively charged side chain carboxylate group of Glu182 might be critical in discrimination against the negatively charged carboxylate moiety of 5caC. Via the introduction of a positively charged arginine at position 182 (E182R), the selectivity is reversed in the following order: 5caC > 5fC > 5hmC > 5mC ≫ C [although the magnitudes of the differences are small among the modifications (<4-fold) and up to 10-fold between 5caC and C (Figure 2j)]. Interestingly, the mutants could accommodate either 5mC or 5caC with similar or smaller side chains compared to that of Glu182 (Q, M, L, T, and A). Structurally, the uncharged amide group of E182Q interacts with the carboxylate group of 5caC. It is possible that multiple changes (surrounding position −4) in the recognition helix might be needed to achieve selectivity (involving structural plasticity and the physical and chemical bases for specificity). The situation is more complicated by a large number of water-mediated secondary contacts, observed in the current atomic-resolution structure in the protein–DNA interface. Such results highlight the much more difficult problem of trying to develop a code that could predict an optimal set of contacts with DNA cytosine modifications.
ACKNOWLEDGMENTS
We thank Brenda Baker at the organic synthesis unit of New England Biolabs for synthesizing the oligonucleotides and Dr. John R. Horton and Dr. Eric Ortlund for comments on the manuscript. X-ray data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source. The Emory University School of Medicine supported the use of SER-CAT beamlines. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
Funding
The National Institutes of Health (Grant GM049245-20) supported this work. X.C. is a Georgia Research Alliance Eminent Scholar.
Footnotes
ASSOCIATED CONTENT
Accession Codes
The X-ray structures (coordinates and structure factor files) of the Zfp57 (E182Q)–5caC DNA complex have been submitted to the Protein Data Bank as entry 4M9V.
Author Contributions
Y.L. performed crystallographic experiments and DNA binding assays. Y.O.O. performed the protein purification of mutant proteins. All authors were involved in analyzing data and preparing the manuscript.
The authors declare no competing financial interest.
REFERENCES
- 1.Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 2.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 3.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 11.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raiber EA, Beraldi D, Ficz G, Burgess HE, Branco MR, Murat P, Oxley D, Booth MJ, Reik W, Balasubramanian S. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase. Genome Biol. 2012;13:R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Z, Terragni J, Borgaro JG, Liu Y, Yu L, Guan S, Wang H, Sun D, Cheng X, Zhu Z, Pradhan S, Zheng Y. High-resolution enzymatic mapping of genomic 5-hydroxymethylcytosine in mouse embryonic stem cells. Cell Rep. 2013;3:567–576. doi: 10.1016/j.celrep.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, Zhang K, Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, Gao J, Liu P, Li L, Xu GL, Jin P, He C. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhasarathy A, Wade PA. The MBD protein family: Reading an epigenetic mark? Mutat. Res. 2008;647:39–43. doi: 10.1016/j.mrfmmm.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto H, Horton JR, Zhang X, Cheng X. UHRF1, a modular multi-domain protein, regulates replicationcoupled crosstalk between DNA methylation and histone modifications. Epigenetics. 2009;4:8–14. doi: 10.4161/epi.4.1.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharif J, Koseki H. Recruitment of Dnmt1: Roles of the SRA protein Np95 (Uhrf1) and other factors. Prog. Mol. Biol. Transl. Sci. 2011;101:289–310. doi: 10.1016/B978-0-12-387685-0.00008-1. [DOI] [PubMed] [Google Scholar]
- 20.Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, Leonhardt H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 Binds to 5hmC Enriched within Active Genes and Accessible Chromatin in the Nervous System. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otani J, Arita K, Kato T, Kinoshita M, Kimura H, Suetake I, Tajima S, Ariyoshi M, Shirakawa M. Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4. J. Biol. Chem. 2013;288:6351–6362. doi: 10.1074/jbc.M112.431098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, Vermeulen M. Dynamic readers for 5-(hydroxy)-methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, Vasanthakumar A, Godley LA, Chang Q, Cheng X, He C, Jin P. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Guan S, Quimby A, Cohen-Karni D, Pradhan S, Wilson G, Roberts RJ, Zhu Z, Zheng Y. Comparative characterization of the PvuRts1I family of restriction enzymes and their application in mapping genomic 5-hydroxymethylcytosine. Nucleic Acids Res. 2011;39:9294–9305. doi: 10.1093/nar/gkr607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto H, Hong S, Bhagwat AS, Zhang X, Cheng X. Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: Its structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012;40:10203–10214. doi: 10.1093/nar/gks845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, Hahnemann JM, Kordonouri O, Masoud AF, Oestergaard E, Storr J, Ellard S, Hattersley AT, Robinson DO, Temple IK. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat. Genet. 2008;40:949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- 31.Collins T, Stone JR, Williams AJ. All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meylan S, Groner AC, Ambrosini G, Malani N, Quenneville S, Zangger N, Kapopoulou A, Kauzlaric A, Rougemont J, Ciuffi A, Bushman FD, Bucher P, Trono D. A gene-rich, transcriptionally active environment and the pre-deposition of repressive marks are predictive of susceptibility to KRAB/KAP1-mediated silencing. BMC Genomics. 2011;12:378. doi: 10.1186/1471-2164-12-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vinogradov AE. Human more complex than mouse at cellular level. PLoS One. 2012;7:e41753. doi: 10.1371/journal.pone.0041753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Toh H, Sasaki H, Zhang X, Cheng X. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26:2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, Trono D. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Zhang X, Blumenthal RM, Cheng X. A common mode of recognition for methylated CpG. Trends Biochem. Sci. 2013;38:177–183. doi: 10.1016/j.tibs.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 38.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 39.Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, Irimia M, Jia P, Ptak C, Xia M, Tice R, Tochigi M, Morera S, Nazarians A, Belsham D, Wong AH, Blencowe BJ, Wang SC, Kapranov P, Kustra R, Labrie V, Klimasauskas S, Petronis A. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol. 2012;19:1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, Behrens MM, Ecker JR. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep. 2012;2:766–773. doi: 10.1016/j.celrep.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Garcia MJ, Shibata M, Anderson KV. Chato, a KRAB zinc-finger protein, regulates convergent extension in the mouse embryo. Development. 2008;135:3053–3062. doi: 10.1242/dev.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 44.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas JH, Emerson RO. Evolution of C2H2-zinc finger genes revisited. BMC Evol. Biol. 2009;9:51. doi: 10.1186/1471-2148-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS One. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krebs CJ, Schultz DC, Robins DM. The KRAB zinc finger protein RSL1 regulates sex- and tissue-specific promoter methylation and dynamic hormone-responsive chromatin configuration. Mol. Cell. Biol. 2012;32:3732–3742. doi: 10.1128/MCB.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chien HC, Wang HY, Su YN, Lai KY, Lu LC, Chen PC, Tsai SF, Wu CI, Hsieh WS, Shen CK. Targeted disruption in mice of a neural stem cell-maintaining, KRAB-Zn finger-encoding gene that has rapidly evolved in the human lineage. PLoS One. 2012;7:e47481. doi: 10.1371/journal.pone.0047481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding G, Lorenz P, Kreutzer M, Li Y, Thiesen HJ. SysZNF: The C2H2 zinc finger gene database. Nucleic Acids Res. 2009;37:D267–D273. doi: 10.1093/nar/gkn782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr. 2003;A59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 51.Evans PR. An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. 2011;D67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. 2013;D69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vandevenne M, Jacques DA, Artuz C, Nguyen CD, Kwan AH, Segal DJ, Matthews JM, Crossley M, Guss JM, Mackay JP. New Insights into DNA Recognition by Zinc Fingers Revealed by Structural Analysis of the Oncoprotein ZNF217. J. Biol. Chem. 2013;288:10616–10627. doi: 10.1074/jbc.M112.441451. [DOI] [PMC free article] [PubMed] [Google Scholar]