Abstract
The process of the colonization of the New World that occurred centuries ago served as a natural experiment, creating unique combinations of genetic material in newly formed admixed populations. The identification and genotyping of ancestry informative markers (AIMs) have allowed for the estimation of proportions of ancestral parental populations among individuals in a sample through the genetic admixture approach. These admixture estimates have been used in different ways to understand the genetic contributions to individual variation in obesity and body composition parameters, particularly among diverse admixed groups known to differ in obesity prevalence within the United States. Although progress has been made through the use of genetic admixture approaches, future investigations are needed in order to explore the interaction of environmental factors with the degree of genetic ancestry in individuals. A challenge to confront at this time would be to further stratify and define environments in progressively more granular terms, including nutrients, muscle biology, stress responses at the cellular level, and the social and built environments.
Keywords: Genetic admixture, obesity, body composition, race/ethnicity, Ancestry Informative Markers
Introduction
The genetic richness of individuals in the XXI Century has emerged from millennia of influential genetic drifting, selection and environmental pressures. The advantages that genes conferred to individuals for survival in diverse geographical regions have been recently challenged by the environmental changes that have occurred through advances in technology, agricultural and agrarian interchanges, accessibility to resources and from the continuous intermixing of individuals that has been recently influenced by intercontinental traveling. Initially, the colonization of the New World served as a natural outbred experiment where long-geographically-separated populations, each with autochthonous genetic makeup, intermated to produce individuals with unique combinations of genetic material. The degree to which the genome of a particular individual from an admixed population is composed of DNA segments descended from one particular parental population relative to others is referred to as genetic admixture.
Today, advances in molecular and population genetics have allowed the identification of ancestry informative markers (AIMs), or DNA variants of differential frequency among those long-geographically-separated parental populations. These AIMs can be quantified as a composite representation of the ancestral information of an individual, based on a finite number of genetic markers. This quantification provides an estimate of an individual's genome that likely corresponds to one ancestral parental population vs. another. The weighted estimation of this proportion, scaled from 0 – 100, is referred to as “estimates” of genetic admixture.
Genetic admixture approaches have been used in research for various purposes: as surrogates of the biological component of race, to elucidate the relationship between ancestry and disease risk, to control for the statistical effects of population stratification in genetic association studies, and to identify genes contributing to phenotypic variation among certain populations. The associations between genetic admixture and disease-related outcomes have been reported in the literature for diseases such as cancer [1-3], cardiovascular disease [4-8] and type 2 diabetes [9-12]. These and other conditions are related to obesity, and given its increased prevalence among admixed populations, an interest has arisen in identifying how admixture may contribute to the etiology of obesity in the population. In this paper, we review the most relevant evidence supporting associations of genetic admixture with obesity, body composition parameters, and obesity-related comorbidities, providing a perspective of the present contributions of admixture toward the understanding of population differences in obesity, and providing some future implications of this approach.
Ancestry Informative Markers and Genetic Admixture Estimation
Initially, the identification of AIMs was based on the difference of the frequency of one allele in a given population from the frequency of the same allele in another population, i.e. delta (δ). AIMs are not limited to autosomal markers, and ancestral markers have also been found in sex chromosomes and in mitochondrial DNA (mtDNA). Given that mtDNA is inherited only through the maternal line and is not subject to recombination, it has been used for assessing population patterns such as sex-biased admixture [13]. Taking into consideration the historical context of an admixed population, AIMs are used to estimate ancestral proportions of each appropriate parental population (e.g. European, West African, and Amerindian) within individuals.
There is not a general consensus about the number of AIMs required to precisely estimate genetic admixture. However, the amount of markers needed seems to be influenced by the type of analysis or overall purpose of the genetic admixture approach. For example, if the purpose is just to estimate parental proportions in individuals to account for the biodiversity of a sample, some investigators have suggested that 15 SNPS can suffice [14]. If the objective is to use admixture estimates to determine the association between a phenotype and admixture proportions or to control against confounding in genetic association studies with admixed samples, it has been suggested that 100 AIMs should be enough [15]. In these cases, the individuals’ estimates of genetic admixture are obtained primarily through the use of maximum likelihood [16] or Bayesian [17] approaches. With technological advances increasing the genotyping of single nucleotide polymorphisms (SNPs), statistical techniques have been applied to create genetic clusters that capture human variation [18]. Consequently, in genome-wide association studies (GWAS) where there is already enough ancestral information in the available genetic data, principal component analyses become an alternative way to account for the confounding effect of ancestry [19]. A relevant use for the inclusion of admixture approaches in statistical analysis for gene mapping is the prevention of Type I and Type II errors. The work of Halder et al. [20] demonstrated that false positive and false negative associations between phenotypes and admixture proportions would be reduced with the use of large AIM panels.
Genetic Admixture and Obesity-Related Traits
Estimating genetic admixture has allowed researchers in the past two decades to identify the relationship between genetic ancestry with obesity-related traits, providing scientific evidence to long-lasting hypotheses of ancestral background influencing disease development in certain populations [21-23]. The most accessible parameter for the evaluation of admixture and obesity has been body mass index (BMI = kg/m2), which has been associated with European, West African and Amerindian admixture in various studies within diverse racial/ethnic groups. Table 1 demonstrates reported associations between BMI with European, West African or Amerindian admixture in the past 10 years, along with the direction of the association and the types of parental populations considered for admixture estimation in each study. It is interesting that some studies demonstrate that admixture estimates from one parental population positively relate to BMI, while others report a negative association. This level of discrepancy appears to be affected by the number of parental populations considered in the study, the ethnicity and gender of the study sample, or the stage of lifespan of participants in the sample.
Table 1.
Associations between BMI (body mass index = kg/m2) and European, African and Amerindian admixture since 2003.
| Admixture Associated with BMI | Study Population | Direction of Association with BMI | Ancestry Measured | Reference | ||
|---|---|---|---|---|---|---|
| Eu | Af | Am | ||||
| European Admixture | Mexican American men and women | + | × | × | × | Tang 2006 [62] |
| Native American men and women | - | × | × | × | Klimentidis 2009 [33] | |
| African American men and women | - | × | × | Cheng 2010 [27] | ||
| Women in Brazil over 60 yrs | + | × | × | × | Lins 2012 [66] | |
| African American and Hispanic American women | - | × | × | × | Nassir 2012 [32] | |
| African Admixture | African American women | + | × | × | Fernandez 2003 [25] | |
| African American men and women | + | × | × | × | Tang 2006 [62] | |
| Women from the Dominican Republic | + | × | × | × | Tajima 2006 [67] | |
| African American and Hispanic American women | + | × | × | × | Nassir 2012 [32] | |
| African American women | + | × | × | × | Nassir 2012 [32] | |
| Amerindian Admixture | Mexican American men and women | - | × | × | × | Tang 2006 [62] |
| African American and Hispanic American women | - | × | × | × | Nassir 2012 [32] | |
*Eu = European, Af = African, Am = Amerindian.
Given that similar levels of BMI among individuals of the same racial/ethnic group do not translate to equivalent levels of body fat [24], researchers have explored the association of admixture estimates with other body composition parameters, particularly in measures related to bone, lean, and fat. Among African Americans (AA) in the US, African admixture has been associated with fat mass, lean mass, and bone mineral density in premenopausal women [25]; and European admixture has been associated with bone mineral density in the femoral neck (women only), bone mineral density in the trabecular spine, appendicular and total lean mass, and total body fat [26]. In addition, levels of European ancestry in AA has been associated with subscapular skinfold thickness and waist circumference in a cohort of AA adults aged 45-64 [27].
The role of genetic admixture in body composition has also been studied in other populations. In a multiethnic sample of children of African-, European-, and Hispanic-American descent, West African admixture was found to be negatively associated with fat mass, total abdominal tissue, intra-abdominal adipose tissue, and subcutaneous abdominal adipose tissue while positively associated with bone mineral content [28]. In a sample of post-menopausal Puerto Rican women, West African admixture was associated with bone mineral density and lower visceral adiposity [29,30]. On the other hand, Amerindian admixture has been associated with higher levels of body fat in children [31], fat distribution in Hispanic-American women [32], and percent body fat in self-identified Native Americans [33].
Estimates of genetic admixture have also been used to evaluate the contributions or the mediation of ancestral background in physiological aspects known to mediate fat accumulation, including insulin sensitivity [10,11,34,35], physical activity and fitness [31,36], dietary factors [37], and IGF-1 [38]. While these studies provide insight into the possible applications of genetic admixture to the complex nature of fat accumulation, much work is still needed to identify the potential mechanistic role of ancestral genetics in obesity-related traits.
Admixture Mapping
The process of admixture has served as a natural experiment that created differences in allele frequencies that can provide variation for mapping of disease traits, as proposed elsewhere [39]. Admixture mapping has focused on the identification of AIMs contributing to disease. Given that the frequencies of particular AIMs differ across admixed populations, genome-wide admixture panels have been designed for AA [24,27,40,41] and Hispanic/Latino populations [30,42]. While groups like Asian American have been less represented in admixture mapping efforts [43], researchers have provided maps for some admixed groups within Asia, such as the Uyghur ethnic group of East/Central Asia [44].
Most of the reported results for admixture mapping have been in AA, likely pioneered by the associations between BMI and genes FY-Null (1q23.2), AT3 (1q25.1), DRD2TAQD (11q23.2) and GNB3 (12p13.31) [25]. This evidence was followed by the identification of chromosome locations 3q29, 5q14 and 15q26 and BMI [45]. Most recently, two loci influencing BMI were identified on Chromosome X (Xq25 and Xq13.1) and a third locus at 5q13.3 in a sample of 15,280 AA [46], and a year later an association was reported between a locus on 2p23.3 and BMI and weight which appeared to decrease BMI by approximately 0.92 kg/m2 in AA [27]. Last year, SNP rs631465 of the gene F2RL1 on chromosome 5 was reported to be associated with BMI, also in AA [47]. Admixture mapping in other populations are scarce; however, an association between BMI and the ABCA1/R230C variant in a Mexican population has been reported by Villareal-Molina et al. [48]
Genetic admixture and environmental factors
Applications of genetic admixture and admixture mapping, as well as heritability estimates for obesity ranging from 30 to 70% [36], clearly support a genetic component predisposing for BMI and body composition parameters. When taking into consideration that obesity prevalence has skyrocketed in recent decades with no evidence of a dramatic change in frequencies or mutations of the population's gene pool, it seems inevitably necessary to contemplate the powerful influence of the environment. Today it is generally believed that a large component of phenotypic variability in obesity traits responds not only to the individual contributions of predisposing genes and obesogenic environments, but also to the extent to which such genes interact with the environment. These interactions are sometimes difficult to interpret due to the conflicting inferences from statistical methodologies that may or may not reflect a biological process, resulting in a mathematical artifact. Consequently, it is imperative that a biological framework is developed prior to statistical evaluation, taking into account that an interaction may have a different meaning according to the variables measured and the context of the experiment.
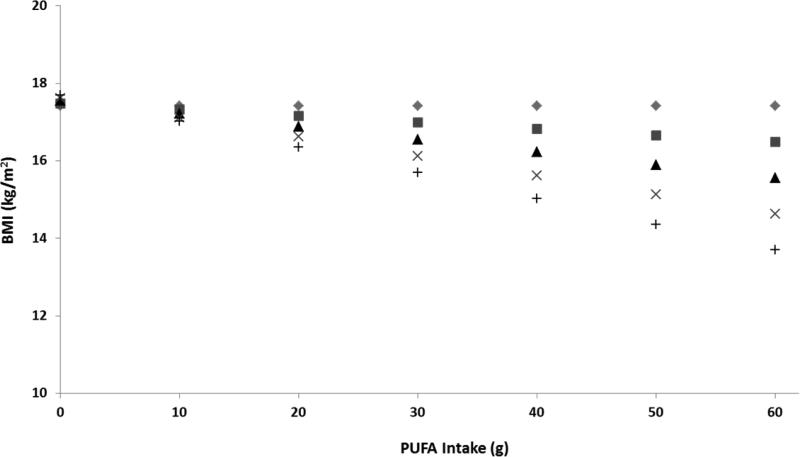
Although the influence of gene-environment interactions in obesity is poorly understood [25], it would be naïve to think that environmental factors would not mediate ancestral-related genetic influences, whether identified through admixture mapping, genetic admixture associations, or a significant association found in a particular admixed group. Insight into the role of environment in common genetic ancestry is best exemplified by the lower BMI of Pima Indians living in traditional environments in Mexico compared to those Pima Indians with high obesity prevalence living in obesogenic environments in the US [49]. More recent research has included environmental parameters in studies evaluating the role of genetic admixture in obesity. For example, Klimentidis et al investigated the interaction among adiposity, genetic admixture, and environmental factors represented by family income, parental education and activity level. Studying a sample of Native Americans and Hispanics in the US, it was found that although family income and admixture were correlated, European admixture better explained the variation in adiposity than family income, parental education, or activity level [33]. As we further stratified and define environments in progressively more granular terms, research has started to explore the interactions of nutrients with genetic admixture. For example, Figure 1 demonstrates our preliminary data of a significant interaction between polyunsaturated fatty acid (PUFA) intake and Amerindian admixture with BMI. As PUFA intake increases, BMI was simultaneously negatively associated with increasing levels of Amerindian admixture.
Figure 1.
Association of BMI and the interaction of PUFA intake and Amerindian admixture in a multiethnic sample of children. Predicted BMI calculated from multiple variable linear regression model including Amerindian admixture, PUFA intake, interaction of Amerindian admixture and PUFA intake controlled for age, sex, ethnicity, tanner stage, total energy intake, and socioeconomic status. PUFA intake and Amerindian admixture interaction was significant (p = 0.0471). Symbols represent the following levels of Amerindian admixture  (0%),
(0%),  (25%),
(25%),  (50%),
(50%),  (75%) and + (100%).
(75%) and + (100%).
The understanding of how genetic admixture interacts with environmental factors to impact obesity-related traits is certainly an area of further exploration. However, investigating these interactions will require (a) a priori identification of which environmental parameters and gene(s) will be considered for the analyses, (b) methodologies to allow the simultaneous consideration of multiple genes and perhaps multiple environmental variables, and/or (c) an effort to overcome the prevalent assumption of genetic and environmental homogeneity mistakenly assumed in many studies evaluating admixed populations.
Genetic admixture and heritability
Although obesity is a heritable trait, results from GWAS have only explained a small portion of the phenotypic variance of BMI given the small effects of identified loci [50]. Research has demonstrated that genetic admixture explains a portion of the heritability of obesity traits [27,33,51]; however, there has not been an approach that estimates heritability by integrating the proportion explained by admixture with that explained by GWAS. Obesity related traits are complex and given their polygenic nature it is probable that a greater proportion of the missing heritability could be explained by approaches that integrate information from multiple SNPS rather than approaches evaluating the contributions of individual SNPS. This principle has been adapted by the implementation of whole-genome marker-enabled prediction (WGP) approaches where GWAS SNP matrices are used to predict the genetic contribution to a trait [52-54]. This approach has been applied to explain 40% of the variance of height compared to the 3% of the variance explained by GWAS alone [55,56], and has been employed to look at other complex traits such as BMI [57] and blood lipids [58]. However, inferences from the prediction of this approach are limited by the need to have a sample that is proportionate to or greater than the number of SNPs utilized for the prediction [59].
Genetic admixture and race/ethnic classification
Throughout human history, individuals have been categorized according to social structures, biological traits, and/or levels of empowerment, mostly by a concept known as race/ethnicity. Race/ethnicity has proven to be a changing concept that has taken numerous permutations, integrating biological and non-biological factors that have traditionally divided individuals into distinct groups [60]. Today, in most epidemiological studies, individuals’ classifications into a particular group rely upon their own self-identification, encompassing a diverse conglomerate of socio-economic status, cultural practices, behavioral preferences, and ancestral backgrounds.
Given the observed differences in obesity prevalence and body composition parameters among racial/ethnic groups, there has been a genuine interest to understand the role of genetic and environmental contributors to racial/ethnic classification. The attempts to “decompose” race/ethnicity into genetic and non-genetic factors promises that understanding the basis of group differences, preventive strategies could be designed to overcome disparities in obesity and its related comorbidities. Several studies have suggested that a synergistic relationship between genetic admixture and the environment may explain observed disparities in disease rates among racial/ethnic groups [61,62]. Although genetic admixture estimates have been used to account for the biological aspects of race/ethnicity in obesity related traits, it is challenging to disentangle the relationships between ancestral genetic background and the historical, cultural, behavioral, and social aspects embedded into racial/ethnic classification. Thoughtful attention must be given toward experimental and statistical designs in order to disentangle the complex factors underlying “race/ethnicity”, as demonstrated by Casazza and colleagues who, when exploring the etiology of differences in certain body composition parameters, showed that genetic admixture and not race/ethnic classification arose as a significant predictor [63].
It is evident that the genetic admixture approach could more accurately explain some of the biological differences in obesity-related traits observed between and among racial/ethnic groups. However, genetic admixture must be used in conjunction with measures of environmental influences and even in conjunction with self-identified race/ethnicity to capture a more holistic view of the biological and non-biological relationships underlying health disparities among populations [38].
Future Implication and Conclusions
The utilization of the genetic admixture approach has provided insights into the understanding of the etiology of obesity-related traits, particularly among different populations. There are, however, some aspects that deserve scientific reflection and consideration for future research initiatives involving the use of genetic admixture in the understanding of the etiology of obesity and body composition parameters:
Given the limited success of admixture mapping in identifying common or rare variants explaining phenotypic variance in obesity-related traits, the investigation of gene and environment interactions becomes pivotal in the understanding of how ancestral genetic markers and/or background may impact obesity [64].
When studying health-related traits, the biodiversity and variability accounted for by the use of admixture estimates should not simply be replaced by the mere classification of race/ethnicity. Although self-identified race/ethnicity could act as a surrogate to provide information about the cultural and environmental factors impacting the phenotypic traits of an admixed population, its sole use negates the historical ancestry of individuals and will confound the understanding of the biological factors underlying obesity, its related traits, and its variation within and among populations.
As the intermating of admixed populations continues to occur, mapping of AIMs influencing obesity may not rely on currently used racial/ethnic classifications because self-identification will become less reflective of true ancestral background. This trend has already been described in a study where, among Hispanic Americans from the Multi-Ethnic Study for Atherosclerosis (MESA), ancestral genetic clusters representing race/ethnicity did not correspond with Hispanic classification in 30% of their Hispanic participants [65].
In order to explore the interaction of genetic ancestry and environmental factors, conscientious consideration will be needed to identify environmental variables that explain the etiology of obesity and its related traits. Admixed individuals in the XXI Century have been placed in an environment unfamiliar to their parental ancestors, providing a unique opportunity to understand how current obesogenic environments impact genetic ancestral makeup in phenotypic variability. The focus of environmental evaluation should expand from dietary intake and physical activity to aspects related to the social and built environment as well as physiological factors that reflect variability in response, such as macronutrients, muscle biology, stress responses at the cellular level, among others.
The utilization of genetic admixture has been limited by its clinical applicability. The future of genetic admixture as a clinical tool may rely on transferring statistical techniques and methods recently developed for gene discovery and gene prediction to genetic panels capturing ancestral background. For example, applying whole genome prediction approaches to genome-wide admixture panels may provide useful information regarding the extent to which genetic ancestry may influence obesity risk. However, such an approach may provide an indicator of the contribution of AIMs to the overall phenotypic variance (increasing prediction), but may not give a clear indication of which ancestral markers are responsible for the variation of the trait, limiting its applicability to admixture mapping, and potentially reducing discovery of genetic mechanisms for prevention. As we move forward into the next generation of genotypic and genomic analyses, it is evident that the genetic admixture approach may be a significant tool in elucidating the obscurities in the complex relationship between genes and environments.
Acknowledgements
We would like to acknowledge the insightful contributions of Dr. Yann Klimentidis to this manuscript.
“The project described was supported in part by Awards Number R01DK067426, T32DK062710, P30DK079626 and P30DK056336 from the National Institute Of Diabetes And Digestive And Kidney Diseases, and Award Number T32HL105349 from the National Heart, Lung, And Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views the National Institutes of Health.”
References
- 1.Schwartz AG, Wenzlaff AS, Bock CH, Ruterbusch JJ, Chen W, Cote ML, Artis AS, Van Dyke AL, Land SJ, Harris CC, Pine SR, Spitz MR, Amos CI, Levin AM, McKeigue PM. Admixture mapping of lung cancer in 1812 African-Americans. Carcinogenesis. 2011;32:312–317. doi: 10.1093/carcin/bgq252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fejerman L, Chen GK, Eng C, Huntsman S, Hu D, Williams A, Pasaniuc B, John EM, Via M, Gignoux C, Ingles S, Monroe KR, Kolonel LN, Torres-Mejia G, Perez-Stable EJ, Burchard EG, Henderson BE, Haiman CA, Ziv E. Admixture mapping identifies a locus on 6q25 associated with breast cancer risk in US Latinas. Hum Mol Genet. 2012;21:1907–1917. doi: 10.1093/hmg/ddr617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ricks-Santi LJ, Apprey V, Mason T, Wilson B, Abbas M, Hernandez W, Hooker S, Doura M, Bonney G, Dunston G, Kittles R, Ahaghotu C. Identification of genetic risk associated with prostate cancer using ancestry informative markers. Prostate Cancer Prostatic Dis. 2012;15:359–364. doi: 10.1038/pcan.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Divers J, Palmer ND, Lu L, Register TC, Carr JJ, Hicks PJ, Hightower RC, Smith SC, Xu J, Cox AJ, Hruska KA, Bowden DW, Lewis CE, Heiss G, Province MA, Borecki IB, Kerr KF, Chen YD, Palmas W, Rotter JI, Wassel CL, Bertoni A, Herrington D, Wagenknecht LE, Langefeld CD, Freedman BI. Admixture mapping of coronary artery calcified plaque in African Americans with type 2 diabetes. Circ Cardiovasc Genet. 2012;6:97–105. doi: 10.1161/CIRCGENETICS.112.964114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allison MA, Peralta CA, Wassel CL, Aboyans V, Arnett DK, Cushman M, Eng J, Ix J, Rich SS, Criqui MH. Genetic ancestry and lower extremity peripheral artery disease in the Multi-Ethnic Study of Atherosclerosis. Vasc Med. 2010;15:351–359. doi: 10.1177/1358863X10375586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz-Narvaez EA, Bare L, Arellano A, Catanese J, Campos H. West African and Amerindian ancestry and risk of myocardial infarction and metabolic syndrome in the Central Valley population of Costa Rica. Hum Genet. 2010;127:629–638. doi: 10.1007/s00439-010-0803-x. [DOI] [PubMed] [Google Scholar]
- 7.Lai CQ, Tucker KL, Choudhry S, Parnell LD, Mattei J, Garcia-Bailo B, Beckman K, Burchard EG, Ordovas JM. Population admixture associated with disease prevalence in the Boston Puerto Rican health study. Hum Genet. 2009;125:199–209. doi: 10.1007/s00439-008-0612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiner AP, Carlson CS, Ziv E, Iribarren C, Jaquish CE, Nickerson DA. Genetic ancestry, population sub-structure, and cardiovascular disease-related traits among African-American participants in the CARDIA Study. Hum Genet. 2007;121:565–575. doi: 10.1007/s00439-007-0350-2. [DOI] [PubMed] [Google Scholar]
- 9.Qi L, Nassir R, Kosoy R, Garcia L, Curb JD, Tinker L, Howard BV, Robbins J, Seldin MF. Relationship between diabetes risk and admixture in postmenopausal African-American and Hispanic-American women. Diabetologia. 2012;55:1329–1337. doi: 10.1007/s00125-012-2486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klimentidis YC, Divers J, Casazza K, Beasley TM, Allison DB, Fernandez JR. Ancestry-informative markers on chromosomes 2, 8 and 15 are associated with insulin-related traits in a racially diverse sample of children. Hum Genomics. 2011;5:79–89. doi: 10.1186/1479-7364-5-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gower BA, Fernandez JR, Beasley TM, Shriver MD, Goran MI. Using genetic admixture to explain racial differences in insulin-related phenotypes. Diabetes. 2003;52:1047–1051. doi: 10.2337/diabetes.52.4.1047. [DOI] [PubMed] [Google Scholar]
- 12.Cheng CY, Reich D, Haiman CA, Tandon A, Patterson N, Elizabeth S, Akylbekova EL, Brancati FL, Coresh J, Boerwinkle E, Altshuler D, Taylor HA, Henderson BE, Wilson JG, Kao WH. African ancestry and its correlation to type 2 diabetes in African Americans: A genetic admixture analysis in three U.S. population cohorts. PLoS One. 2012;7:e32840. doi: 10.1371/journal.pone.0032840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makowsky R, Yan Q, Wiener HW, Sandel M, Aissani B, Tiwari HK, Shrestha S. The utility of mitochondrial and y chromosome phylogenetic data to improve correction for population stratification. Front Genet. 2012;3:301. doi: 10.3389/fgene.2012.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kodaman N, Aldrich MC, Smith JR, Signorello LB, Bradley K, Breyer J, Cohen SS, Long J, Cai Q, Giles J, Bush WS, Blot WJ, Matthews CE, Williams SM. A small number of candidate gene SNPs reveal continental ancestry in African Americans. Ann Hum Genet. 2013;77:56–66. doi: 10.1111/j.1469-1809.2012.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Divers J, Vaughan L, Padilla M, Fernandez J, Allison D, Redden D. Correcting for measurement error in individual ancestry estimates in structured association tests. Genetics. 2007;176:1823–1833. doi: 10.1534/genetics.107.075408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanis CL, Chakraborty R, Ferrell RE, Schull WJ. Individual admixture estimates: Disease associations and individual risk of diabetes and gallbladder disease among Mexican-Americans in Starr County, Texas. Am J Phys Anthropol. 1986;70:433–441. doi: 10.1002/ajpa.1330700404. [DOI] [PubMed] [Google Scholar]
- 17.McKeigue PM, Carpenter JR, Parra EJ, Shriver MD. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: Application to African-American populations. Ann Hum Genet. 2000;64:171–186. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]
- 18.Shriver MD, Mei R, Parra EJ, Sonpar V, Halder I, Tishkoff SA, Schurr TG, Zhadanov SI, Osipova LP, Brutsaert TD, Friedlaender J, Jorde LB, Watkins WS, Bamshad MJ, Gutierrez G, Loi H, Matsuzaki H, Kittles RA, Argyropoulos G, Fernandez JR, Akey JM, Jones KW. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genomics. 2005;2:81–89. doi: 10.1186/1479-7364-2-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum Mutat. 2008;29:648–658. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- 21.Samet JM, Coultas DB, Howard CA, Skipper BJ, Hanis CL. Diabetes, gallbladder disease, obesity, and hypertension among Hispanics in New Mexico. Am J Epidemiol. 1988;128:1302–1311. doi: 10.1093/oxfordjournals.aje.a115083. [DOI] [PubMed] [Google Scholar]
- 22.Zimmet P, Kirk RL, Serjeantson SW, King H. Genetic and environmental influence in the epidemiology of noninsulin-dependent diabetes mellitus: A global perspective. Ann Acad Med Singapore. 1985;14:347–353. [PubMed] [Google Scholar]
- 23.King H, Heywood P, Zimmet P, Alpers M, Collins V, Collins A, King LF, Raper LR. Glucose tolerance in a highland population in Papua New Guinea. Diabetes Res. 1984;1:45–51. [PubMed] [Google Scholar]
- 24.Chen G, Shriner D, Zhou J, Doumatey A, Huang H, Gerry N, Herbert A, Christman M, Chen Y, Dunston G, Faruque M, Rotimi C, Adeyemo A. Development of admixture mapping panels for African Americans from commercial high-density SNP arrays. BMC Genomics. 2010;11:417. doi: 10.1186/1471-2164-11-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez JR, Shriver MD, Beasley TM, Rafla-Demetrious N, Parra E, Albu J, Nicklas B, Ryan AS, McKeigue PM, Hoggart CL, Weinsier RL, Allison DB. Association of African genetic admixture with resting metabolic rate and obesity among women. Obes Res. 2003;11:904–911. doi: 10.1038/oby.2003.124. [DOI] [PubMed] [Google Scholar]
- 26.Shaffer JR, Kammerer CM, Reich D, McDonald G, Patterson N, Goodpaster B, Bauer DC, Li J, Newman AB, Cauley JA, Harris TB, Tylavsky F, Ferrell RE, Zmuda JM. Genetic markers for ancestry are correlated with body composition traits in older African Americans. Osteoporos Int. 2007;18:733–741. doi: 10.1007/s00198-006-0316-6. [DOI] [PubMed] [Google Scholar]
- 27.Cheng CY, Reich D, Coresh J, Boerwinkle E, Patterson N, Li M, North KE, Tandon A, Bailey-Wilson JE, Wilson JG, Kao WH. Admixture mapping of obesity-related traits in African Americans: The Atherosclerosis Risk In Communities (ARIC) study. Obesity (Silver Spring) 2010;18:563–572. doi: 10.1038/oby.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardel M, Higgins PB, Willig AL, Keita AD, Casazza K, Gower BA, Fernandez JR. African genetic admixture is associated with body composition and fat distribution in a cross-sectional study of children. Int J Obes (Lond) 2011;35:60–65. doi: 10.1038/ijo.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonilla C, Shriver MD, Parra EJ, Jones A, Fernandez JR. Ancestral proportions and their association with skin pigmentation and bone mineral density in Puerto Rican women from New York City. Hum Genet. 2004;115:57–68. doi: 10.1007/s00439-004-1125-7. [DOI] [PubMed] [Google Scholar]
- 30.Mao X, Bigham AW, Mei R, Gutierrez G, Weiss KM, Brutsaert TD, Leon-Velarde F, Moore LG, Vargas E, McKeigue PM, Shriver MD, Parra EJ. A genomewide admixture mapping panel for Hispanic/Latino populations. Am J Hum Genet. 2007;80:1171–1178. doi: 10.1086/518564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willig AL, Hunter GR, Casazza K, Heimburger DC, Beasley TM, Fernandez JR. Body fat and racial genetic admixture are associated with aerobic fitness levels in a multiethnic pediatric population. Obesity (Silver Spring) 2011;19:2222–2227. doi: 10.1038/oby.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nassir R, Qi L, Kosoy R, Garcia L, Allison M, Ochs-Balcom HM, Tylavsky F, Manson JE, Shigeta R, Robbins J, Seldin MF. Relationship between adiposity and admixture in African-American and Hispanic-American women. Int J Obes (Lond) 2012;36:304–313. doi: 10.1038/ijo.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klimentidis YC, Miller GF, Shriver MD. The relationship between European genetic admixture and body composition among Hispanics and Native Americans. Am J Hum Biol. 2009;21:377–382. doi: 10.1002/ajhb.20886. [DOI] [PubMed] [Google Scholar]
- 34.Casazza K, Willig AL, Gower BA, Nagy TR, Hunter GR, Wallace S, Amaya M, Franklin F, Beasley M, Fernandez JR. The role of European genetic admixture in the etiology of the insulin resistance syndrome in children: Are the effects mediated by fat accumulation? J Pediatr. 2010;157:50–56. e51. doi: 10.1016/j.jpeds.2010.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casazza K, Phadke RP, Fernandez JR, Watanabe RM, Goran MI, Gower BA. Obesity attenuates the contribution of African admixture to the insulin secretory profile in peripubertal children: A longitudinal analysis. Obesity (Silver Spring) 2009;17:1318–1325. doi: 10.1038/oby.2008.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alfredo Martinez J, Enriquez L, Moreno-Aliaga MJ, Marti A. Genetics of obesity. Public Health Nutr. 2007;10:1138–1144. doi: 10.1017/S1368980007000626. [DOI] [PubMed] [Google Scholar]
- 37.Bogardus C. Missing heritability and GWAS utility. Obesity (Silver Spring, Md) 2009;17:209–210. doi: 10.1038/oby.2008.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caulfield T, Fullerton S, Ali-Khan S, Arbour L, Burchard E, Cooper R, Hardy B-J, Harry S, Hyde-Lay R, Kahn J, Kittles R, Koenig B, Lee S, Malinowski M, Ravitsky V, Sankar P, Scherer S, Séguin B, Shickle D, Suarez-Kurtz G, Daar A. Race and ancestry in biomedical research: exploring the challenges. Genome Medicine. 2009;1:8. doi: 10.1186/gm8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKeigue PM. Mapping genes underlying ethnic differences in disease risk by linkage disequilibrium in recently admixed populations. Am J Hum Genet. 1997;60:188–196. [PMC free article] [PubMed] [Google Scholar]
- 40.Smith M, Patterson N, Lautenberger J, Truelove A, McDonald G, Waliszewska A, Kessing B, Malasky M, Scafe C, Le E, De Jager P, Mignault A, Yi Z, De The G, Essex M, Sankale J-L, Moore J, Poku K, Phair J, Goedert J, Vlahov D, Williams S, Tishkoff S, Winkler C, De La Vega F, Woodage T, Sninsky J, Hafler D, Altshuler D, Gilbert D, O'Brien S, Reich D. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian C, Hinds DA, Shigeta R, Kittles R, Ballinger DG, Seldin MF. A genomewide single-nucleotide-polymorphism panel with high ancestry information for African American admixture mapping. Am J Hum Genet. 2006;79:640–649. doi: 10.1086/507954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, Tandon A, Schirmer C, Neubauer J, Bedoya G, Duque C, Villegas A, Bortolini MC, Salzano FM, Gallo C, Mazzotti G, Tello-Ruiz M, Riba L, Aguilar-Salinas CA, Canizales-Quinteros S, Menjivar M, Klitz W, Henderson B, Haiman CA, Winkler C, Tusie-Luna T, Ruiz-Linares A, Reich D. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80:1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu S. Human population admixture in Asia. Genomics Inform. 2012;10:133–144. doi: 10.5808/GI.2012.10.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu S, Jin L. A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery. Am J Hum Genet. 2008;83:322–336. doi: 10.1016/j.ajhg.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basu A, Tang H, Arnett D, Gu CC, Mosley T, Kardia S, Luke A, Tayo B, Cooper R, Zhu X, Risch N. Admixture mapping of quantitative trait loci for BMI in African Americans: Evidence for loci on chromosomes 3q, 5q, and 15q. Obesity (Silver Spring) 2009;17:1226–1231. doi: 10.1038/oby.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng CY, Kao WH, Patterson N, Tandon A, Haiman CA, Harris TB, Xing C, John EM, Ambrosone CB, Brancati FL, Coresh J, Press MF, Parekh RS, Klag MJ, Meoni LA, Hsueh WC, Fejerman L, Pawlikowska L, Freedman ML, Jandorf LH, Bandera EV, Ciupak GL, Nalls MA, Akylbekova EL, Orwoll ES, Leak TS, Miljkovic I, Li R, Ursin G, Bernstein L, Ardlie K, Taylor HA, Boerwinckle E, Zmuda JM, Henderson BE, Wilson JG, Reich D. Admixture mapping of 15,280 African Americans identifies obesity susceptibility loci on chromosomes 5 and X. PLoS Genet. 2009;5:e1000490. doi: 10.1371/journal.pgen.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shetty PB, Tang H, Tayo BO, Morrison AC, Hanis CL, Rao DC, Young JH, Fox ER, Boerwinkle E, Cooper RS, Risch NJ, Zhu X. Variants in CXADR and F2RL1 are associated with blood pressure and obesity in African-Americans in regions identified through admixture mapping. J Hypertens. 2012;30:1970–1976. doi: 10.1097/HJH.0b013e3283578c80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villarreal-Molina MT, Aguilar-Salinas CA, Rodriguez-Cruz M, Riano D, Villalobos-Comparan M, Coral-Vazquez R, Menjivar M, Yescas-Gomez P, Konigsoerg-Fainstein M, Romero-Hidalgo S, Tusie-Luna MT, Canizales-Quinteros S. The ATP-binding cassette transporter A1 R230C variant affects HDL cholesterol levels and BMI in the Mexican population: Association with obesity and obesity-related comorbidities. Diabetes. 2007;56:1881–1887. doi: 10.2337/db06-0905. [DOI] [PubMed] [Google Scholar]
- 49.Ravussin E, Valencia ME, Esparza J, Bennett PH, Schulz LO. Effects of a traditional lifestyle on obesity in Pima Indians. Diabetes Care. 1994;17:1067–1074. doi: 10.2337/diacare.17.9.1067. [DOI] [PubMed] [Google Scholar]
- 50.Loos RJF. Genetic determinants of common obesity and their value in prediction. Best Pract Res Clin Endocrinol Metab. 2012;26:211–226. doi: 10.1016/j.beem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meuwissen T, Goddard M. Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics. 2010;185:623–631. doi: 10.1534/genetics.110.116590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makowsky R, Pajewski NM, Klimentidis YC, Vazquez AI, Duarte CW, Allison DB, de los Campos G. Beyond missing heritability: Prediction of complex traits. PLoS Genet. 2011;7:e1002051. doi: 10.1371/journal.pgen.1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de los Campos G, Gianola D, Allison DB. Predicting genetic predisposition in humans: The promise of whole-genome markers. Nat Rev Genet. 2010;11:880–886. doi: 10.1038/nrg2898. [DOI] [PubMed] [Google Scholar]
- 55.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann JS, Vollenweider P, Waterworth DM, Mooser V, Palmer CN, Morris AD, Ouwehand WH, Zhao JH, Li S, Loos RJ, Barroso I, Deloukas P, Sandhu MS, Wheeler E, Soranzo N, Inouye M, Wareham NJ, Caulfield M, Munroe PB, Hattersley AT, McCarthy MI, Frayling TM. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proenca C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grassler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jorgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, Konig IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaloy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimaki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O'Donnell CJ, O'Rahilly S, Ong KK, Oostra B, Pare G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstrale M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tonjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kahonen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Gronberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O'Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr., Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gianola D. Priors in whole-genome regression: The Bayesian alphabet returns. Genetics. doi: 10.1534/genetics.113.151753. DOI: 10.1534/genetics.113.151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cooper R, Kaufman J, Ward R. Race and genomics. New Eng J Med. 2003;348:1166–1170. doi: 10.1056/NEJMsb022863. [DOI] [PubMed] [Google Scholar]
- 61.Schulz LO, Weidensee RC. Non-insulin-dependent diabetes mellitus in Mexico. Prog Food Nutr Sci. 1993;17:99–117. [PubMed] [Google Scholar]
- 62.Tang H, Jorgenson E, Gadde M, Kardia SL, Rao DC, Zhu X, Schork NJ, Hanis CL, Risch N. Racial admixture and its impact on BMI and blood pressure in African and Mexican Americans. Hum Genet. 2006;119:624–633. doi: 10.1007/s00439-006-0175-4. [DOI] [PubMed] [Google Scholar]
- 63.Casazza K, Thomas O, Dulin-Keita A, Fernandez JR. Adiposity and genetic admixture, but not race/ethnicity, influence bone mineral content in peripubertal children. J Bone Miner Metab. 2010;28:424–432. doi: 10.1007/s00774-009-0143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaiser J. Human genetics. Genetic influences on disease remain hidden. Science (New York, NY) 2012;338:1016–1017. doi: 10.1126/science.338.6110.1016. [DOI] [PubMed] [Google Scholar]
- 65.Divers J, Redden DT, Rice KM, Vaughan LK, Padilla MA, Allison DB, Bluemke DA, Young HJ, Arnett DK. Comparing self-reported ethnicity to genetic background measures in the context of the Multi-Ethnic Study of Atherosclerosis (MESA). BMC Genet. 2011;12:28. doi: 10.1186/1471-2156-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lins TC, Pires AS, Paula RS, Moraes CF, Vieira RG, Vianna LG, Nobrega OT, Pereira RW. Association of serum lipid components and obesity with genetic ancestry in an admixed population of elderly women. Genet Mol Biol. 2012;35:575–582. doi: 10.1590/S1415-47572012005000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tajima A, Hamaguchi K, Terao H, Oribe A, Perrotta VM, Baez CA, Arias JR, Yoshimatsu H, Sakata T, Horai S. Genetic background of people in the Dominican Republic with or without obese type 2 diabetes revealed by mitochondrial DNA polymorphism. J Hum Genet. 2004;49:495–499. doi: 10.1007/s10038-004-0179-7. [DOI] [PubMed] [Google Scholar]