Abstract
A multiscale investigation was carried out to study the dark and light-enhanced bactericidal mechanisms of poly(phenylene ethynylene) (PPE)-based cationic conjugated polyelectrolytes (CPEs) and oligo-phenylene ethynylenes (OPEs). On the morphological scale, Gram-negative E. coli cells exposed to CPE and OPE compounds in the dark show damage to the cell envelope, plasma membrane, and in some cases the cytoplasm, while with UV-irradiation, E. coli sustained catastrophic damages to both the cell envelope and cytoplasm. In contrast, the Gram-positive S. epi bacteria appeared intact when exposed to CPE and OPE compounds in the dark but showed damages to the cell envelope with UV-irradiation. To better understand the molecular basis of CPE and OPE induced morphological changes and damages to bacteria, we investigated the effect of these compounds on model bacterial plasma membrane, and bacterial proteins and plasmid DNA. Measurements of dark membrane perturbation activity of the CPEs and OPEs using model lipid membranes support a carpet or detergent-like mechanism by which the antimicrobial compounds induce membrane collapse and phase transitions. Under UV-irradiation, E. coli bacteria exposed to CPEs and OPEs showed covalent modifications and damages to both cellular protein and plasmid DNA, likely through oxidative pathways mediated by singlet oxygen and subsequent reactive oxygen species sensitized by the CPE and OPE compounds. Our finding thus show that the antimicrobial polymers and oligomers exert toxicity towards Gram-negative bacteria by disrupting the morphology and structures of cell envelope and cytoplasm, including cellular components such as proteins and DNA while exert toxicity towards Gram-positive bacteria by binding to and disrupting just the cell wall.
Introduction
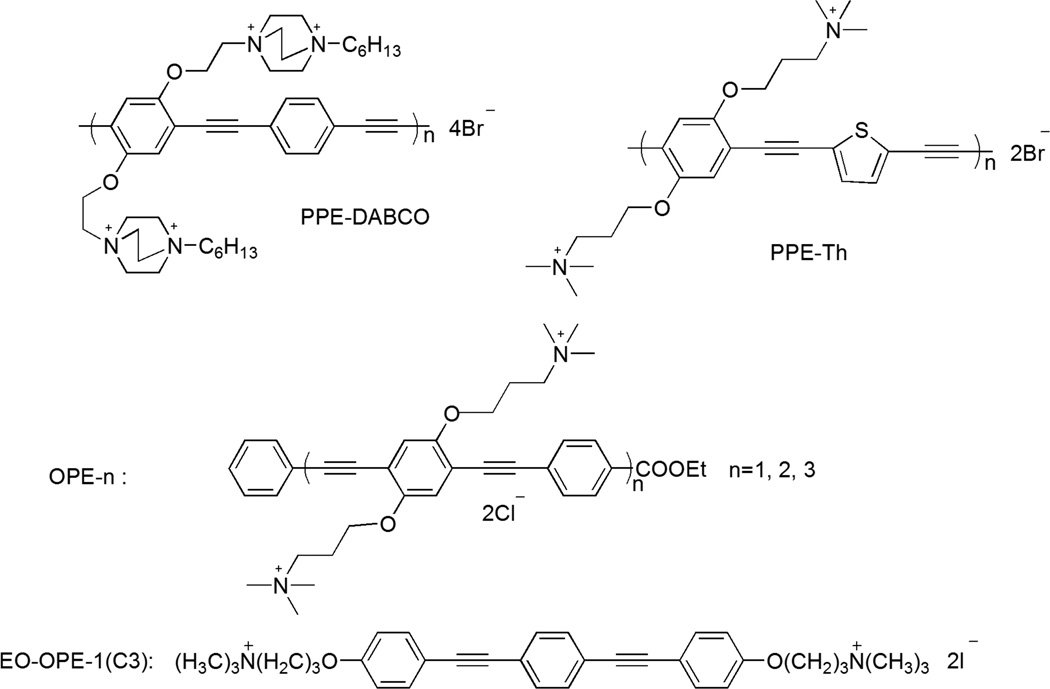
During the past fifty years, synthetic antimicrobial agents have raised much attention as alternatives to traditional biocides and antibiotics.1–4 In general, these materials are able to non-specifically disrupt the outer envelope of the microbes and result in cell lysis and subsequent death. Recently, we have developed a series of poly(p-phenylene ethynylene) (PPE)-based conjugated polyelectrolytes (CPEs) and oligo-(p-phenylene ethynylene)s (OPEs) with controlled chain lengths and functional side groups that exhibit significant light-activated biocidal activity and efficient killing efficacy in the dark against a broad spectrum of pathogens.5–8 Investigations of the biocidal mechanisms of the CPEs and OPEs revealed that these cationic and amphiphilic compounds are membrane-active, capable of inducing disruptions to the membrane structure. For example, the small oligomeric EO-OPE-1(C3) (Scheme 1) can disrupt model bacterial membranes and induce a phase transition from a lamellar to a hexagonal phase.9 Scanning and transmission electron microscopy (SEM and TEM) imaging gave further insights on how different CPEs and OPEs interact with the Gram-negative Escherichia coli (E. coli) bacteria in the dark.9 To gain a deeper mechanistic understanding of the observed broad spectrum bactericidal activity of these compounds, we carried out a multiscale investigation in this study, from morphological scale to molecular level, to characterize the interactions of the CPE and OPE compounds with both Gram-positive and Gram-negative bacteria in the dark and with UV-irradiation.
Scheme 1.
Structures of the antimicrobial compounds used in this study. n denotes the number of repeat units.
A set of structurally diverse PPE-based CPEs and OPEs were used in this study (Scheme 1). The CPEs and OPEs are water-soluble and exhibit high affinity towards bacteria due to their cationic side or end groups. The polymeric CPEs10 tend to form aggregates in aqueous environments via intra- or inter-chain stacking of the hydrophobic backbone.11–12 In comparison, the oligomeric OPE-n compounds, where n denotes the number of repeat units, can retain a planar or near planar conformation with increasing n and side functional groups.13 In particular, the smallest oligomer EO-OPE-1(C3), with a linear backbone, charged end groups, and no side-chains, has a needle-like conformation. The conjugated hydrophobic backbone of the CPE and OPE compounds are chromophores, which absorb light in the visible/near-UV region that results in low-lying singlet and triplet states and generate singlet oxygen. It has been demonstrated that the light-activated killing can be correlated to the generation of singlet oxygen species (1O2) and subsequent secondary reactive oxygen species (ROS).5, 14–15 In the absence of oxygen, the light-activated biocidal activities of these PPE-based compounds will be significantly reduced.14 In addition, the amphiphilic nature of these compounds gives rise to the compounds’ dark killing efficacy, as they can interact with various cellular targets, including the lipid membrane. Comparing the membrane perturbation and biocidal activities of the CPEs and OPEs can provide insights into the structure-function relationship of this novel class of compounds.
The bacterial cell envelope is anionic due to the presence of lipopolysaccharides (LPS), teichoic and lipoteichoic acids, and negatively charged phospholipids.16 Since the cell surface serves as the first point of contact for biocidal agents, structure of the cell envelope is important in determining bacterial susceptibility to the CPEs and OPEs. In Gram-positive bacteria, the cell envelope has a two-layer structure: a thick (20–80 nm) cell wall primarily composed of peptidoglycans and the cytoplasmic membrane.16 Specifically, the peptidoglycans in Gram-positive bacteria are cross-linked and give rise to the strong and rigid cell wall. In contrast, the Gram-negative bacterial envelope is a three-layer structure: an outer membrane, a thin (1–3 nm) peptidoglycan cell wall, and a cytoplasmic membrane. Since there are no advanced organelles in the bacteria, the cell wall and membrane serve many essential biological functions, including structural support against osmotic pressure gradients, nutrient and waste transport, metabolic reactions, and synthesis.16 Therefore, compromises to the structural integrity of the bacterial cell envelope may trigger a lethal effect. For example, ethylenediaminetetraacetic acid (EDTA) has been demonstrated to cause the release of LPS molecules from E. coli outer membrane by chelating Mg2+ or Ca2+ ions, which destabilizes the LPS assembly and increases the permeability of the bacterial outer envelope.17
As reported previously in a letter,9 due to the large differences in molecular weights, the polymeric CPEs and oligomeric OPEs exert dark antimicrobial activity against the Gram-negative E. coli bacteria via different mechanisms. For example, the large polymeric PPE-Th (Scheme 1) primarily damages the bacterial cell surface. In contrast, the small oligomeric EO-OPE-1(C3) exerts a strong bacteriolytic effect by disrupting the cytoplasmic membrane, implying that the outer membrane and cell wall in Gram-negative bacteria do not serve as efficient barriers to these OPEs. However, as described above, the cell wall in Gram-positive bacteria is much thicker and made of cross-linked peptidoglycans and anionic teichoic and lipoteichoic acids. The biological functions of these acids are not fully understood; some of their proposed functions include ( i ) binding of divalent cations (particularly Mg2+), ( ii ) regulation of autolytic enzymes, and (iii) barrier to control the diffusion of nutrients and wastes.18–19 Although the cell wall of Gram-positive bacteria is believed to be an open network and accessible to solutes with a broad range of molecular weights,20 the cationic CPEs and OPEs may bind with the negatively charged cell wall components, thus reducing their penetration through the cell envelope. As such, toxicity of CPEs and OPEs against Gram-positive bacteria may stem from different a mechanism than those towards Gram-negative bacteria. 1O2 and the secondary ROS sensitized by the CPE and OPE compounds have shown to be to be highly toxic to bacteria by possibly inducing damages to proteins, RNA, DNA, and unsaturated lipids.21–22 Due to the different structures and components of the bacterial cell envelopes, Gram-positive and Gram-negative bacteria have been found to exhibit different susceptibilities towards chemical damages induced by pure 1O2.23 Briefly, for Gram-negative bacteria, 1O2 can react with the components of the outer membrane (e.g., LPS) and generate secondary ROS so that the damage to the bacteria is the culmination of 1O2 damage of the cytoplasmic membrane and the lethal effect caused by the ROS products on the outer membrane. However, for Gram-positive bacteria, 1O2 can rapidly diffuse through the cell wall and cause lethal damages directly on the cytoplasmic membrane.23 Aside from the intrinsic diffusing rate and reactivity of 1O2, the light-enhanced biocidal activity of CPEs and OPEs is also expected to be highly dependent on the penetrating ability or the location of the compounds in bacteria.
It has been reported that Gram-positive bacteria were more susceptible to the CPEs and OPEs than Gram-negative bacteria in both dark and UV-light irradiated conditions.6–7 To build on our previous work,9, 24 we employed a suite of characterization techniques herein to gain a more complete understanding of both the light-activated and dark toxicity mechanisms for the CPE and OPE compounds, from morphological to molecular scales. Disruptions to model membranes were visualized by single giant vesicle and cryogenic-transmission electron microscopy (cryo-TEM) imaging. Small angle x-ray scattering (SAXS) and solid state NMR (SS-NMR) were employed to elucidate molecular scale membrane perturbations. Damages to the morphology of the Gram-negative E. coli and Gram-positive Staphylococcus epidermidis (S. epi) cells exposed to the biocidal compounds were visualized by scanning and transmission electron microscopies (SEM and TEM). Damages to E. coli proteins and plasmid DNA was investigated by SDS-PAGE and DNA gel electrophoresis methods.
Experimental Methods
Materials
The antimicrobial compounds (Scheme 1) used in this study were synthesized as reported.7, 11, 13 Lipids and lipid vesicle extrusion supplies were purchased from Avanti Polar Lipids (Alabaster, AL) and used as received. Other chemicals were purchased either from Sigma-Aldrich (St. Louis, MO) or BD Biosciences (Franklin Lakes, NJ). E. coli strain ATCC 11303 and S. epi strain ATCC 14990 were obtained from American Type Culture Collection (ATCC, Manassas, VA). E. coli strain BL21(DE3)pLysS (with plasmid pET20b) was a generous gift from Dr. Graves’ lab at the University of New Mexico. Ultrapure water was used throughout the study (Milli-Q, 18.2 MΩ cm−1 resistivity).
Bacterial Culture
All bacterial cells were grown in standard Luria broth (LB). A fresh bacterial culture was inoculated from an overnight culture followed by approximately three hours of incubation at 37°C to the exponential growth phase. During this growth phase, the cells were collected by centrifugation and washed twice with 10 mM PBS (138 mM NaCl and 2.7 mM KCl at pH 7.4). The cell pellet was resuspended with PBS buffer for future use.
Single Giant Vesicles Observation and X-Ray Scattering
Giant vesicles were prepared and observed by a method as reported previously.9 Rhodamine tagged lipid 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (DMPE-Rh) was used and gives rise to the fluorescence of the giant vesicles when imaged by a fluorescence microscope (Zeiss Imager A2, Excitation filter BP 545/25 nm, Emission filter BP 605/70 nm, Thornwood, NY). CPEs and OPEs were not excited under these conditions. SAXS experiments were performed at the Stanford Synchrotron Radiation Laboratory (Palo Alto, CA) (Beam Line 4–2) as described in the literature.9, 25 The OPEs used in the SAXS assays were prepared in water containing 10 vol% DMSO.
NMR spectroscopy
31P solid-state NMR spectra were recorded on a Bruker Avance III Widebore 300 NMR spectrometer as described in the literature.26 The reported Hahn echo pulse sequence was used in the current study. The NMR samples were prepared by hydrating a 25 mg dry phospholipid film with 100 µL water or EO-OPE-1(C3) solution at appropriate concentrations by vigorous vortexing. In order to thoroughly hydrate the lipid sample and mix the test compounds with lipid membrane, the samples were subjected to 10 freeze-thaw-vortex cycles. 7 mm CPMASS probe was used in the NMR spectroscopy.
Electron Microscopy
Fresh bacterial cells in exponential growth phase (1~4×108 colony forming units (CFU)/mL)) were incubated with various amounts of CPEs or OPEs in the dark at 37°C or under UV-light irradiation for various periods and imaged by SEM and TEM as previously described.9, 27 The UV-light irradiation experiments were carried out in a photoreactor (LZC-ORG, Luzchem Research Inc., Ottawa, Canada) at room temperature. Different illumination sources were used according to the different light-absorbing properties of the CPEs and OPEs. UVA (centered at ~350 nm) and UV-420 (centered at ~420 nm) were used to irradiate OPEs and CPEs, respectively.8
Cryo-TEM imaging was performed by a modified method from literature28 at The Scripps Research Institute. Large unilamellar vesicles (LUV) composed of DOPE:DOPG29 (8:2, molar ratio) were made by an extrusion method30 to a final concentration of 10 mg/ml in water at room temperature and mixed with the CPEs or OPEs at various molar ratios. A 3 µL aliquot of each sample was applied to the grid (Protochips, Raleigh, NC, CF-2/0.5-4C) and then vitrified in liquid ethane using a Vitrobot (FEI Company, Hillsboro, Oregon). Cryo-TEM imaging was carried out on an FEI TF20 microscope operating at a low lose, 200 kV condition using a Gatan cryo-transfer holder and the Leginon data collection software.31
SDS-PAGE and DNA Electrophoresis
SDS-PAGE and DNA electrophoretic methods were used to characterize protein and DNA damage in E. coli cells. Fresh E. coli (ATCC 11303) cells in exponential growth phase (4×108 CFU/mL) were incubated with 25 µg/ml EO-OPE-1(C3) in PBS in the dark or under UV irradiation for 1 hour. A 12 µL aliquot of each E. coli sample was mixed with 6 µL of 3X standard SDS-PAGE sample loading buffer and heated in boiling water for 10 min followed by centrifugation. The supernate of denatured cell samples were loaded directly onto the 12% precast polyacrylamide-gel (BIO-RAD, 456–1043S). Electrophoresis was performed at 200 V for approximately 30 min, after which the gel was stained with Coomassie brilliant blue R. For DNA electrophoresis, fresh bacterial cells (E. coli BL21(DE3), 1×108 CFU/mL) were incubated with 1 µg/mL EO-OPE-1(C3) in PBS in the dark or under UV irradiation for 1 hour. The plasmid DNA was extracted from each bacterial sample using a Miniprep Kit (QIAGEN Inc., Valencia, CA) according to the manufacturer’s manual. A 10 µL aliquot of each plasmid sample was mixed with 2 µL of 6X loading buffer and loaded onto a 1% agarose-gel. Electrophoresis was carried out at 100 V for approximately 40 min.
Results and Discussion
Previously, we have shown that the cytoplasmic membrane in Gram-negative bacteria is one of the main targets for oligomeric OPEs.9, 27 Although the penetration ability of the polymeric CPEs through the outer membrane and peptidoglycan layer in E. coli cell is largely hindered by their bulky sizes, these compounds may still interact with the bacterial cytoplasmic membrane. Herein, further characterizations on membrane perturbation mechanisms have been performed with representative CPEs and OPEs against model E. coli membranes.
Multiscale membrane perturbations induced by CPE and OPE compounds
In order for cell lysis to occur, the permeability barrier, which includes the cytoplasmic membrane, of the E. coli cells need to be compromised. Since this membrane is located underneath the cell surface, it is difficult to visualize its disruption. Therefore, we used model phospholipid membranes32–33 composed of DOPG/DOPE or E. coli lipid extracts as alternatives in this study, both of which are routinely used as models of the E. coli plasma membrane.34 In this study, we investigated membrane binding and perturbation activities of the CPE and OPE compounds in the dark. Morphological changes induced by CPE and OPE compounds were characterized by fluorescence imaging of single giant vesicles and cryo-TEM imaging of large unilamellar vesicles (LUVs). Molecular level changes to bilayer lipid membrane structure were determined by small angle X-ray scattering (SAXS) and solid-state NMR (SS-NMR) experiments.
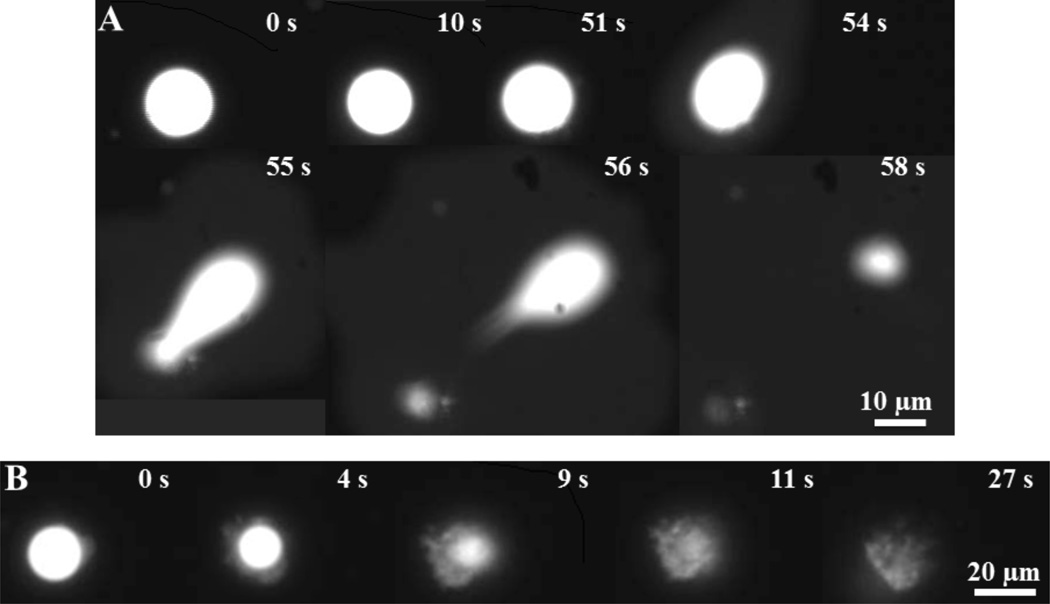
Fluorescence imaging of single giant vesicles has been previously used to visualize membrane disruptions induced by oligomeric OPE-1 and EO-OPE-1(C3).9 In the current study, the same technique is used to image membrane morphological changes induced by polymeric PPE-DABCO and oligomeric OPE-3. As shown in Figure 1A, upon the addition of PPE-DABCO to a giant vesicle composed of E. coli total lipids, the vesicle appeared relatively intact until it suddenly ruptured 55 – 56 s after PPE-DABCO addition and then mostly disappeared at around 58 s. In contrast, after the addition of OPE-3, the giant vesicle immediately started to change, shrinking in size and becoming diffuse and eventually losing its structural integrity by 27 s (Figure 1B). During the process, the well-defined and bright lipid membrane became amorphous in shape and inhomogeneous in brightness, indicating that lipid-OPE-3 complexes may have been formed (Figure 1B). These images show that both PPE-DABCO and OPE-3 exhibit strong disruptive activities against the model bacterial membrane and that the two compounds cause membrane disruptions via different mechanisms. However, this assay does not give further insights into the molecular scale structural transformations of the bilayer induced by the two biocidal compounds.
Figure 1.
Time lapse fluorescence microscopy images showing the damage of a giant vesicle caused by the addition of PPE-DABCO (10 µg/mL) (A) and OPE-3 (50 µg/mL) (B) at room temperature. The vesicle is composed of E. coli total lipids and labeled with 0.5 mol% of DMPE-Rh for imaging and 2.5 mol% of Biotin-PEG-DSPE for localization to the slide surface.9 The elapsed time after the addition of the antimicrobial agent is labeled in each image.
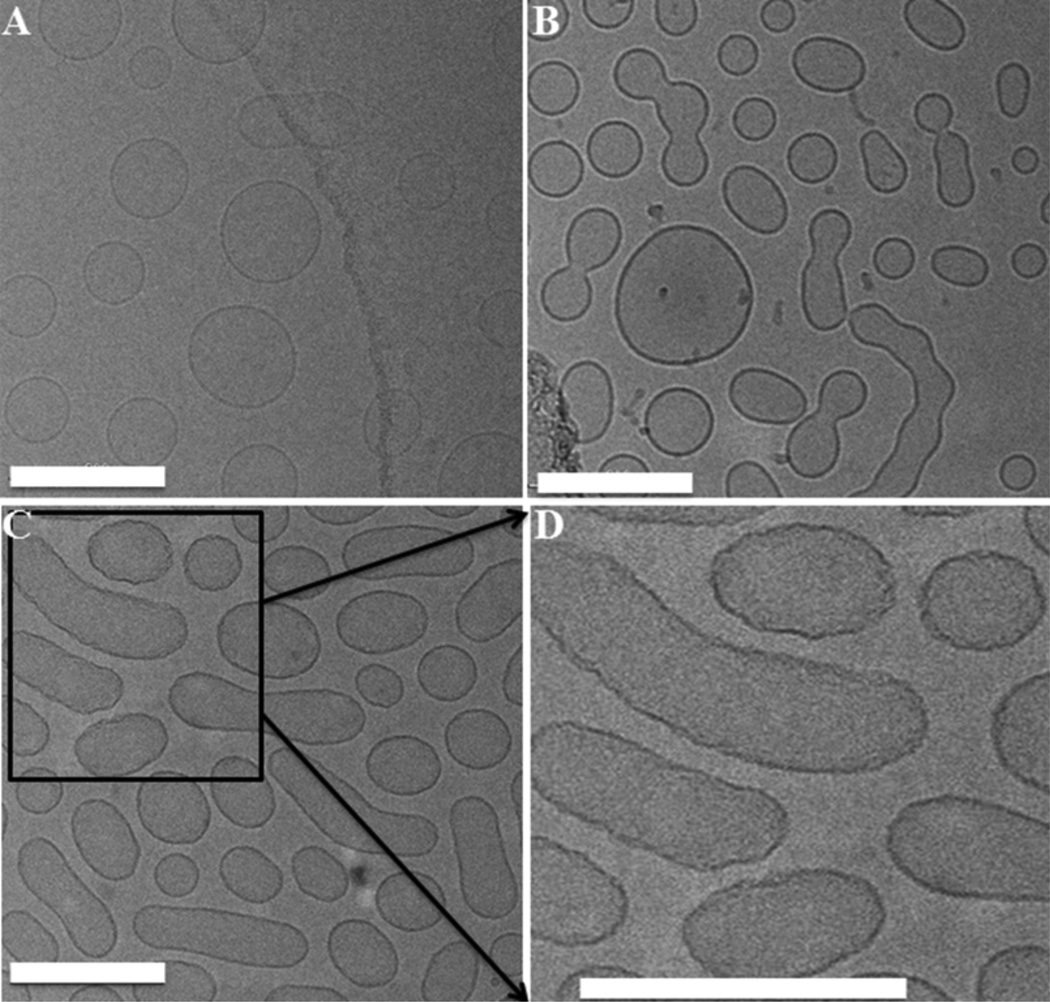
To more closely examine membrane changes induced by the biocidal compounds, cryo-TEM was used to image LUVs before and after exposure to two different OPEs, OPE-2 and EO-OPE-1(C3). The cryo-TEM image of LUVs (Figure 2A) showed largely round vesicles with diameters of around 100 nm. The addition of OPE-2 appeared to induce vesicle fusion, resulting in the formation of many dumbbell-like bilayer structures (Figure 2B). The vesicle fusion process may proceed by the attachment of OPE-2 to vesicle surface, thereby changing its charge distribution and reducing vesicle-vesicle electrostatic repulsion, resulting in vesicle fusion. Similarly, the addition of EO-OPE-1(C3) to vesicles also caused vesicle fusion (Figure 2C). In addition, EO-OPE-1(C3) induced significant roughness to a number of vesicles (Figure 2C and D), which could be an early stage of membrane failure/collapse. Although cryo-TEM is a powerful technique in visualizing assemblies of soft biological materials with minimal disturbance to the sample, as compared to conventional TEM where samples are dried on a substrate, it does have the limitation that only a very thin section of the samples are visualized. Structures larger than sample thickness, for example, visible aggregates of OPE-membrane complexes formed during the experiment, were not imaged. As such, cryo-TEM imaging does not capture the full range of OPE and CPE-induced membrane changes.
Figure 2.
Cryo-TEM micrographs of DOPG/DOPE vesicles (10 mg/mL) alone (A), incubated with 2.16 mg/mL OPE-2 (B) 1.53 mg/mL EO-OPE-1(C3) (C) for 30 min in the dark. A closer view for the EO-OPE-1(C3) treated vesicles shown in D. The lipid to OPE molar ratio is 25:1. The scale bars represent 200 nm.
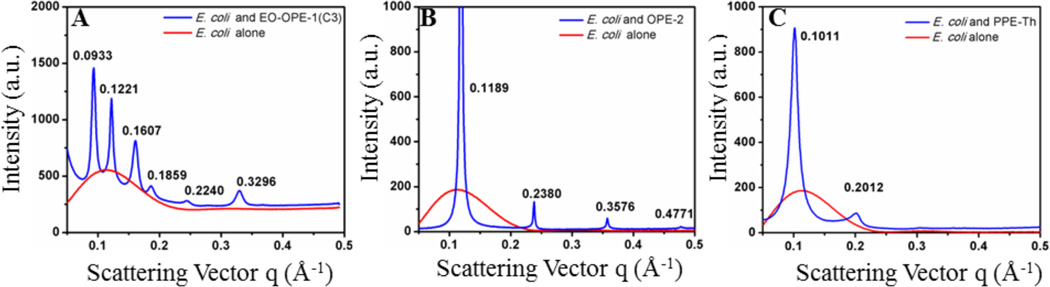
To resolve molecular level changes to bilayer lipid structure induced by CPEs and OPEs, SAXS and SS-NMR experiments were carried out. SAXS can resolve Å-scale structural details of ordered lipid phases in bilayer membranes and has been used to detect the presence of lipid ordered structures in a bulk background.35 We have shown previously that the oligomeric EO-OPE-1(C3) can induce the formation of a hexagonal or cubic lipid phase from model E. coli membranes in a lamellar phase (Figure 3A).9 In this study, we used SAXS to characterize the effects of oligomeric OPE-2 and polymeric PPE-Th on lipid membrane structure. As shown in Figure 3, lipid vesicles comprised of E. coli total lipids alone gave rise to one broad peak with low scattering intensity, which is indicative of a lamellar phase for the vesicles.25 After exposure to OPE-2 and PPE-Th, scattering profiles of the vesicles changed dramatically (Figures 3B and 3C). A number of new and sharp scattering peaks were observed. The peak positions (q values) of the model membranes treated by OPE-2 and PPE-Th have the characteristic ratio of 1:2:3:4, indicative of a new multi-lamellar structure.36 As a control, DMSO, which was used to increase the solubility of the OPEs, had negligible effect on the membrane structure (see Figure S1 in Supporting Information). Therefore, the emergence of the new peaks due to the formation of multilamellar structures, were solely caused by the interactions between the CPE and OPE compounds with the model membrane. The SAXS experiments demonstrate that both polymeric and oligomeric biocidal compounds are capable of inducing structural reorganization of the lipid membrane on the molecular scale.
Figure 3.
SAXS data for EO-OPE-1(C3) (3.8 mg/mL) (A), OPE-2 (5.78 mg/mL) (B) and PPE-Th (2.59 mg/mL) (C) complexed with E. coli total lipid (12.5 mg/mL) model membranes. Figure A is reprinted with permission from ref 9.
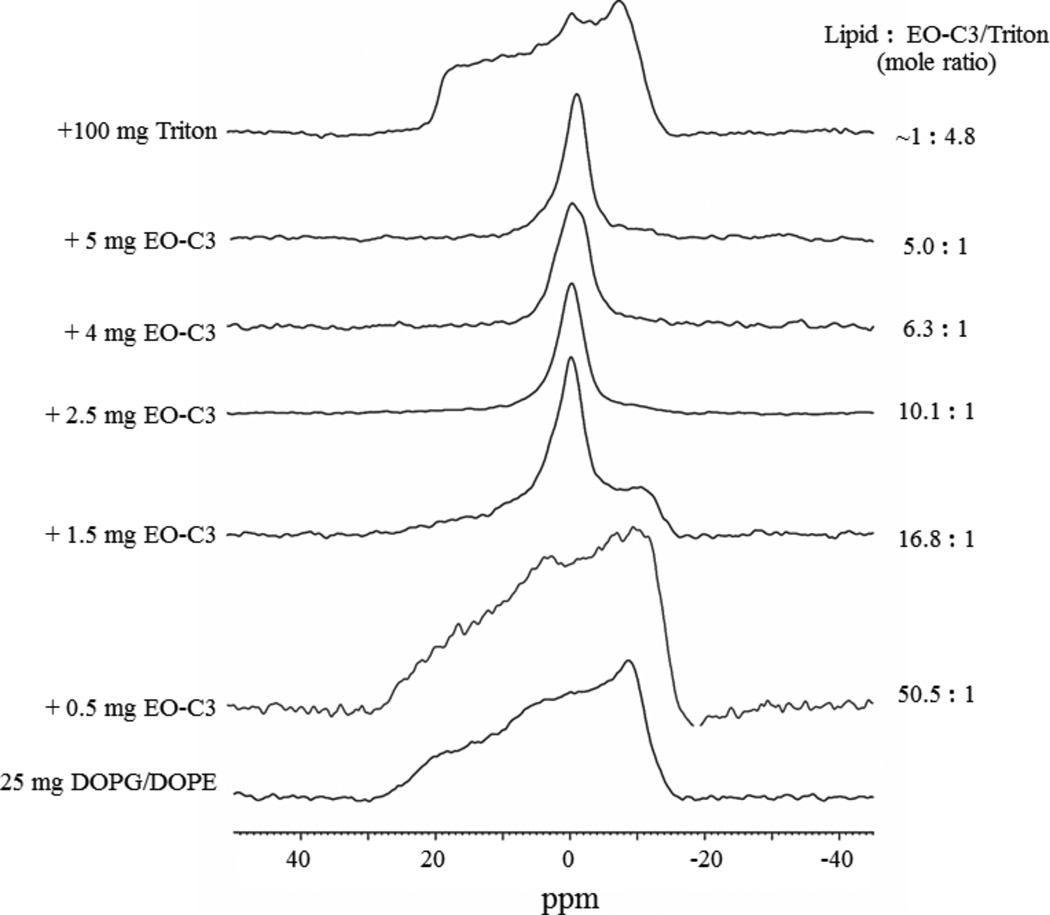
In addition to SAXS, 31P SS-NMR spectroscopy was also used to investigate the bulk phase behavior of model membranes in the presence of increasing amounts of EO-OPE-1(C3). In these experiments, the anisotropy (orientation of the phosphate head groups) of the lipid self-assemblies exhibit distinct NMR line shapes for different phases. The 31P NMR signal of the bacterial mimic DOPG/DOPE membrane is characteristic of a randomly dispersed lipid bilayer (bottom spectrum in Figure 4).26, 37 The addition of 1.5 mg EO-OPE-1(C3) (to 25 mg of lipid) caused a significant decrease in the signal intensity in the high field and induced an additional peak at the isotropic chemical shift position, indicating the formation of isotropic non-bilayer phases, such as micelles, inverted micelles, or various cubic phases.37 With increasing concentrations of EO-OPE-1(C3) (2.5–5 mg), the lipid sample showed a single sharp isotropic peak, indicating that EO-OPE-1(C3) did not induce the formation of new structures other than the isotropic phases in this concentration range. The addition of Triton X-100, a well-known non-ionic detergent that is widely used as a lipid membrane solubilizing agent, also gave rise to a weak isotropic peak, which implies that the two compounds induced similar bulk phase changes to the lipid membranes (Figure 4). SS-NMR data were also collected from vesicles composed of E. coli total lipid extract. The appearance of non-bilayer phases caused by the addition of EO-OPE-1(C3) to this model membrane was also observed (see Figure S2 in Supporting Information). However, since the exact composition of the total lipid extract is unknown, the NMR spectra could not be further resolved.
Figure 4.
31P SS-NMR spectra of 25 mg DOPG/DOPE (molar ration 2:8) model membranes mixed with various amounts of EO-OPE-1(C3) and 100 mg Triton.
Results from this study, combined with our previous work,9, 24 further confirm that the CPE and OPE compounds are membrane active and induce membrane changes from morphological to molecular scale. Giant vesicle imaging shows disintegration of vesicles while cryo-TEM imaging provided clear evidence of OPE and CPE-induced membrane fusion and roughening. Our results also provide the molecular scale structural basis to the observed membrane morphological and functional changes (i.e., membrane permeabilization from vesicle leakage studies30). The CPE and OPE compounds can disrupt the lamellar membrane structure and induce the formation of non-bilayer structures, such as hexagonal and cubic phases and micellar structures. It is important to note that, due to different sample requirements by the different analytical methods, such as concentration, sample thickness, hydration state, and lamellarity, each method probes the interactions between the biocidal compounds and model membranes under a specific set of conditions that give insights to specific aspects of biocide-induced membrane perturbation.32 Recent all-atom molecular dynamics simulations on the micro-second timescale reveal that OPE-3 can cause membrane damage that results in water leakage, which may be the initial step that subsequently lead to membrane failure.38 Our multi-scale characterization of the dark membrane perturbation activity of the CPEs and OPEs using model E. coli plasma membranes so far support a carpet or detergent-like mechanism9, 24 by which these antimicrobial compounds induce membrane collapse and phase transitions.
Visualization of dark and light-enhanced antimicrobial actions against Gram-negative and Gram-positive bacteria
Although interactions with the plasma membrane are necessary in the bactericidal actions of CPE and OPE compounds, interactions of these compounds with the bacterial cell envelope are also crucial since the cell envelope serves as the first point-of-contact for exogenous materials. As described earlier, the cell envelopes of Gram-positive and Gram-negative bacteria are compositionally and structurally different. Understanding the interactions of the CPE and OPE compounds with the different cell surfaces will not only provide a deeper and more complete understanding of the toxicity mechanism, but will also give us insights to the susceptibilities of the two different classes of bacteria. The complexities of the cell envelopes make such biological entities difficult to mimic with model systems. In this study, the interactions between the CPE and OPE compounds with Gram-negative E. coli bacteria and Gram-positive S. epi bacteria were studied by visualizing cell morphology using SEM and TEM. The effect of UV-irradiation in the presence of CPE and OPE compounds on cellular morphology was also investigated.
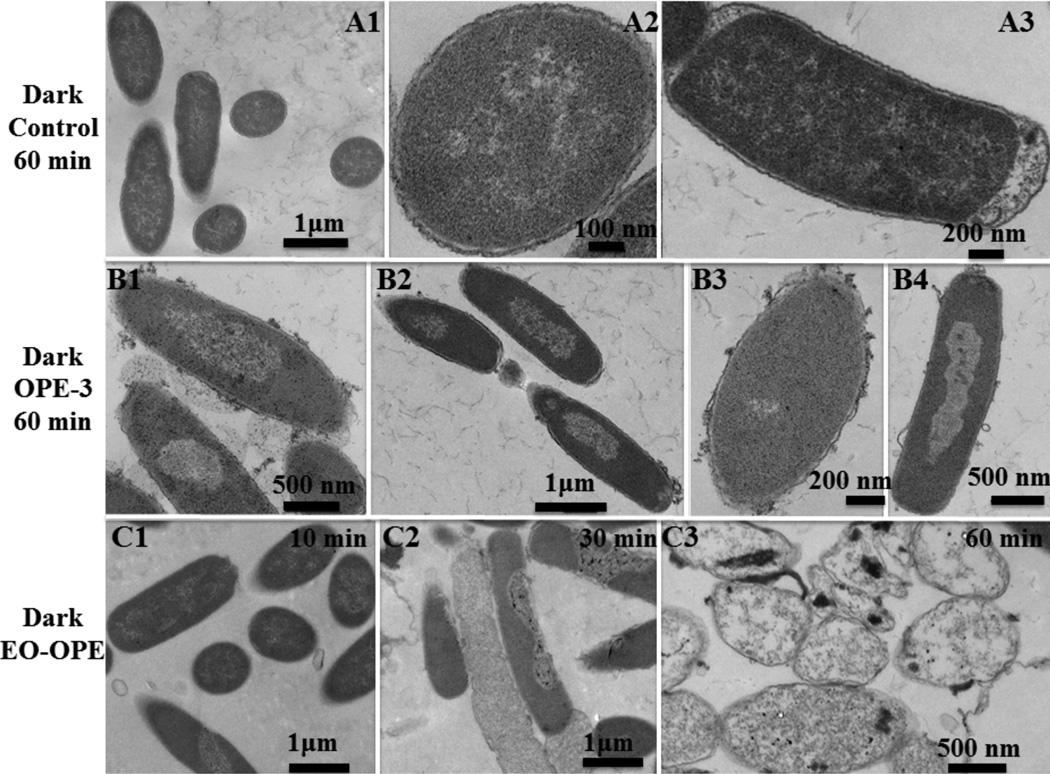
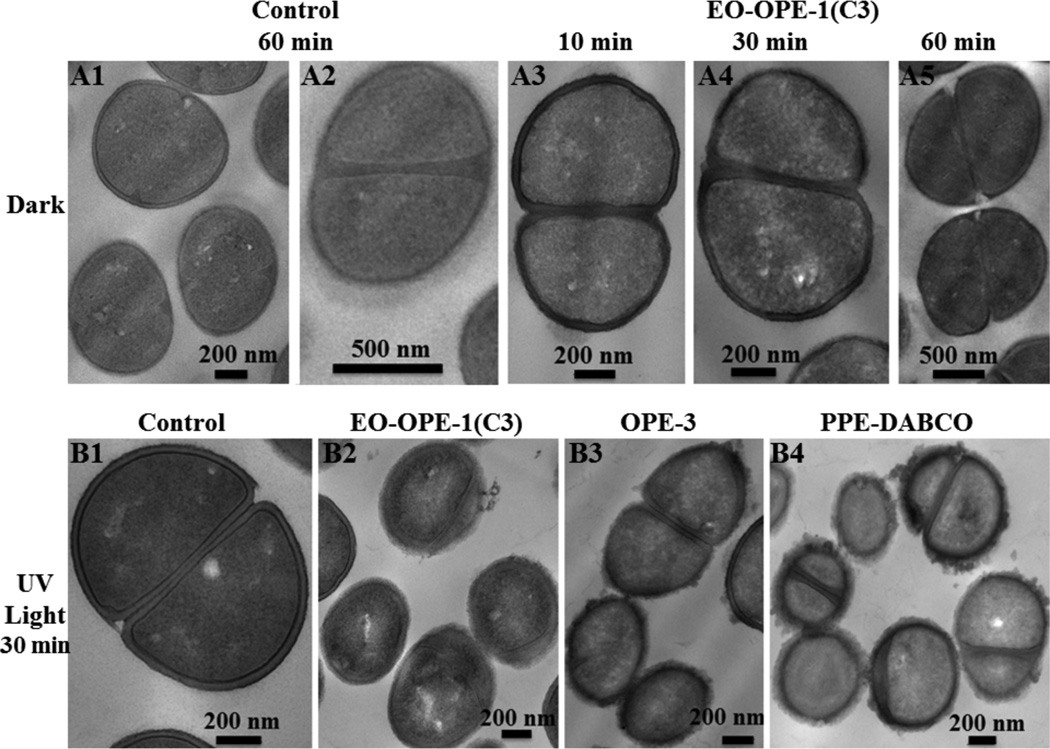
TEM was used to image morphological damages to bacterial cells upon incubation with OPEs in the dark. As a control, untreated E. coli cells (1×108 CFU/mL) in PBS appear structurally intact and the outer envelopes are clearly visible (Figure 5A1, A2, and A3). After 1 hour incubation with OPE-3 or EO-OPE-1(C3) in the dark, the E. coli cells show remarkable structural damages (Figure 5B and 5C). The attachment of OPE-3 to the bacterial cells extensively remodeled the outer membrane, leading to the roughening of the cell surface and formation of blebs (Figure 5B). In addition, the cytoplasm density of the majority of OPE-3 treated cells decreased, which implies that the cytoplasm contents were being released during incubation through damaged cell envelopes. Disruption and permeabilization of the cell envelope can also lead to the penetration of OPE-3 into the bacteria cytoplasm. The cationic and amphiphilic oligomer can then bind to and disrupt other cellular components, for example, proteins and nucleic acids. Likewise, incubation with the oligomeric EO-OPE-1(C3) also led to significant disruptions to the cell envelope (Figure 5C), which is consistent with previous findings that EO-OPE-1(C3) can permeabilize cell envelopes and cause cell lysis9, 27 Furthermore, the TEM images in of cells incubated with EO-OPE-1(C3) (Figure 5C) also showed time-dependent release of the cell cytoplasm, where only a small population of the cells show partial cytoplasm leakage/damage after exposure to the oligomer for 10 min (Figure 5C1), whereas most cells were empty and/or collapsed after one hour (Figure 5C3). This finding is consistent with a previous observation that EO-OPE-1(C3) exerts time-dependent biocidal activity against E. coli cells in the dark.6–7
Figure 5.
TEM micrographs of Gram-negative E. coli (ATCC 11303) cells (108 CFU/mL) alone (A1, A2 and A3), incubated with 10 µg/mL OPE-3 (B1, B2, B3 and B4) and EO-OPE-1(C3) (C1, C2 and C3) for different time intervals in the dark. Figure A3 is reprinted with permission from ref 9.
The outer membrane of Gram-negative bacteria, such as E. coli, is permeable to solutes with a molecular weight smaller than 600 Da due to the presence of porin channels.39 In terms of molecular weight and chain length, PPE-Th and EO-OPE-1(C3) represent two extremes in our current antimicrobial agent library (Scheme 1), whereas OPE-3 falls in between. PPE-Th, with a high molecular weight, is believed to exert toxicity towards Gram-negative bacteria by disrupting the outer membrane (LPS assembly) through an “ion-exchange” process.9, 17, 40 However, PPE-Th is not expected to penetrate through the bacterial envelop and get into the cytoplasm due to its high molecular weight.9 In contrast, EO-OPE-1(C3) with its needlelike structure may easily penetrate through the outer membrane and the thin peptidoglycan layer in the bacteria without causing serious structural damages. Subsequently, it can reach, perturb and even penetrate the bacterial cytoplasmic membrane, leading to cell lysis. The molecular weight of the intermediate sized OPE-3 exceeds the permeability limit of the porin channel. Thus, similar to PPE-Th, the oligomer may exert toxicity against E. coli by disrupting the outer membrane. However, due to its rod-like structure and moderate molecular weight, OPE-3 may penetrate through the peptidoglycan layer, disturb the cytoplasmic membrane and trigger the release of cell content. Therefore, both molecular weight and architecture of the CPEs and OPEs are key factors controlling their interactions and toxicities with E. coli cells.
Generally, the antimicrobial activities of CPE and OPE compounds are greatly enhanced with the irradiation of UV or visible light such that they exhibit rapid and efficient toxicities at very low doses. The light-enhanced toxicities of the compounds are in part contributed by the dark biocidal mechanisms of the CPE and OPE compounds, such as those visualized in Figure 5. Light enhanced biocidal actions of CPEs and OPEs, however, have not been directly visualized. In this study, Gram-negative E. coli cells incubated with oligomeric OPE-3 and EO-OPE-1(C3) and polymeric PPE-DABCO under UV-irradiation have been imaged with TEM and SEM.
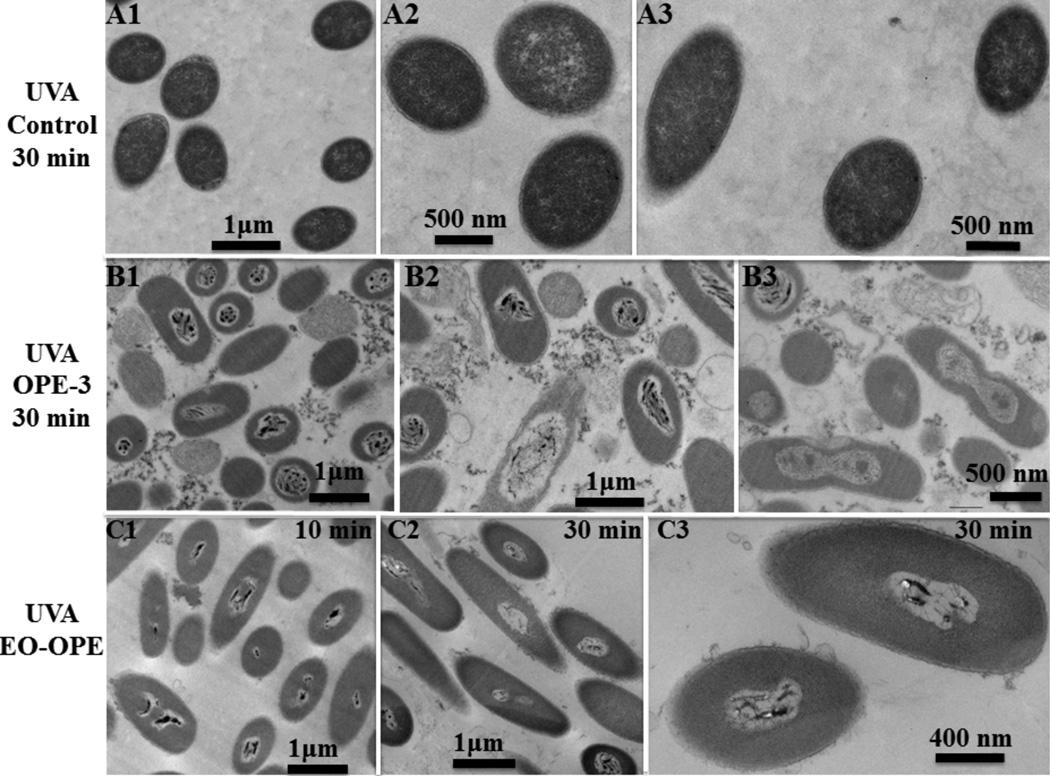
Similar to untreated cells (Figure 5A), a control sample of E. coli cells irradiated for 30 min without the addition of biocidal compounds appeared intact with unperturbed cytoplasm (Figure 6A). Thus UV-irradiation alone did not cause obvious damages to cell morphology, consistent with our previous findings that UV-irradiation alone causes very low-level toxicity to E. coli cells.6 However, the addition of OPE-3 or EO-OPE-1(C3) with UV-irradiation caused catastrophic damages to the bacteria (Figure 6B and 6C). In addition to the disruptions to the cell surfaces similar to those seen in cells incubated with the oligomers in the dark (Figure 5B and 5C), the cytoplasm of the UV-irradiated cells is also clearly damaged. In the OPE-3 and UV-light treated sample, a large amount of amorphous materials outside the cells was observed, which may be complexes of OPE-3 with cell envelope components such as LPS and/or released cell content. In contrast to the cells incubated with EO-OPE-1(C3) in the dark (Figure 5C1) where the cytoplasm remained relative intact after 10 minutes, UV-irradiation in the presence of EO-OPE-1(C3) for the same duration of time caused significant damages to the cell, including decreased density of the cytoplasm. However, the time dependent bacteriolytic effect as observed by the loss of cell cytoplasm, was not observed under UV irradiation (Figure 6C). This may be partly explained by the appearance of the dark, therefore dense, features in the UV-irradiated cells. Although the nature of these dark inclusions is not known, they could be oxidatively damaged and cross-linked cytoplasm components, such as proteins and nucleic acids.
Figure 6.
TEM micrographs of Gram negative E. coli (ATCC 11303) cells (108 CFU/mL) alone (A1, A2 and A3), incubated with 10 µg/mL OPE-3 (B1, B2 and B3) and EO-OPE-1(C3) (C1, C2 and C3) for different time intervals under UVA irradiation.
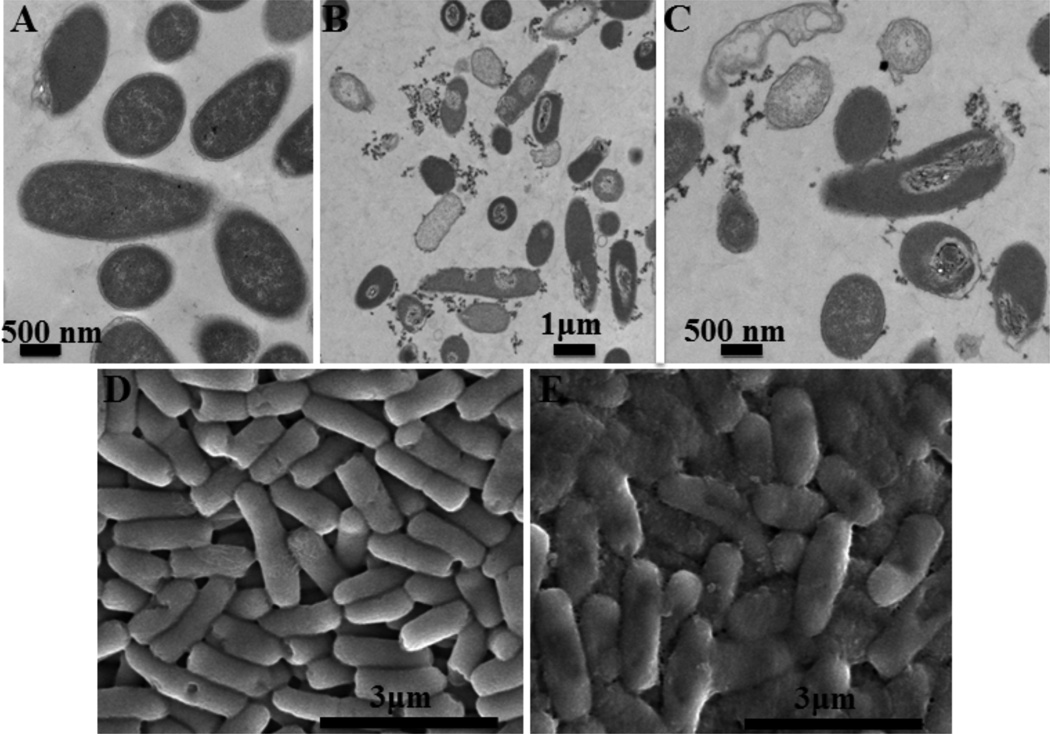
Polymeric CPEs have been observed to strongly bind to and remodel the outer membrane (previous findings of TEM imaging) of E. coli cells in the dark.9 Although their high molecular weights attenuate their ability to penetrate through the cell envelope, the oxygen radicals generated by the CPE compounds under UV-irradiation may cause sufficient damages and defects on the cell envelope to allow these polymeric agents to reach the cell interior and/or cell cytoplasm to be released. Similar to OPE-3, under UV-light irradiation, PPE-DABCO caused catastrophic damages to the bacteria cell envelope as well as induced the leakage of cell cytoplasm as evidenced by the empty (lighter colored) cells and the appearance of amorphous materials outside the cells (Figure 7B and C). These morphological changes are induced by the biocidal polymer as cells irradiated by UV-light alone are intact and smooth (Figure 7A). Damages induced to the cell envelope by PPE-DABCO under UV-irradiation are further confirmed by the drastic roughening of the cell surface imaged by SEM. Additionally, SEM imaging also confirmed the presence of amorphous materials outside the cells, which could be leaked cytoplasm and material complexed with the polymer (Figure 7D and E).
Figure 7.
TEM (A, B and C) micrographs of Gram-negative E. coli (ATCC 11303) cells (108 CFU/mL) alone (A), incubated with 10 µg/mL PPE-DABCO (B and C) under UV-420 irradiation for 30 min. SEM (D and E) micrographs of E. coli cells alone (D) and incubated with 1 µg/mL PPE-DABCO (E) under UV-420 irradiation for 60 min.
It is evident from our results that UV-irradiation causes further damages to the E. coli cell morphology in the presence of oligomeric and polymeric compounds. 1O2 has a relative long lifetime (10−6−10−5s) and diffusion distance in pure water. However, in cells, both lifetime and diffusion range of 1O2 must be significantly reduced due to its high reactivity towards biomolecules in the cytoplasm.21, 41 As a result, the damage caused by 1O2 is likely related to the positions of the sensitizers in the cells, whether at the cell surface or in the cell interior. Likewise, locations of damage of the bacteria under UV-irradiation also confirm the presence of the CPEs or OPEs, either at the cell surface or inside the cells. This “self-promoted uptake” mechanism42, which has been extensively studied for antimicrobial peptides, may also apply for the CPE and OPE compounds where defects created by the CPEs and OPEs on the bacterial outer membrane facilitate their entrance into the cell interior.
The CPE and OPE compounds have been shown to exert toxicity towards both Gram-negative (e.g., E coli.) and Gram-positive bacteria, which have compositionally and structurally different cell envelopes. Bactericidal actions of the compounds on Gram-positive bacteria have not been directly visualized. In this study, the dark and light-enhanced antimicrobial actions of oligomers and polymers on the Gram-positive S. epi bacteria were visualized for the first time via TEM and SEM imaging. S. epi cells incubated with oligomeric EO-OPE-1(C3) in the dark for 10–60 minutes do not appear damaged, with intact cell envelope and cytoplasm, compared with the control sample (Figure 8A). UV-irradiation alone also did not cause any obvious damages to the cells (Figure 8B1). However, UV-irradiation in the presence of CPE and OPE compounds caused obvious damage to the cell surfaces with PPE-DABCO inducing the highest level of roughness to the cell surface (Figure 8B2, 8B3, and 8B4). In contrast to extensive damage induced in Gram-negative E. coli cells, however, damage induced by the biocidal compounds in the Gram-positive S. epi cells under UV irradiation seems to be localized only to the cell surface. The compounds do not appear to have caused sufficient defects that extend through the cell envelope to cause leakage of the cytoplasm, for example. SEM imaging further verifies the cell surface damage caused by PPE-DABCO in similar conditions (see Figure S3 in Supporting Information).
Figure 8.
TEM micrographs of S.epi (ATCC 14990) cells (4×108 CFU/mL) alone incubated with 25 µg/mL antimicrobial agents in the dark or under UV-light irradiation for various periods.
It has been determined that the CPEs and OPEs exhibit both dark and light-enhanced antimicrobial activities against Gram-positive bacteria at concentrations lower than that used for the electron microscopy experiments in this study.6–7, 43 In addition, at similar concentrations of the CPE and OPE compounds, UV irradiation always increases the toxicities of the compounds. As shown in Figure 8B, the cytoplasm of the S. epi cells do not appear damaged, even under UV-irradiation with the biocidal compounds, which implies that the tested compounds were not capable of penetrating through the Gram-positive cell envelop. Boix and co-workers have found that the eosinophil cationic protein can induce significant morphological damages to the Gram-negative E. coli cells, but the protein does not induce any damage to the morphology of Gram-positive Staphylococcus aureus (S. aureus) cells under the same conditions.44 One explanation of this phenomenon is that the eosinophil protein can penetrate the E. coli cell envelope via the “self-promoted uptake” mechanism and then release the cell content. However, the thick and tough peptidoglycan layer in the Gram-positive bacterial cell walls provides a sufficient barrier to prevent damage to the cytoplasm. Meanwhile, the eosinophil protein exhibits high affinity toward bacterial peptidoglycan. The same principle may also account for the similar observations of the effect of the CPEs and OPEs on Gram-positive S. epi cells. The thick and negatively charged Gram-positive bacteria cell wall serves as the main binding site for the cationic CPE and OPE compounds but can also prevent the penetration of the compounds into the cell interior which may cause further damages to the cell. However, since the CPE and OPE compounds are toxic toward Gram-positive bacteria, disruption of the structure, and thereby function, of the peptidoglycan layer and anionic teichoic/lipoteichoic acid, seems to be sufficient for inducing cell death and serves as the toxicity mechanism for these compounds against Gram-positive bacteria.
Damages to E. coli proteins and plasmid DNA
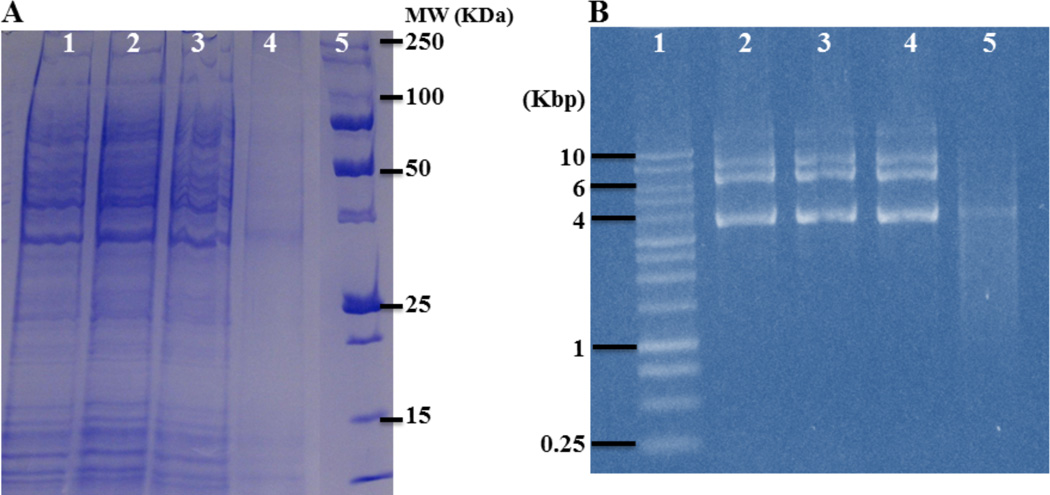
It is evident that the CPE and OPE compounds can induce damage to multiple cellular targets. Proteins in the cytoplasm are believed to be another target, particularly for 1O2 species generated under UV-irradiation21 as amino acids are readily oxidized by 1O2 with a rate constant in the range 107−108 M−1s−1.21, 45 Moreover, secondary ROS could react with a broader range of targets in the cell,46 including the DNA and RNA. The ROS sensitized by the CPE and OPE compounds can directly or indirectly covalently modify biomolecules in the cytoplasm, thereby inducing toxicity. As shown earlier, the CPEs and OPEs can cause severe damage inside the Gram-negative bacteria in the presence of UV-light. To better understand the nature of the changes, we investigated the effect of the oligomeric EO-OPE-1(C3) under UV-irradiation on two classes of potential targets in E. coli, proteins and plasmid DNA. As shown in Figure 9A, SDS-PAGE of E. coli cells under different exposure conditions to EO-OPE-1(C3) showed no significant differences in the electrophoretic mobilities or intensities of protein bands from cells treated with EO-OPE-1(C3) in the dark (Lane A2) or UV-irradiation alone (Lane A3) compared to untreated cells (Lane A1), indicating that the proteins did not undergo covalent modifications, such as cross linking or degradation. In contrast, protein bands from E. coli cells incubated with EO-OPE-1(C3) under UVA irradiation show significantly decreased intensities (Figure 9, Lane A4), indicating that 1O2 and ROS generated by the irradiation of EO-OPE-1(C3) had induced significant covalent modifications to the proteins that either caused the formation of insoluble aggregates that are too large to enter the electrophoresis gel or degradation of proteins into fragments too small to be detected by the technique. Similarly, the ROS sensitized by EO-OPE-1(C3) caused considerable decreases to the intensities of the E. coli plasmid DNA bands (Lane B5) compared to untreated cells (Lane B2), whereas the bands remained relatively unchanged from E. coli cells incubated with EO-OPE-1(C3) for an hour in the dark (Lane B3) or irradiated for an hour (Lane B4). Cross-linking and subsequent aggregation are the primary protein chemical degradations induced by 1O2 and secondary ROS.21 Likewise, 1O2 and secondary ROS could also induce the formation of covalent DNA-protein complexes.47 However, protein-DNA complexes and aggregates may not be soluble in water or identified from electrophoresis. In addition, ROS can also induce protein backbone fragmentation and DNA cleavage, which may also contribute to the changes observed in protein and plasmid DNA gel electrophoresis and the toxic functions of EO-OPE-1(C3). In addition to disrupting membrane lipid bilayer structure, damage to the proteins in bacterial cytoplasmic membranes by light-induced ROS, which serve critical functions, also compromise the membrane integrity and promote the release of cell content as well as the entrance of the CPEs and OPEs.
Figure 9.
(A) SDS-PAGE gels of the E. coli (4×108 CFU/mL) cells incubated with EO-OPE-1(C3) (25 µg/mL) for 1 hour. Lane A1: E. coli in the dark; Lane A2: E. coli incubated with EO-OPE for 1 hour in the dark; Lane A3: E. coli irradiated with UVA for 1 hour; Lane A4: E. coli incubated with EO-OPE under UVA-irradiation for 1 hour; Lane A5: Protein ladder. (B) Agarose-gel of the pET-20b plasmid extracted from E. coli BL21(DE3) (1×108 CFU/mL) incubated with EO-OPE-1(C3) (1 µg/mL) for 1 hour. Lane B1: DNA ladder; Lane B2: E. coli in the dark; Lane B3: E. coli incubated with EO-OPE in the dark for 1 hour; Lane B4: E. coli irradiated with UVA for 1 hour; Lane B5: E. coli incubated with EO-OPE under UVA-irradiation for 1 hour.
Summary
It is clear from our results that the polymeric CPE and oligomeric OPE compounds exert toxicity towards Gram-negative and Gram-positive bacteria through different mechanisms. While the compounds cause visible damage toward only the cell walls of Gram-positive bacteria, they damage the cell wall, plasma membrane, proteins, and plasmid DNA in Gram-negative bacteria. Our results indicate that the structures of the antimicrobial agents and bacterial outer envelope control their interactions as well as the biocidal mechanisms. In terms of bacteria cell envelope structures, the relative thin and soft cell envelope in Gram-negative E. coli cells does not serve as an efficient barrier for the oligomeric OPEs in the dark, but can impair the penetrating ability of bulky polymeric CPEs. Under UV-irradiation, all of the tested antimicrobial compounds can cross the cell envelope of the Gram-negative E. coli cells and cause damage to the cytoplasm, including oxidative and covalent modifications of proteins and plasmid DNA. In contrast, the thicker and tougher cell envelope in Gram-positive bacteria seems to be an efficient permeability barrier for the CPEs and OPEs both in the dark and under UV-irradiation. The cell envelope is also the main target of the CPEs and OPEs.
Damages to E. coli cells in the dark reveal the important role molecular structure of the CPE and OPE compounds play toward their toxicity mechanism. The large polymeric CPEs with high charge density may sequester and remove molecules from the bacterial surface and destabilize the cell envelope and outer membrane through an “ion-exchange” process, while the small oligomeric EO-OPE-1(C3) may easily cross the outer membrane without causing serious damages and directly disturb the cytoplasmic membrane and cytoplasm. The intermediate sized OPE-n compounds can induce damages to both bacterial surface and cytoplasm. The membrane activity of the CPEs and OPEs are affected by many factors, such as molecular conformation, size, side functional groups, and membrane composition. Furthermore, molecular level characterization of lipid membrane structural changes induced by the OPEs and CPEs point to a carpet or detergent-like mechanism for the membrane perturbation activities for these cationic and amphiphilic antimicrobial agents.
Supplementary Material
Acknowledgments
This research is financially supported by the Defense Threat Reduction Agency (Contract No. HDTRA 1-11-1-0004). Eva Y. Chi and David G. Whitten would like to acknowledge support from NSF (Award number 1207362). Ying Wang would like to thank the University of New Mexico (UNM) for the Graduate Research Supplement Grants. The authors gratefully acknowledge help from Dr. Yanli Tang and Dr. Zhijun Zhou of the Center for Biomedical Engineering (CBME) at UNM for providing the antimicrobial agents. We appreciate the kind help to the experimental work from Dr. Steve W. Graves, Mrs. Jingshu Zhu, Dr. Thomas S. Corbitt and Dr. Linnea K. Ista of CBME at UNM. The cryo-TEM work was performed at the National Resource for Automated Molecular Microscopy (NRAMM), which supported by grants from the National Center for Research Resources (2P41RR017573-11) and the National Institute of General Medical Sciences (9 P41 GM103310-11) from the National Institutes of Health. We also thank Dr. Bridget Carragher and Clint Potter from NRAMM for their assistance with cryo-TEM imaging. Bacterial TEM data were generated in the UNM Electron Microscopy Shared Facility supported by the UNM Health Sciences Center and the UNM Cancer Center. The Bruker Avance 300 NMR spectrometer was purchased with funds from NSF and ARRA (grant # 0840523). We thank Dr. Karen Ann Smith at UNM for assistance with NMR data collection.
We thank Dr. Tsutomu Matsui and Dr. Thomas Weiss for assistance with SAXS data collection, which was carried out at the Stanford Synchrotron Radiation Lightsource (SSRL), a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS, NCRR or NIH.
Footnotes
Supporting Information.
SAXS data for E. coli total lipid model membrane with DMSO. 31P SS-NMR spectra of E. coli model membranes with EO-OPE-1(C3). SEM micrographs of S. epi irradiated with PPE-DABCO. This material is available free of charge via the Internet at http://pubs.acs.org.
Reference
- 1.Kenawy ER, Worley SD, Broughton R. The chemistry and applications of antimicrobial polymers: A state-of-the-art review. Biomacromolecules. 2007;8(5):1359–1384. doi: 10.1021/bm061150q. [DOI] [PubMed] [Google Scholar]
- 2.Siedenbiedel F, Tiller JC. Antimicrobial Polymers in Solution and on Surfaces: Overview and Functional Principles. Polymers. 2012;4:46–71. [Google Scholar]
- 3.Muñoz-Bonilla A, Fernández-García M. Polymeric materials with antimicrobial activity. Prog. Polym. Sci. 2012;37(2):281–339. [Google Scholar]
- 4.Timofeeva L, Kleshcheva N. Antimicrobial polymers: mechanism of action, factors of activity, and applications. Appl. Microbiol. Biotechnol. 2011;89(3):475–492. doi: 10.1007/s00253-010-2920-9. [DOI] [PubMed] [Google Scholar]
- 5.Ji E, Corbitt TS, Parthasarathy A, Schanzes KS, Whitten DG. Light and Dark-Activated Biocidal Activity of Conjugated Polyelectrolytes. ACS Appl. Mater. Interfaces. 2011;3(8):2820–2829. doi: 10.1021/am200644g. [DOI] [PubMed] [Google Scholar]
- 6.Tang YL, Corbitt TS, Parthasarathy A, Zhou ZJ, Schanze KS, Whitten DG. Light-Induced Antibacterial Activity of Symmetrical and Asymmetrical Oligophenylene Ethynylenes. Langmuir. 2011;27(8):4956–4962. doi: 10.1021/la105018g. [DOI] [PubMed] [Google Scholar]
- 7.Zhou ZJ, Corbitt TS, Parthasarathy A, Tang YL, Ista LF, Schanze KS, Whitten DG. “End-Only” Functionalized Oligo(phenylene ethynylene)s: Synthesis, Photophysical and Biocidal Activity. J. Phys. Chem. Lett. 2010;1(21):3207–3212. [Google Scholar]
- 8.Wang Y, Canady TD, Zhou ZJ, Tang YL, Price DN, Bear DG, Chi EY, Schanze KS, Whitten DG. Cationic Phenylene Ethynylene Polymers and Oligomers Exhibit Efficient Antiviral Activity. ACS Appl. Mater. Interfaces. 2011;3(7):2209–2214. doi: 10.1021/am200575y. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Corbitt TS, Jett SD, Tang Y, Schanze KS, Chi EY, Whitten DG. Direct visualization of bactericidal action of cationic conjugated polyelectrolytes and oligomers. Langmuir. 2012;28(1):65–70. doi: 10.1021/la2044569. [DOI] [PubMed] [Google Scholar]
- 10.The exact molecular weights are currently unavailable, but the number average molar mass (Mn) values are estimated to be within the range of 20–30 kD [Google Scholar]
- 11.Zhao XY, Pinto MR, Hardison LM, Mwaura J, Muller J, Jiang H, Witker D, Kleiman VD, Reynolds JR, Schanze KS. Variable band gap poly(arylene ethynylene) conjugated polyelectrolytes. Macromolecules. 2006;39(19):6355–6366. [Google Scholar]
- 12.Wang Y, Jones EM, Tang YL, Ji EK, Lopez GP, Chi EY, Schanze KS, Whitten DG. Effect of Polymer Chain Length on Membrane Perturbation Activity of Cationic Phenylene Ethynylene Oligomers and Polymers. Langmuir. 2011;27(17):10770–10775. doi: 10.1021/la201820k. [DOI] [PubMed] [Google Scholar]
- 13.Tang YL, Hill EH, Zhou ZJ, Evans DG, Schanze KS, Whitten DG. Synthesis, Self-Assembly, and Photophysical Properties of Cationic Oligo(p-phenyleneethynylene)s. Langmuir. 2011;27(8):4945–4955. doi: 10.1021/la1050173. [DOI] [PubMed] [Google Scholar]
- 14.Chemburu S, Corbitt TS, Ista LK, Ji E, Fulghum J, Lopez GP, Ogawa K, Schanze KS, Whitten DG. Light-induced biocidal action of conjugated polyelectrolytes supported on colloids. Langmuir. 2008;24(19):11053–11062. doi: 10.1021/la8016547. [DOI] [PubMed] [Google Scholar]
- 15.There may be other mechanisms involved in the light-activated biocidal actions, such as the halide counter ion could be oxidized by the CPEs/OPEs’ excited state, leading to the formation of corrosive halogen atoms [Google Scholar]
- 16.Cowan MK, Talaro KP. Microbiology: A Systems Approach. 2 ed. New York: McGraw-Hill Science; 2008. [Google Scholar]
- 17.Amro NA, Kotra LP, Wadu-Mesthrige K, Bulychev A, Mobashery S, Liu GY. High-Resolution Atomic Force Microscopy Studies of the Escherichia coli Outer Membrane: Structural Basis for Permeability. Langmuir. 2000;16(6):2789–2796. [Google Scholar]
- 18.Rogers HG, Perkins HR, Ward JB. Microbial Cell Walls and Membranes. 1 ed. London: Chapman and Hall; 1980. [Google Scholar]
- 19.Schäffer C, Messner P. The structure of secondary cell wall polymers: how Gram-positive bacteria stick their cell walls together. Microbiology. 2005;151(3):643–651. doi: 10.1099/mic.0.27749-0. [DOI] [PubMed] [Google Scholar]
- 20.Lambert PA. Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J. Appl. Microbiol. 2002;92:46S–54S. [PubMed] [Google Scholar]
- 21.Davies MJ. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Bioph. Res. Co. 2003;305(3):761–770. doi: 10.1016/s0006-291x(03)00817-9. [DOI] [PubMed] [Google Scholar]
- 22.Xing CF, Xu QL, Tang HW, Liu LB, Wang S. Conjugated Polymer/Porphyrin Complexes for Efficient Energy Transfer and Improving Light-Activated Antibacterial Activity. J. Am. Chem. Soc. 2009;131(36):13117–13124. doi: 10.1021/ja904492x. [DOI] [PubMed] [Google Scholar]
- 23.Dahl TA, Midden WR, Hartman PE. Comparison of killing of gram-negative and gram-positive bacteria by pure singlet oxygen. J. Bacteriol. 1989;171(4):2188–94. doi: 10.1128/jb.171.4.2188-2194.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Chi EY, Schanze KS, Whitten DG. Membrane activity of antimicrobial phenylene ethynylene based polymers and oligomers. Soft Matter. 2012;8:8547–8558. [Google Scholar]
- 25.Mishra A, Gordon VD, Yang LH, Coridan R, Wong GCL. HIV TAT forms pores in membranes by inducing saddle-splay curvature: Potential role of bidentate hydrogen bonding. Angew. Chem. Int. Edit. 2008;47(16):2986–2989. doi: 10.1002/anie.200704444. [DOI] [PubMed] [Google Scholar]
- 26.Bechinger B. Detergent-like properties of magainin antibiotic peptides: a 31P solid-state NMR spectroscopy study. Biochim. Biophys. Acta. 1712;(1):101–108. doi: 10.1016/j.bbamem.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Zhou ZJ, Zhu JS, Tang Y, Canady TD, Chi EY, Schanze KS, Whitten DG. Dark Antimicrobial Mechanisms of Cationic Phenylene Ethynylene Polymers and Oligomers against Escherichia coli. Polymers. 2011;3:1199–1214. [Google Scholar]
- 28.Wu G, Khant HA, Chiu W, Lee KYC. Effects of bilayer phases on phospholipid-poloxamer interactions. Soft Matter. 2009;5:1496–1503. [Google Scholar]
- 29.Phospholipid abbreviation: 1,2-Dioleoyl-sn-Glycero-3-Phosphoethanolamine=DOPE; 1,2-Dioleoyl-sn-Glycero-3-[Phospho-rac-(1-glycerol)] (Sodium Salt)=DOPG [Google Scholar]
- 30.Wang Y, Tang YL, Zhou ZJ, Ji E, Lopez GP, Chi EY, Schanze KS, Whitten DG. Membrane Perturbation Activity of Cationic Phenylene Ethynylene Oligomers and Polymers: Selectivity against Model Bacterial and Mammalian Membranes. Langmuir. 2010;26(15):12509–12514. doi: 10.1021/la102269y. [DOI] [PubMed] [Google Scholar]
- 31.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Wimley WC, Hristova K. Antimicrobial peptides: successes, challenges and unanswered questions. J. Membrane Biol. 2011;239(1–2):27–34. doi: 10.1007/s00232-011-9343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wimley WC. Describing the Mechanism of Antimicrobial Peptide Action with the Interfacial Activity Model. ACS Chem. Biol. 2010;5(10):905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang LH, Gordon VD, Mishra A, Sorn A, Purdy KR, Davis MA, Tew GN, Wong GCL. Synthetic antimicrobial, oligomers induce a composition-dependent topological transition in membranes. J. Am. Chem. Soc. 2007;129(40):12141–12147. doi: 10.1021/ja072310o. [DOI] [PubMed] [Google Scholar]
- 35.Turner DC, Gruner SM. X-ray diffraction reconstruction of the inverted hexagonal (HII) phase in lipid-water systems. Biochemistry. 1992;31(5):1340–1355. doi: 10.1021/bi00120a009. [DOI] [PubMed] [Google Scholar]
- 36.Alexandridis P, Olsson U, Lindman B. A Record Nine Different Phases (Four Cubic, Two Hexagonal, and One Lamellar Lyotropic Liquid Crystalline and Two Micellar Solutions) in a Ternary Isothermal System of an Amphiphilic Block Copolymer and Selective Solvents (Water and Oil) Langmuir. 1998;14(10):2627–2638. [Google Scholar]
- 37.Strandberg E, Ulrich AS. NMR methods for studying membrane-active antimicrobial peptides. Concepts Magn. Reson. A. 2004;23A(2):89–120. [Google Scholar]
- 38.Hill EH, Stratton K, Whitten DG, Evans DG. Molecular Dynamics Simulation Study of the Interaction of Cationic Biocides with Lipid Bilayers: Aggregation Effects and Bilayer Damage. Langmuir. 2012 doi: 10.1021/la303158c. [DOI] [PubMed] [Google Scholar]
- 39.Epand RM, Epand RF. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochem. Biophys. Acta, Biomembr. 2009;1788(1):289–294. doi: 10.1016/j.bbamem.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 40.Lenoir S, Pagnoulle C, Galleni M, Compere P, Jerome R, Detrembleur C. Polyolefin matrixes with permanent antibacterial activity: preparation, antibacterial activity, and action mode of the active species. Biomacromolecules. 2006;7(8):2291–2296. doi: 10.1021/bm050850c. [DOI] [PubMed] [Google Scholar]
- 41.Redmond RW, Kochevar IE. Spatially resolved cellular responses to singlet oxygen. Photochem. Photobiol. 2006;82(5):1178–1186. doi: 10.1562/2006-04-14-IR-874. [DOI] [PubMed] [Google Scholar]
- 42.Powers J-PS, Hancock REW. The relationship between peptide structure and antibacterial activity. Peptides. 2003;24(11):1681–1691. doi: 10.1016/j.peptides.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 43.Corbitt TS, Ding LP, Ji EY, Ista LK, Ogawa K, Lopez GP, Schanze KS, Whitten DG. Light and dark biocidal activity of cationic poly(arylene ethynylene) conjugated polyelectrolytes. Photochem. Photobiol. Sci. 2009;8(7):998–1005. doi: 10.1039/b902646k. [DOI] [PubMed] [Google Scholar]
- 44.Torrent M, Navarro S, Moussaoui M, Nogues MV, Boix E. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry. 2008;47(11):3544–3555. doi: 10.1021/bi702065b. [DOI] [PubMed] [Google Scholar]
- 45.Rabek JF. Photochemistry and Photophysics. Vol. 3. Boca Raton: CRC Press; 1991. [Google Scholar]
- 46.Imlay JA. Pathways of oxidative damage. Annu. Rev. Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 47.Meffert R, Dose K, Rathgeber G, Schefer HJ. Ultraviolet crosslinking of DNA protein complexes via 8-azidoadenine. Methods Mol. Biol. 2009;543:389–402. doi: 10.1007/978-1-60327-015-1_24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.