The exoribonuclease Rrp6 forms a complex with Rrp47 that functions in RNA processing and turnover pathways in eukaryotic cells. Using a novel experimental strategy we call DECOID (decreased expression of complexes by overexpression of interacting domains), Rrp47 is resolved from the Rrp6 exoribonuclease, and the two proteins are shown to have separable functions. This study demonstrates a general role for Rrp47 in Rrp6-mediated processing reactions prior to catalysis and reveals a redundant requirement for the two exoribonucleases Rrp6 and Rex1 in the 3′ maturation of box C/D snoRNAs.
Keywords: RNA surveillance, RNA processing, yeast, synthetic lethality, protein overexpression, exosome
Abstract
The eukaryotic exosome exoribonuclease Rrp6 forms a complex with Rrp47 that functions in nuclear RNA quality control mechanisms, the degradation of cryptic unstable transcripts (CUTs), and in the 3′ end maturation of stable RNAs. Stable expression of Rrp47 is dependent upon its interaction with the N-terminal domain of Rrp6 (Rrp6NT). To address the function of Rrp47 independently of Rrp6, we developed a DECOID (decreased expression of complexes by overexpression of interacting domains) strategy to resolve the Rrp6/Rrp47 complex in vivo and employed mpp6Δ and rex1Δ mutants that are synthetic lethal with loss-of-function rrp47 mutants. Strikingly, Rrp47 was able to function in mpp6Δ and rex1Δ mutants when separated from the catalytic and exosome-binding domains of Rrp6, whereas a truncated Rrp47 protein lacking its C-terminal region caused a block in cell growth. Northern analyses of the conditional mutants revealed a specific block in the 3′ maturation of box C/D snoRNAs in the rex1 rrp47 mutant and widespread inhibition of Rrp6-mediated RNA surveillance processes in the mpp6 rrp47 mutant. In contrast, growth analyses and RNA northern blot hybridization analyses showed no effect on the rrp47Δ mutant upon overexpression of the Rrp6NT domain. These findings demonstrate that Rrp47 and Rrp6 have resolvable functions in Rrp6-mediated RNA surveillance and processing pathways. In addition, this study reveals a redundant requirement for Rrp6 or Rex1 in snoRNA maturation and demonstrates the effective use of the DECOID strategy for the resolution and functional analysis of protein complexes.
INTRODUCTION
The degradation of RNA is a fundamentally important biological process. Firstly, cellular RNAs are typically generated by the accurate, limited degradation of larger precursor molecules. RNA fragments released through such processing reactions are also subjected to degradation. In addition, mRNA transcripts have defined functional lifetimes and are degraded in a temporally controlled manner. Furthermore, both mRNAs and noncoding RNAs are subjected to quality control systems that eliminate incorrectly processed transcripts or misassembled ribonucleoprotein particles (Houseley and Tollervey 2009; Wolin et al. 2012). A significant proportion of transcripts are degraded by such “RNA surveillance” processes, even in normally growing, healthy cells (Gudipati et al. 2012). Finally, low level antisense and inter-genic transcription generates a large number of noncoding RNAs, many of which are subjected to rapid degradation (Wyers et al. 2005).
A major source of 3′→5′ ribonucleolytic activity in eukaryotic cells is the exosome ribonuclease complex (Mitchell et al. 1997; Allmang et al. 1999b). The exosome functions in both the limited trimming of RNA precursors in RNA processing pathways and in the complete destruction of RNAs as part of RNA surveillance mechanisms, the removal of RNA fragments generated during RNA processing, and the elimination of transiently expressed, cryptic unstable transcripts (CUTs) (Chlebowski et al. 2013). The core of the eukaryotic exosome is itself catalytically inert, ribonuclease activity being provided by two additional associated proteins; Rrp44 (also known as Dis3) is bound to the base of the core, while Rrp6 is associated with the cap region (Bonneau et al. 2009; Makino et al. 2013). Rrp44 is a member of the RNase II family of exoribonucleases that also has an N-terminal PIN domain with associated endonucleolytic activity and is found tightly associated with the core exosome in both the nucleus and cytoplasmic regions. Rrp6 is an RNase D-related 3′→5′ exoribonuclease (Burkard and Butler 2000) that contains an N-terminal PMC2NT domain, a “DEDD” catalytic domain, an HRDC domain, and a C-terminal region that is required for interaction with the exosome complex. A C-terminal truncation of Rrp6 that prevents its stable association with the exosome does not inhibit Rrp6-dependent processes (Callahan and Butler 2008), suggesting that Rrp6 can function largely independently of its association with the exosome.
The activity of the nuclear exosome is dependent upon additional proteins or complexes, including the Trf/Air/Mtr4 polyadenylation complex TRAMP, the Nrd1/Nab3/Sen1 complex, and the nuclear RNA-binding proteins Rrp47 (also known as Lrp1) and Mpp6 (Chlebowski et al. 2013). The Nrd1/Nab3/Sen1 complex functions in the transcription termination of short RNA polymerase II transcripts (Steinmetz et al. 2001), copurifies together with RNA polymerase II and exosomes in large transcription/processing complexes, and promotes exosome-mediated degradation of transcripts containing Nrd1/Nab3 binding sites (Vasiljeva and Buratowski 2006). TRAMP promotes Nrd1/Nab3-dependent transcription termination of snoRNAs (Grzechnik and Kufel 2008) and adds poly(A) tails onto RNA substrates, which promotes their subsequent degradation (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005). Rrp47 interacts directly with Rrp6 and functions specifically to promote Rrp6-mediated processes (Mitchell et al. 2003; Peng et al. 2003). In contrast, the exosome-associated protein Mpp6 has been proposed to function as an Rrp44-specific cofactor (Milligan et al. 2008).
Functional redundancy is a common feature of biological systems. Yeast rrp6 and rrp44 mutants have synergistic phenotypes, and both Rrp6 and Rrp44 function in the processing of stable RNAs, in RNA surveillance pathways, and in the degradation of CUTs (Gudipati et al. 2012; Schneider et al. 2012). Redundancy between Rrp6 and an Mpp6-stimulated activity, presumably that of Rrp44, is supported by the synthetic lethality observed for rrp6Δ mpp6Δ and rrp47Δ mpp6Δ double mutants (Milligan et al. 2008). Yeast rrp6Δ and rrp47Δ mutants are also synthetic lethal with loss-of-function mutations in REX1/RNA82 (van Hoof et al. 2000a; Peng et al. 2003), a gene that encodes another RNase D-related 3′→5′ exoribonuclease (Ozanick et al. 2009) that is not physically associated with the exosome. Like rrp6 mutants, rex1 strains are defective in the 3′ end maturation of stable RNAs and accumulate polyadenylated RNAs (Piper et al. 1983; van Hoof et al. 2000a).
Rrp6 and Rrp47 interact directly through their N-terminal PMC2NT and Sas10/C1D domains, respectively, and normal Rrp47 expression levels are strongly dependent upon this interaction. In contrast, Rrp6 expression and nuclear localization are not significantly affected in the rrp47Δ mutant (Stead et al. 2007; Feigenbutz et al. 2013). Both Rrp47 and the homologous human protein C1D show nucleic acid binding activity in vitro, with a specificity for structured substrates (Schilders et al. 2007; Stead et al. 2007). Notably, purified recombinant Rrp6 exonuclease shows poor processivity through structured RNA (Burkard and Butler 2000; Liu et al. 2006). This led to the proposal that Rrp47 may promote the Rrp6-mediated degradation of structured RNA. However, Rrp6 lacking the PMC2NT domain shows comparable exonucleolytic activity to the full-length protein on the substrates tested in vitro (Assenholt et al. 2008; Wasmuth and Lima 2012). Furthermore, the N-terminal Sas10/C1D domain is able to complement the synthetic lethality of an rrp47Δ rex1Δ mutant but is itself insufficient for the RNA-binding activity of Rrp47 (Costello et al. 2011).
In these studies, we sought to address whether the physical association of Rrp47 with the Rrp6 exonuclease is critical for its function in RNA processing and degradation. Demonstration that Rrp6 and Rrp47 can function when the proteins are divorced would provide prima facie evidence that Rrp47 functions in substrate recognition or activation, before catalysis. Rrp47 stability is dependent upon its interaction with the Rrp6NT domain (Feigenbutz et al. 2013). We therefore used an overexpression system to induce expression of the Rrp6NT domain and titrate Rrp47 out of Rrp6 complexes. We refer to this approach as DECOID (decreased expression of complexes by overexpression of interacting domains).
Our results reveal that Rrp47 is functional as a full-length protein when separated from the catalytic and exosome-binding domains of Rrp6 but that its ability to function is lost upon deletion of the C-terminal region of the protein. This region of Rrp47 has previously been shown to be required for its interaction with the snoRNP proteins Nop56 and Nop58 and for binding to RNA (Costello et al. 2011). Genetic assays and Northern analyses show that the resolved Rrp6/Rrp47 complex functions in the processing or degradation of all Rrp6 substrates tested. Intriguingly, conditional rrp47 rex1 mutants showed a specific defect in snoRNA production, while rrp47 mpp6 mutants were blocked in RNA turnover pathways. These findings are consistent with a function for Rrp47 in the recognition of most, if not all, substrates prior to RNA processing or degradation by Rrp6.
RESULTS AND DISCUSSION
Overexpression of the Rrp6NT domain segregates Rrp47 from full-length Rrp6
To determine whether Rrp47 could be titrated out of complexes containing full-length Rrp6 by overexpression of the Rrp6NT domain, a yeast strain expressing an Rrp47-GFP fusion protein from its own promoter (Feigenbutz et al. 2013) was transformed with plasmids that encode either a GAL-inducible GST-Rrp6NT fusion protein or just the GST domain. Lysates from cells grown in galactose-based medium were fractionated by ultracentrifugation through glycerol gradients, and the sedimentation behavior of Rrp47 was compared to the total cellular protein profiles and to protein markers that were resolved in parallel gradients.
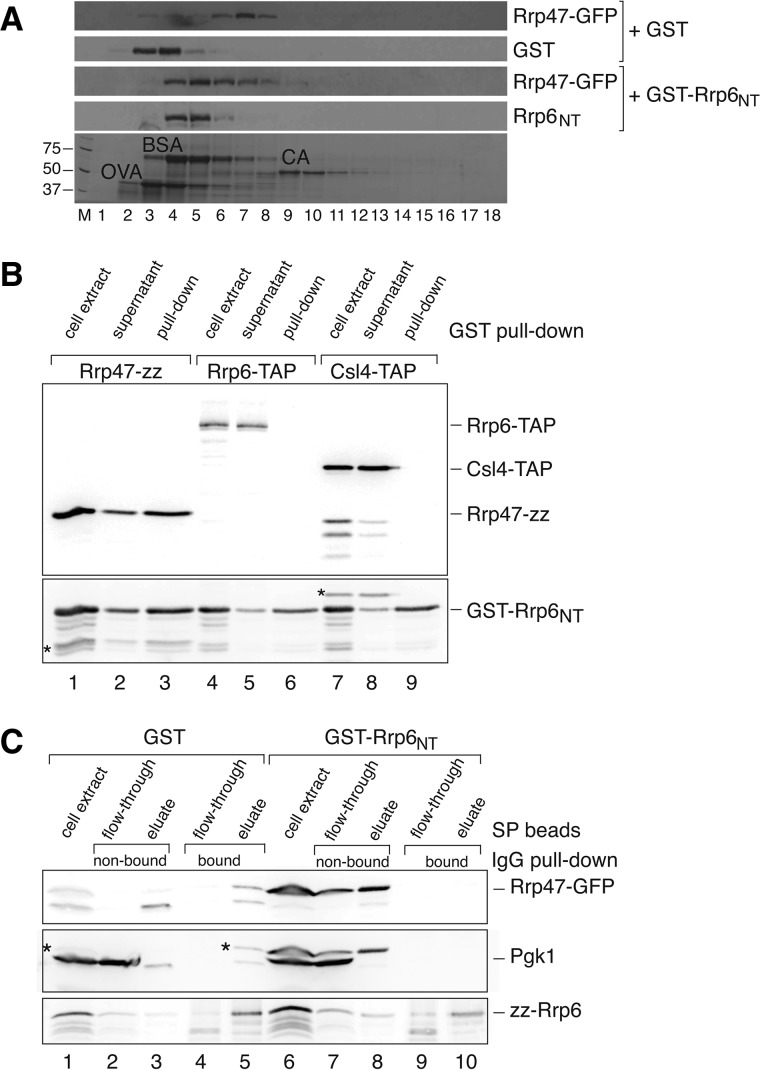
In control cells expressing GST, Rrp47 sedimented as part of a moderately sized complex with a sedimentation coefficient between that of BSA (4.6 S) and catalase (11.3 S) (Fig. 1A). A similar distribution of Rrp47, with a peak in fraction 7, was observed in lysates from nontransformed cells grown in glucose-based minimal medium (data not shown). GST sedimented in a distinct region of the gradient from Rrp47-GFP, being observed in fractions 3 and 4. Upon overexpression of GST-Rrp6NT, Rrp47 exhibited a slower sedimentation rate, and the two proteins showed a clear overlap, with a peak in fraction 5 (Fig. 1A). The observed shift in sedimentation profile for Rrp47 upon induction of GST-Rrp6NT was supported by completely reproducible protein patterns of these gradient fractions upon SDS-PAGE analyses.
FIGURE 1.
Rrp6NT induction leads to disruption of the Rrp6/Rrp47 complex. (A) Glycerol gradient analyses of extracts from strains expressing epitope-tagged Rrp47-GFP after induction of GST-Rrp6NT or GST. Gradient fractions 1–18 were resolved by SDS-PAGE and analyzed by Western blotting. Bovine serum albumin (BSA), ovalbumin (OVA), and catalase (CA) markers were resolved in a parallel gradient and fractions analyzed by SDS-PAGE; molecular weight markers (masses indicated in kDa) are resolved in lane M. (B) Pull-down analysis of Rrp47-zz, Rrp6-TAP, and Csl4-TAP fusion proteins with GST-Rrp6NT. The upper panel shows a Western analysis of Rrp47-zz, Rrp6-TAP, and Csl4-TAP proteins in equivalent aliquots of the cell extract, supernatant, and pull-down fractions. The lower panel shows a subsequent Western analysis of the same blot using a GST antibody. Asterisks indicate bands due to residual signal from the initial Western. (C) Pull-down analysis of Rrp47-GFP binding to zz-Rrp6 upon induction of either GST or GST-Rrp6NT. Cell extracts were initially resolved into “flow-through” and “eluate” fractions by SP sepharose chromatography. Both fractions were then incubated with IgG-sepharose beads, giving “bound” and “nonbound” fractions. Equivalent aliquots of each fraction were then analyzed by Western blotting for the presence of Rrp47-GFP (upper panel), Pgk1 (center panel), and zz-Rrp6 (lower panel). Asterisks in the center panel indicate bands due to residual signal from the Rrp47-GFP Western.
To determine whether the observed cosedimentation of GST-Rrp6NT and Rrp47 reflects an interaction between these two proteins, GST pull-down assays were performed on lysates from strains expressing an Rrp47-zz fusion protein from its own promoter (Mitchell et al. 2003) after induction of GST-Rrp6NT overexpression. Parallel assays were also performed on strains expressing C-terminal TAP-tagged fusion proteins of Rrp6 or the core exosome subunit Csl4, both expressed from their own promoter. The pull-down data shown in Figure 1B clearly demonstrate that Rrp47, but not full-length Rrp6 or Csl4, is associated with the induced GST-Rrp6NT protein (Fig. 1B). We have previously shown that no interaction is observed between Rrp47-zz and GST (see also Fig. 1A; Stead et al. 2007). Depletion of Rrp47-zz and GST-Rrp6NT from the cell lysates upon incubation with glutathione sepharose beads occurred at comparable efficiencies (cf. the band intensities for Rrp47-zz and GST-Rrp6NT in Fig. 1B, lanes 1–3), suggesting that the majority of Rrp47 is associated with the GST fusion protein.
We then determined whether Rrp47 could still be detected in complexes with Rrp6 upon overexpression of GST-Rrp6NT. GST or the GST-Rrp6NT protein was induced in an rrp47-GFP rrp6Δ strain expressing a plasmid-encoded zz-Rrp6 fusion protein, and Rrp6 complexes were captured in IgG pull-down assays. We have previously shown that the zz-Rrp6 fusion protein, which is expressed from the RRP4 promoter in this construct (Allmang et al. 1999b), is expressed at a level comparable to that of the Rrp6-TAP fusion protein (Stead et al. 2007). To reduce the degree of Rrp6 proteolysis observed during pull-down reactions, Rrp6 complexes were initially enriched from whole cell lysates by ion exchange chromatography using SP-sepharose beads (Allmang et al. 1999b). IgG pull-down assays were performed on both the flow-through and eluate fractions obtained by ion exchange chromatography, and Western analyses were performed on equivalent aliquots of the cell extracts, flow-through, and eluate fractions.
Western analyses revealed that Rrp47 efficiently copurified with Rrp6 from lysates of strains overexpressing the GST protein (Fig. 1C; cf. “nonbound” lanes 2 and 3 with “bound” lanes 4 and 5), demonstrating that Rrp47 and Rrp6 remained stably associated under the experimental conditions used in the pull-down assay. In contrast, the interaction between Rrp47 and endogenous Rrp6 was barely detectable upon overexpression of the GST-Rrp6NT fusion protein (Fig. 1C; cf. lanes 7 and 8 with 9 and 10). A significantly higher level of expression of Rrp47-GFP was observed in the cells expressing GST-Rrp6NT, presumably because the increased availability of this domain serves to stabilize Rrp47 (Feigenbutz et al. 2013). Comparison of the amount of Rrp47 present in the Rrp6 pull-down samples upon induction of GST or GST-Rrp6NT was therefore standardized to the expression level of Pgk1 in the cell extracts. Pgk1 is clearly visible as the major band in the cell extract and the SP sepharose flow-through fractions in the center panel of Figure 1C and is absent from the upper panel. Quantitative analyses revealed that the amount of Rrp47 associated with Rrp6 upon overexpression of GST-Rrp6NT was ∼4% of that observed upon overexpression of GST. Comparable amounts of Rrp6 protein were observed in the pull-downs in both samples (Fig. 1C, lower panel; cf. lanes 4 and 5 with 9 and 10). Taken together, the data from glycerol gradient analyses and pull-down experiments demonstrate that Rrp47 is effectively titrated out of full-length Rrp6-containing complexes upon overexpression of the Rrp6NT “decoy” domain.
Rrp47 is functional when physically segregated from the catalytic and exosome-binding domains of Rrp6
To test genetically whether Rrp47 can function when physically separated from full-length Rrp6, mpp6Δ and rex1Δ mutants were transformed with plasmids expressing either GST or GST-Rrp6NT from the GAL promoter, and the resultant transformants were assayed for growth on glucose- and galactose-based media. Isogenic wild-type and rrp47Δ strains were also transformed with the same plasmids and assayed in parallel. Strikingly, transformants of both mpp6Δ and rex1Δ mutants expressing GST-Rrp6NT were viable on galactose-based medium. We then compared the growth rates of transformants expressing either GST or GST-Rrp6NT during incubation in galactose-based medium. There was only a very slight decrease in growth rate seen in the wild-type strain upon induction of the GST-Rrp6NT protein. In contrast, mpp6Δ and rex1Δ strains overexpressing GST-Rrp6NT clearly grew slower than transformants overexpressing the GST domain (Fig. 2A). No detectable difference was observed in the growth rate of transformants of the rrp47Δ mutant, demonstrating that induction of the GST-Rrp6NT domain does not elicit an Rrp47-independent dominant negative growth phenotype. Plasmid shuffle assays showed that overexpression of GST-Rrp6NT in itself did not suppress the synthetic lethality of the mpp6Δ rrp47Δ double mutant (data not shown). These analyses demonstrate that the function of Rrp47 is impaired, but nevertheless not blocked, when Rrp47 is segregated from full-length Rrp6.
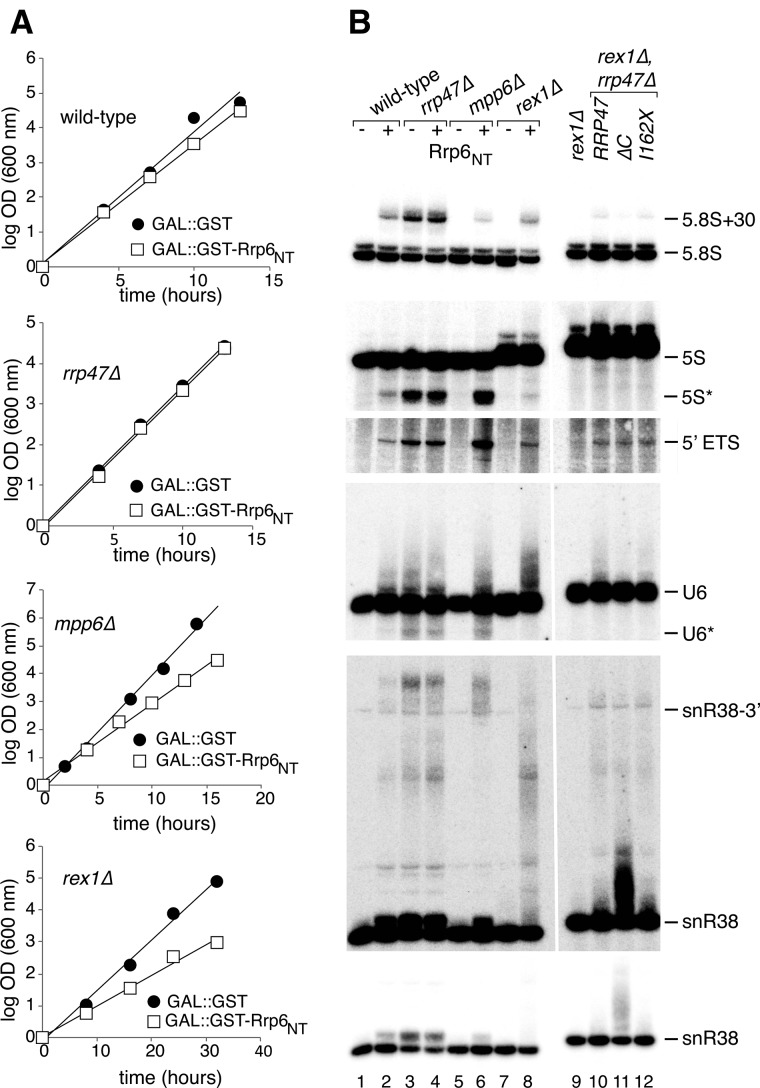
FIGURE 2.
Growth rate and RNA analyses of mpp6Δ and rex1Δ mutants upon Rrp6NT induction. (A) Growth rate analyses of wild-type, rrp47Δ, mpp6Δ, and rex1Δ strains after induction of GST or GST-Rrp6NT. Cultures were grown in galactose-based minimal medium at 30°C and maintained in early exponential growth by dilution with pre-warmed medium. The increase in OD600 is expressed as log2 values. All plotted lines of best fit have Rf values greater than 0.98. (B) Lanes 1–8, acrylamide gel Northern analyses of RNA from wild-type, rrp47Δ, mpp6Δ, or rex1Δ strains expressing either GST (−) or GST-Rrp6NT (+). RNA was isolated from cultures grown in selective galatose-based medium. Lanes 9–12, Northern analyses of RNA from a rex1Δ mutant and from a rex1Δ rrp47Δ mutant expressing either the full-length RRP47 gene, the rrp47ΔC allele, or the rrp47I162X allele. RNA was isolated from cultures grown in glucose-based selective minimal medium. The blot was successively hybridized with probes complementary to the RNA species indicated. Identical hybridizations and exposure times are shown for lanes 1–8 and 9–12.
To address the nature of the slow growth phenotypes in mpp6Δ and rex1Δ strains observed upon induction of GST-Rrp6NT expression, Northern analyses were performed on RNA isolated from transformants of a wild-type strain and from rrp47Δ, mpp6Δ, or rex1Δ mutants (Fig. 2B). RNA processing and degradation phenotypes were observed upon induction of GST-Rrp6NT in the wild-type strain that are similar in nature to those observed in rrp47Δ mutants (Mitchell et al. 2003; Costello et al. 2011) but considerably weaker, consistent with a mild inhibition of Rrp47 function in wild-type cells upon segregation of Rrp47 from full-length Rrp6 (Fig. 2B; cf. lanes 1–3). Specifically, induction of GST-Rrp6NT in the wild-type strain led to a detectable accumulation of the “5.8S + 30” rRNA processing intermediate and 3′ extended forms of the snoRNA snR38, a modest accumulation of the excised RNA polymerase I 5′ external transcribed spacer (5′ ETS) fragment, and the accumulation of extended forms and shorter degradation intermediates of U6 snRNA (the degradation product is denoted U6* in Fig. 2B). A discrete, truncated 5S rRNA fragment was observed (also labeled with an asterisk in Fig. 2B) that was not detected with a probe complementary to the 3′ terminus and, hence, is shortened from the 3′ end; a similarly sized, 3′ truncated 5S rRNA degradation fragment has previously been reported in rrp44, rrp6Δ, and trf4 mutants and is proposed to reflect an RNA surveillance substrate (Kadaba et al. 2006; Callahan and Butler 2008).
In contrast to the wild-type strain, induction of GST-Rrp6NT expression in the rrp47Δ strain had no significant discernible effect. Thus, GST-Rrp6NT expression induced no detectable Rrp47-independent “off-target” effects in either growth rate or the RNA processing and degradation pathways analyzed.
Notably, induction of GST-Rrp6NT in the mpp6Δ mutant caused a greater accumulation of the truncated 5S rRNA and the 5′ ETS fragment than was observed in the rrp47Δ mutant, while the 3′ end maturation of 5.8S rRNA was only modestly affected (Fig. 2B; cf. lanes 4–6). This strain also accumulated the U6 snRNA degradation fragments seen in the rrp47Δ mutant. Furthermore, hybridization with a probe against the box C/D snoRNA snR38 revealed a specific accumulation of the longest extended forms seen in the rrp47Δ mutant. Overexpression of GST-Rrp6NT in the rex1Δ mutant largely caused the modest defects observed in the wild-type strain. However, there was a specific accumulation of extended snR38 and snR50 RNAs (Fig. 2B; data not shown).
The data shown in Figures 1 and 2 strongly supports the conclusion that Rrp47 is able to function effectively in Rrp6-dependent RNA processing and degradation pathways when the two proteins are physically uncoupled from one another. We cannot formally eliminate the possibility that the small fraction of Rrp47 that remains associated with full-length Rrp6 upon GST-Rrp6NT induction is sufficient for Rrp47 function. However, further analyses with an rrp47 mutant detailed below provide additional support for our interpretation. These data also suggest that Rrp47 and Mpp6 function in redundant pathways that largely target RNA for degradation, while Rex1 and Rrp47 are required for redundant pathways that mediate productive processing of snoRNA.
The C-terminal region of Rrp47 is critical for its function when segregated from the catalytic and exosome-binding domains of Rrp6
As noted above, expression of the N-terminal Sas10/C1D domain of Rrp47 complements the synthetic lethality of rex1Δ rrp47Δ strains (Costello et al. 2011). We performed Northern analyses on RNA from a rex1Δ rrp47ΔC strain, which encodes essentially only the Sas10/C1D domain of Rrp47, and from a rex1Δ rrp47I162X mutant, which encodes a mutant Rrp47 protein lacking a cluster of basic residues at the C terminus of the protein that is required for RNA binding activity in vitro (Costello et al. 2011). The rex1Δ rrp47ΔC double mutant, but not the rex1Δ rrp47I162X double mutant or either rex1Δ or rrp47Δ single mutants, showed a strong accumulation of a heterogeneous population of snR38 molecules with short 3′ extensions (Fig. 2B, lane 11). Again, similar results were observed for snR50 (data not shown). Thus, either segregation of Rrp47 from full-length Rrp6 or deletion of the C-terminal region of Rrp47 causes a specific defect in snoRNA 3′ end maturation in the rex1Δ mutant.
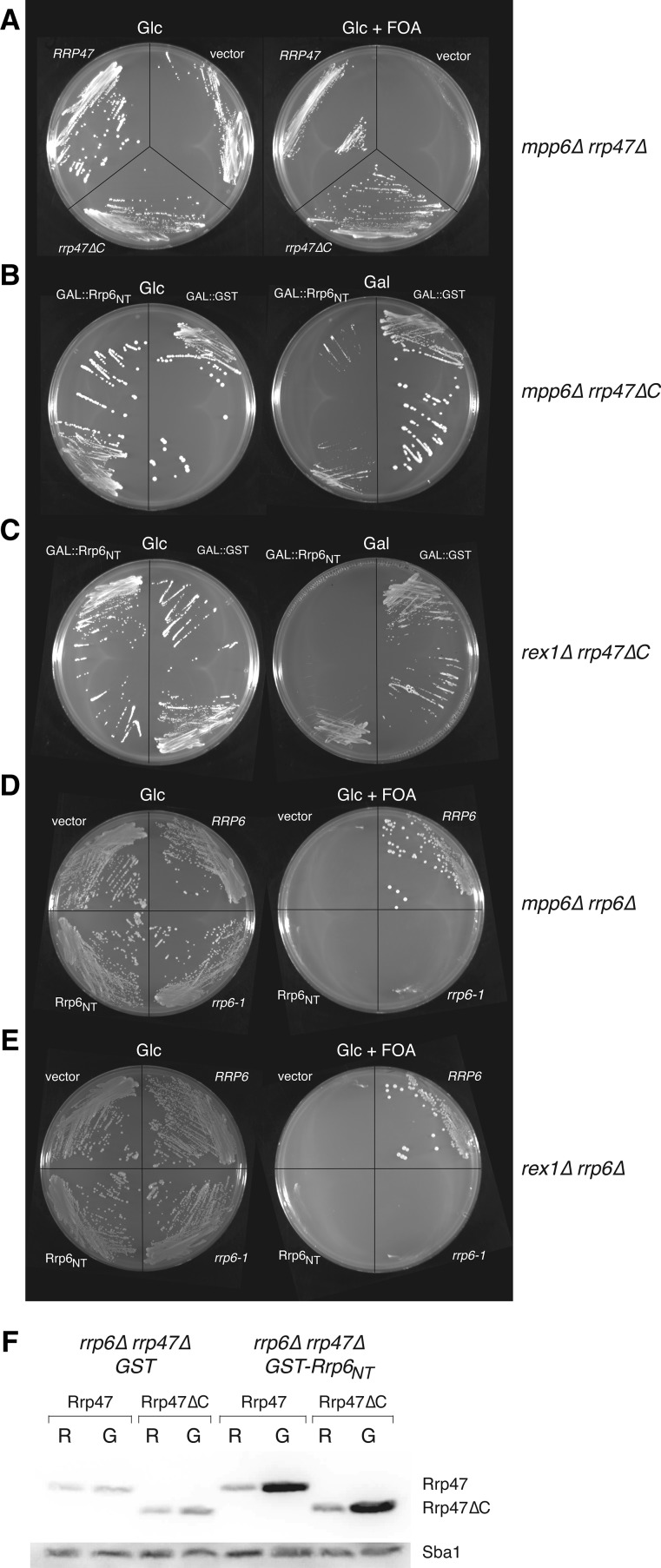
The observation that similar snoRNA processing defects are observed in rex1Δ mutants expressing the rrp47ΔC mutant or upon GST-Rrp6NT induction suggested that the C-terminal region of Rrp47 may be important for the function of the protein when it is segregated from full-length Rrp6. To test this hypothesis, a rex1Δ rrp47ΔC double mutant was transformed with plasmids encoding GAL-regulated GST or GST-Rrp6NT, and the transformants were assayed for growth on glucose- and galactose-based medium. Strikingly, induction of GST-Rrp6NT in the rex1Δ rrp47ΔC mutant caused a strong block in growth, while a transformant harboring plasmids encoding GAL-regulated GST grew on both glucose- and galactose-based medium (Fig. 3C). A plasmid shuffle assay demonstrated that the rrp47ΔC allele also complements the synthetic lethality of the mpp6Δ rrp47Δ double mutant (Fig. 3A). Similarly, mpp6Δ rrp47ΔC mutants transformed with a plasmid encoding GAL-regulated GST-Rrp6NT showed growth inhibition on galactose-based medium, while the transformant expressing GST grew on both glucose- and galactose-based medium (Fig. 3B).
FIGURE 3.
Full-length Rrp47, but not the Rrp47ΔC mutant, is functional when divorced from the catalytic and exosome-binding domains of Rrp6. (A–E) Plate growth assays of yeast strains on permissive glucose-based medium (Glc, left-hand side) and on galactose-based medium (Gal) or medium containing 5-FOA (Glc + FOA) (right-hand side). Relevant genotypes of the yeast strains are indicated on the right. (A) Plasmid shuffle assay for complementation of the mpp6Δ rrp47Δ strain by the rrp47ΔC allele. (B) Growth of the mpp6Δ rrp47ΔC strain bearing a plasmid encoding either GAL-regulated GST or GST-Rrp6NT. (C) Growth of the rex1Δ rrp47ΔC strain bearing a plasmid encoding either GAL-regulated GST or GST-Rrp6NT. (D) Plasmid shuffle assay for complementation of the mpp6Δ rrp6Δ mutant by the rrp6-1 allele or expression of Rrp6NT domain. (E) Plasmid shuffle assay for complementation of the rex1Δ rrp6Δ mutant by the rrp6-1 allele or expression of Rrp6NT domain. (F) Western analysis of Rrp47 and Rrp47ΔC protein expression. Yeast rrp6Δ rrp47Δ strains harboring plasmids expressing C-terminal TAP-tagged fusion proteins of wild-type Rrp47, or the C-terminal truncation mutant and the galactose-inducible GST, or GST-Rrp6NT proteins were lysed under denaturing conditions and analyzed by SDS-PAGE/Western blotting. Rrp47 and Rrp47ΔC fusion proteins were detected with the PAP antibody (upper panel). An antiserum against Sba1 was used as a loading control on the same blot (lower panel).
Since the Rrp47ΔC protein is functional when expressed in a complex with Rrp6 (Costello et al. 2011), these data argue against the interpretation that the residual Rrp47/Rrp6 complex upon GST-Rrp6NT induction is sufficient for growth. Based on previous work (Costello et al. 2011), we envisage that the C-terminal region of Rrp47 engages with RNP substrates of Rrp6 through contacts with protein and/or RNA and promotes their subsequent processing or degradation, even when Rrp47 is divorced from catalytically active Rrp6.
A caveat to this model is that the truncated Rrp47ΔC protein is stably expressed when associated with the GST-Rrp6NT protein. We generated TAP-tagged alleles of full-length Rrp47 and the C-terminal truncated mutant and compared their expression levels in rrp6Δ mutants before and after induction of GST-Rrp6NT protein or the GST control (Fig. 3F). As previously observed (Feigenbutz et al. 2013), expression of full-length Rrp47 was increased in the rrp6Δ mutant upon expression of the Rrp6NT fusion protein. In contrast, no difference in the Rrp47 expression level was observed upon induction of GST. Importantly, expression of the Rrp47ΔC mutant was increased upon expression of GST-Rrp6NT protein in a comparable manner to that observed for the full-length protein.
Since rrp6Δ mutants express very low levels of Rrp47 (Feigenbutz et al. 2013), it is formally possible that the synthetic lethality observed in rrp6Δ mpp6Δ and rrp6Δ rex1Δ mutants is due to the lack of Rrp47 protein rather than a requirement for Rrp6. We, therefore, tested whether expression of the Rrp6NT domain was sufficient to complement the synthetic lethality of mpp6Δ rrp6Δ and rex1Δ rrp6Δ mutants, using a plasmid shuffle assay. No complementation was observed upon transformation with a plasmid encoding only the N-terminal region of Rrp6 (Fig. 3, panels D and E). Moreover, the rrp6-1 mutation also failed to allow growth of the plasmid shuffle strain on 5-FOA medium, whereas complementation was observed for the wild-type RRP6 gene. Thus, separable activities provided by both Rrp47 and Rrp6 are required in the absence of Mpp6 or Rex1.
Processing and degradation defects in rrp47 mpp6 and rrp47 rex1 mutants
To determine the basis of the loss-of-growth phenotypes upon induction of GST-Rrp6NT in the mpp6Δ rrp47ΔC and rex1Δ rrp47ΔC mutants, changes in the levels of specific RNAs upon transfer from raffinose-based medium to galactose-based medium were assayed by Northern blot hybridization. The analyzed strains also harbored a high-copy-number plasmid encoding the well-characterized NEL025c gene (Wyers et al. 2005; Arigo et al. 2006b); a previous study has shown that plasmid-based expression of the NEL025c gene recapitulates the expression and degradation pattern of the chromosomal transcripts (Thiebaut et al. 2006). Control strains expressing GST were harvested during growth in raffinose- and galactose-based medium and analyzed in parallel.
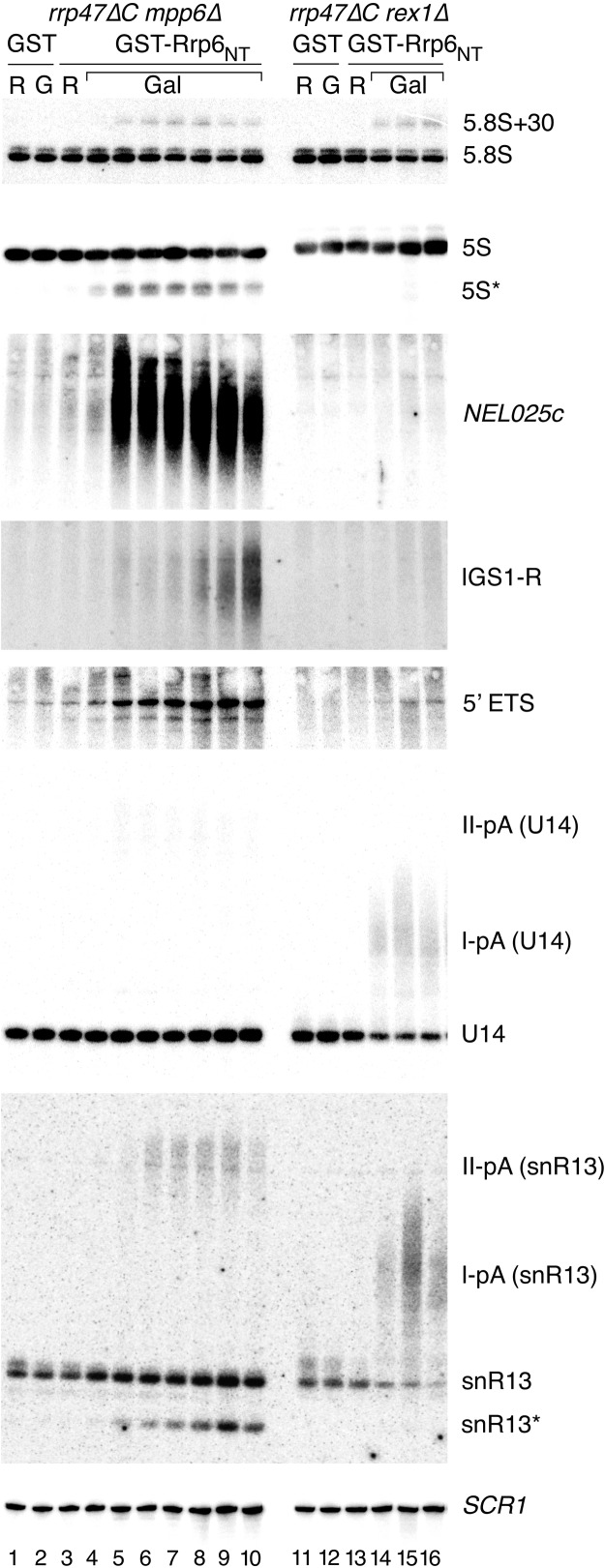
The mpp6Δ rrp47ΔC mutant showed slowed growth before the fourth doubling after induction of the Rrp6NT domain. Northern hybridization analyses revealed a dramatic accumulation of the NEL025c CUT well before this time-point, with the length of the CUT becoming less heterogeneous and shorter after the onset of slow growth (Fig. 4, lanes 4–10). The IGS1-R CUT from the inter-genic region of the rDNA repeat (Houseley et al. 2007; Vasiljeva et al. 2008) also accumulated upon transfer to galactose-based medium, with a strong increase at later time-points. The mpp6Δ rrp47ΔC mutant also showed defects in the degradation of the 5′ ETS fragment and truncated fragments of 5S, snR13, U3, and U6 at early time-points (Fig. 4; data not shown; truncated 5S and snR13 species are denoted with asterisks in Fig. 4).
FIGURE 4.
Northern analyses of conditional rrp47 mpp6 and rrp47 rex1 mutants. RNA was isolated from mpp6Δ rrp47ΔC and rex1Δ rrp47ΔC mutants expressing GST-Rrp6NT from the GAL promoter. Cells were harvested during growth in raffinose-based medium (lanes labeled R) and at each doubling of cell density after transfer to galactose-based medium (lanes labeled Gal). RNA was also isolated from cells expressing GAL-regulated GST during growth in raffinose- or galactose-based medium (lanes 1,2,11, and 12; labeled R or G, respectively). RNA was resolved through denaturing acrylamide gels and analyzed by Northern blot hybridization, using probes complementary to the RNAs indicated. Lanes 1–10, RNA isolated from the mpp6Δ rrp47ΔC mutant. Lanes 11–16, RNA from the rex1Δ rrp47ΔC mutant. Asterisks denote truncated fragments of the full-length, mature 5S rRNA, and snR13. Dispersed bands labeled I-pA and II-pA denote snoRNA precursors that are polyadenylated at termination sites I or II, respectively.
Transcription termination of snoRNA genes occurs at a proximal Nrd1-dependent site I and a more distal, “fail-safe” mRNA 3′ processing site II (Steinmetz and Brow 2003; Steinmetz et al. 2006). In rrp6Δ mutants, heterogeneous 3′ extended forms of snR13, U14, and snR50 accumulate due to polyadenylation after termination at both sites I and II (Allmang et al. 1999a; van Hoof et al. 2000b; Grzechnik and Kufel 2008), denoted I-pA and II-pA. Hybridization with probes complementary to the snoRNAs U14, snR13, and snR50 revealed that the mpp6Δ rrp47ΔC mutant specifically accumulated snoRNAs before inhibition of growth that correspond in length to transcripts terminating at the distal site II (Fig. 4; data not shown; denoted II-pA in Fig. 4). The level of mature snoRNA was not affected. A weak accumulation of the 5.8S + 30 fragment equivalent to that seen upon induction in the presence of full-length Rrp47 was also observed (cf. Figs. 2B and 4). Agarose gel Northern analyses of the mpp6 rrp47ΔC mutant revealed the accumulation of a range of apparent pre-rRNA degradation intermediates of diverse sizes upon induction of the Rrp6NT domain that are not seen in rrp47Δ or rrp6Δ mutants (data not shown). In sum, these data are consistent with the loss of redundant RNA surveillance pathways upon inactivation of both Rrp47 and Mpp6 (Milligan et al. 2008).
The rex1Δ rrp47ΔC mutant was impeded in growth more rapidly upon induction of the Rrp6NT domain than the mpp6Δ rrp47ΔC mutant, with a clear effect within less than two doublings after transfer to galactose-based medium. There was no clear defect in the degradation of CUTs or truncated stable RNAs in this mutant, and the defects in the processing of the 5.8S + 30 species, U6 processing, or the degradation of the 5′ ETS fragment were equivalent to those seen upon induction of the Rrp6NT domain in the rex1Δ mutant expressing full-length Rrp47 (see Fig. 2B). In contrast, there was a striking, rapid depletion of U14, snR13, snR38, and snR50 snoRNAs and an associated accumulation of longer, heterogeneous species corresponding in length to transcripts that are terminated at site I and subsequently polyadenylated (Fig. 4, lanes 13–16; data not shown; denoted I-pA in Fig. 4). These data support the model that normal productive snoRNA synthesis is mediated by termination at site I followed by polyadenylation (Grzechnik and Kufel 2008) and requires either Rrp6 or Rex1 activity for snoRNA 3′ end maturation. In contrast, fail-safe termination at site II generates RNAs that can apparently be targeted to exosome-mediated RNA surveillance by either Rrp6 or an Mpp6-dependent activity. The 5S rRNA and dimeric tRNAArg-tRNAAsp processing defects of rex1Δ mutants (Piper et al. 1983; van Hoof et al. 2000a) were not exacerbated (Fig. 4; data not shown).
Widespread inhibition of the degradation of CUTs and RNA surveillance substrates may well significantly contribute to the block in cell growth. In contrast, it is less obvious how the depletion of snoRNAs could cause rapid growth inhibition. A small number of snoRNAs are essential for viability in yeast and function in pre-rRNA processing (Watkins and Bohnsack 2011). However, agarose gel Northern blot analyses did not reveal a pre-rRNA processing defect upon Rrp6NT induction in the rex1Δ rrp47ΔC strain, and conditional ribosome synthesis mutants are slow-acting because the pre-existing pool of ribosomes must be depleted through growth. We envisage that a significant accumulation of polyadenylated snoRNAs or CUTs could lead to the depletion of the free pool of nuclear poly(A)-binding proteins such as Pab1 and Nab2, or the Nrd1/Nab3 termination complex, and have widespread effects on gene expression profiles.
Our data are consistent with the model that Rrp47 functions in the recognition of Rrp6 substrates, prior to RNA degradation or processing. Rrp47, Rrp6, and the TRAMP component Trf4 have been proposed to promote Nrd1/Nab3-dependent transcription termination (Arigo et al. 2006a; Grzechnik and Kufel 2008; Castelnuovo et al. 2013), in addition to subsequent RNA hydrolysis. We envisage that the Rrp47/Rrp6NT complex, perhaps in combination with TRAMP, retains the ability to recognize Nrd1/Nab3-mediated termination events when separated from full-length Rrp6. Deletion of the C-terminal region of Rrp47, which is known to mediate interactions with RNA and RNA-bound proteins (Costello et al. 2011), would be predicted to weaken this activity and thereby impede subsequent recognition of RNP substrates by Rrp6. In strains expressing the truncated Rrp47 protein bound to full-length Rrp6, substrate recognition may be more effective due to contacts between the TRAMP complex and both Nrd1/Nab3 (Arigo et al. 2006a) and Rrp6 (Callahan and Butler 2010).
The DECOID approach provides a simple method to determine whether proteins that are normally expressed within a complex are able to function when resolved from one another. Most proteins function as part of larger complexes, and the majority of proteins are predicted to be significantly overexpressed when placed under the control of the GAL promoter. Use of a neutral N-terminal tag such as GST would generally allow stable expression of individual protein domains. Application of the technique requires only knowledge of the interacting domain within one of the partner proteins. It is of note that we were able to obtain far tighter conditional growth phenotypes using the DECOID strategy than we observed with GAL::rrp47 rex1Δ and GAL::mpp6 rrp47Δ mutants. Given the accelerating wealth of detailed structural biology data, DECOID could provide a widely applicable, additional approach for the functional dissection of protein complexes.
MATERIALS AND METHODS
Plasmids
The URA3 marker gene in the GAL::rrp6NT construct (p280) (Stead et al. 2007) was inactivated by restriction digestion with ApaI and StuI, treatment with Klenow fragment, and religation. Selection of the resultant plasmid (p601) in yeast was dependent upon the leu2-d marker. The NEL025c gene was amplified from wild-type yeast genomic DNA using primers o636 (catggatccatatgctgtctttaagcc) and o637 (catgtcgacgaacgtaacgacttttcc) and cloned into the 2μ URA3 vector pRS426 (Christianson et al. 1992) after digestion with BamHI and SalI, giving p532. The MPP6 ORF and downstream ∼500 nt were amplified from wild-type yeast genomic DNA using primers o597 (acggaattcaatgagtgctaacaatgg) and o598 (cgcaagcttggcgtgcatgagacg) and cloned in-frame into p44 (Mitchell et al. 1996) by digestion with EcoRI and HindIII. The resultant plasmid (p593) encodes an N-terminal Mpp6 protein fusion bearing two copies of the z domain of protein A from Staphylococcus aureus that is expressed under the control of the RRP4 promoter and also bears the URA3 marker. Analogous constructs expressing a functional N-terminal full-length Rrp6 fusion protein (zz-Rrp6, p263) or rrp6 mutants expressing only the N-terminal 196 aa residues (Rrp6NT, p287) or expressing the catalytically inactive D238N mutant (Burkard and Butler 2000) (p389) have been described (Allmang et al. 1999b; Feigenbutz et al. 2013). Constructs encoding the full-length Rrp47 protein (p262) or a C-terminally truncated mutant that encodes the first 120 aa (denoted Rrp47ΔC) (p293) have also been described previously (Costello et al. 2011). For plasmid shuffle assays, inserts from RRP6 or RRP47 constructs were transposed into pRS313 (Sikorski and Hieter 1989), which contains the HIS3 marker. Plasmids expressing C-terminal TAP-tagged fusion proteins of full-length Rrp47 or the first 120 aa were generated by homologous recombination in yeast, as follows. The pRS313-based RRP47 genomic clone p425 was linearized downstream from the RRP47 ORF by digestion with SalI and transformed into an rrp6Δ rrp47Δ strain, together with amplicons generated from pBS1479 (Puig et al. 2001) that comprise the TAP tag and the downstream TRP1 marker into an rrp6Δ rrp47Δ strain harboring the GAL::rrp6NT construct. PCR was performed using the reverse primer o879 (tataagcatttttgcatttgtgctctcacatcacctttaatctacgactcactataggg) and the forward primer o878 (aaaagtaaaagattggataaagttggaaaaaagaaaggagggaagaagtccatggaaaagagaag) or o877 (caggcagagcaagaaaaagctaagaatatcatttccaatgttttggactccatggaaaagagaag) for the full-length clone (p670) or truncated clone (p671), respectively. Transformants were isolated on selective medium and screened for expression of the zz fusion protein by Western analyses. Positive clones were recovered from yeast and confirmed by sequence analysis of the ORF/TAP tag junction.
Strains
Strains were grown at 30°C in selective medium comprising 2% glucose, raffinose, or galactose, 0.5% ammonium sulphate, and 0.17% yeast nitrogen base, supplemented with appropriate amino acids and bases. Isogenic wild-type, rrp47Δ::kanMX4, rex1Δ::kanMX4, and mpp6Δ::kanMX4 strains were obtained from Euroscarf (University of Frankfurt, Germany). The rrp47Δ::kanMX4 disruption allele was converted to an rrp47Δ::hphMX4 allele by PCR-mediated homologous recombination of the rrp47Δ::kanMX4 recipient strain, using the hygromycin B resistance gene in plasmid pAG32 (Goldstein and McCusker 1999). An mpp6 rrp47 plasmid shuffle strain was generated by transforming the mpp6Δ::kanMX4 strain with the MPP6 expression construct p593 and disruption of the RRP47 gene by transposing the rrp47Δ::hphMX4 allele. The mpp6Δ rrp6Δ and rex1Δ rrp6Δ shuffle strains were made by targeting the kanMX4 cassette to the MPP6 or REX1 locus in an rrp6Δ::TRP1 strain bearing the plasmid p263, which encodes the zz-Rrp6 fusion protein and the URA3 gene (Allmang et al. 1999b). The rrp6Δ::kanMX4 rrp47Δ::hphMX4 strain was generated by PCR-mediated gene disruption of the RRP6 locus in the rrp47Δ::hphMX4 strain. Csl4-TAP and Rrp6-TAP strains were obtained from Open Biosystems. The rrp47-zz, rrp47-GFP, and rrp47-GFP::HIS3 rrp6Δ::kanMX4 strains have been described (Mitchell et al. 2003; Feigenbutz et al. 2013). The chromosomally encoded Csl4-TAP, Rrp6-TAP, Rrp47-zz, and Rrp47-GFP fusion protein and the plasmid-encoded Rrp47-TAP fusion proteins were expressed from their homologous promoters. Expression of the plasmid-encoded zz-Rrp6 fusion protein, which is driven from the RRP4 promoter, is comparable to that of the Rrp6-TAP fusion protein (Stead et al. 2007).
RNA analyses
For Rrp6NT induction analyses, cultures were harvested during growth in permissive, raffinose-based medium and upon every doubling of OD600 after transfer to medium containing 2% galactose. RNA was isolated by glass bead extraction in the presence of phenol and guanidinium isothiocyanate (Tollervey and Mattaj 1987). Total cellular RNA was resolved through 8% polyacrylamide gels containing 50% urea or 1.2% agarose gels and transferred to Hybond-N+ (GE Healthcare) for hybridization analyses. The sequences of the oligonucleotide probes used were as follows: 5.8S, gcgttgttcatcgatgc; 5S, ctactcggtcaggctc; 5′ ETS, cgctgctcaccaatgg; U6, atctctgtattgtttcaaattgaccaa; snR38, gagaggttacctattattacccattcagacagggataactg; NEL025c, ggcttctacagaacaagttgtatcgaaatgattgttggcgac; IGS1-R, gatgtaagagacaagtgaacagtgaacagtgaacagtggggaca; U14, tcactcagacatcctagg; snR13 3′, ggtcagataaaagtaaaaaaaggtagc; SCR1, aaggacccagaactaccttg. Hybridized blots were placed under phosphor storage screens, analyzed using a Personal Molecular Imager FX machine (BioRad), and nonsaturated images were obtained using the ImageJ64 package (NIH).
Protein analyses
Yeast cell extracts for gradient centrifugation analyses and pull-down assays were made by glass bead disruption in TMN150 buffer (10 mM Tris, pH 7.6, 150 mM NaCl, 5 mM MgCl2, 1 mM PMSF) (Mitchell et al. 1996), while the protein expression analyses in Figure 3 were done on alkaline denatured cell lysates (Motley et al. 2012). Samples were analyzed by Western blotting, using the peroxidase-antiperoxidase conjugate (PAP, Sigma), mouse anti-GFP (Roche), mouse anti-Pgk1 (Invitrogen), rabbit anti-GST (Sigma), and rabbit anti-Sba1 (Mollapour and Piper 2012) primary antibodies, and HRP-conjugated goat anti-mouse (BioRad) or anti-rabbit (Sigma) secondary antibodies. Proteins were detected by ECL and visualized using a G:Box iChemi XL system running GeneSnap and GeneTools software packages (Syngene).
For sedimentation analyses, the rrp47-GFP strain was transformed with plasmids encoding either GST or GST-Rrp6NT under the control of the GAL promoter. Strains were grown in galactose-based medium, and cell lysates were resolved through 12 mL 10%–30% glycerol gradients in TMN150 buffer at 36,000 rpm for 24 h using an SW40 rotor (Beckman Coulter). 0.65 mL fractions were collected and analyzed by Western blotting. Protein markers (BSA, 4.6 S; chicken ovalbumin, 3.6 S; bovine catalase, 11.3 S) (Erickson 2009) were resolved in parallel gradients. Protein profiles of glycerol gradients were routinely assessed by SDS-PAGE analysis of the fractions and staining with InstantBlue colloidal Coomassie Blue stain (Expedion).
For Rrp6NT pull-down experiments, yeast strains expressing Rrp47-zz, Rrp6-TAP, or Csl4-TAP fusion proteins were transformed with a plasmid expressing GAL-driven GST-Rrp6NT and grown in galactose-based medium. Clarified lysates were passed over glutathione beads, and retained protein was eluted with lysis buffer containing 25 mM glutathione. For the Rrp6 pull-down experiments, the rrp47-GFP::HIS3 rrp6Δ::kanMX4 strain was transformed with plasmids encoding zz-Rrp6 and either GAL-driven GST or GST-Rrp6NT. Cell extracts were initially fractionated by binding with SP sepharose beads; retained protein was eluted with buffer containing 500 mM NaCl. After adding NaCl to the flow-though fraction to a final concentration of 500 mM, both flow-through and eluate fractions were incubated with IgG sepharose beads (GE Healthcare) to trap Rrp6 complexes. The beads were washed with buffer containing 500 mM NaCl, and retained protein was recovered by elution with 0.5 M acetic acid and lyophilized.
ACKNOWLEDGMENTS
This work was supported by a grant from the Wellcome Trust to P.M. (08836/Z/09/Z) and BBSRC-funded PhD studentships to W.G. and M.T. We thank Stefan Millson and Peter Piper (University of Sheffield) for the Sba1 antibody.
Footnotes
Freely available online through the RNA Open Access option.
REFERENCES
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. 1999a. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. 1999b. The yeast exosome and human PM–Scl are related complexes of 3′→5′ exonucleases. Genes Dev 13: 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigo JT, Carroll KL, Ames JM, Corden JL. 2006a. Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell 21: 641–651. [DOI] [PubMed] [Google Scholar]
- Arigo JT, Eyler DE, Carroll KL, Corden JL. 2006b. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell 23: 841–851. [DOI] [PubMed] [Google Scholar]
- Assenholt J, Mouaikel J, Andersen KR, Brodersen DE, Libri D, Jensen TH. 2008. Exonucleolysis is required for nuclear mRNA quality control in yeast THO mutants. RNA 14: 2305–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. 2009. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 139: 547–559. [DOI] [PubMed] [Google Scholar]
- Burkard KT, Butler JS. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol 20: 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan KP, Butler JS. 2008. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res 36: 6645–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan KP, Butler JS. 2010. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem 285: 3540–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelnuovo M, Rahman S, Guffanti E, Infantino V, Stutz F, Zenklusen D. 2013. Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat Struct Mol Biol 20: 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A, Lubas M, Jensen TH, Dziembowski A. 2013. RNA decay machines: The exosome. Biochim Biophys Acta 1829: 552–560. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Costello JL, Stead JA, Feigenbutz M, Jones RM, Mitchell P. 2011. The C-terminal region of the exosome-associated protein Rrp47 is specifically required for box C/D small nucleolar RNA 3′-maturation. J Biol Chem 286: 4535–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. 2009. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online 11: 32–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigenbutz M, Jones R, Besong TMD, Harding SE, Mitchell P. 2013. Assembly of the yeast exoribonuclease Rrp6 with its associated cofactor Rrp47 occurs in the nucleus and is critical for the controlled expression of Rrp47. J Biol Chem 288: 15959–15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Grzechnik P, Kufel J. 2008. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol Cell 32: 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Xu Z, Lebreton A, Séraphin B, Steinmetz LM, Jacquier A, Libri D. 2012. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell 48: 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. 2009. The many pathways of RNA degradation. Cell 136: 763–776. [DOI] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, Hage El A, Tollervey D. 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J 26: 4996–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT. 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724. [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237. [DOI] [PubMed] [Google Scholar]
- Makino DL, Baumgärtner M, Conti E. 2013. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 495: 70–75. [DOI] [PubMed] [Google Scholar]
- Milligan L, Decourty L, Saveanu C, Rappsilber J, Ceulemans H, Jacquier A, Tollervey D. 2008. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol Cell Biol 28: 5446–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Tollervey D. 1996. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev 10: 502–513. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. 1997. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91: 457–466. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. 2003. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol Cell Biol 23: 6982–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Piper PW. 2012. Activity of the yeast zinc-finger transcription factor War1 is lost with alanine mutation of two putative phosphorylation sites in the activation domain. Yeast 29: 39–44. [DOI] [PubMed] [Google Scholar]
- Motley AM, Nuttall JM, Hettema EH. 2012. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J 31: 2852–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick SG, Wang X, Costanzo M, Brost RL, Boone C, Anderson JT. 2009. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res 37: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W, et al. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113: 919–933. [DOI] [PubMed] [Google Scholar]
- Piper PW, Bellatin JA, Lockheart A. 1983. Altered maturation of sequences at the 3′ terminus of 5S gene transcripts in a Saccharomyces cerevisiae mutant that lacks a RNA processing endonuclease. EMBO J 2: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. 2001. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218–229. [DOI] [PubMed] [Google Scholar]
- Schilders G, van Dijk E, Pruijn GJM. 2007. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res 35: 2564–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D. 2012. Transcriptome-wide analysis of exosome targets. Mol Cell 48: 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead JA, Costello JL, Livingstone MJ, Mitchell P. 2007. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res 35: 5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol Cell Biol 23: 6339–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Ng SBH, Cloute JP, Brow DA. 2006. cis- and trans-Acting determinants of transcription termination by yeast RNA polymerase II. Mol Cell Biol 26: 2688–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. 2006. Transcription termination and nuclear degradation of cryptic unstable transcripts: A role for the Nrd1-Nab3 pathway in genome surveillance. Mol Cell 23: 853–864. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Mattaj IW. 1987. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J 6: 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R. 2000a. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J 19: 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R. 2000b. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol 20: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Buratowski S. 2006. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell 21: 239–248. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S. 2008. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell 29: 313–323. [DOI] [PubMed] [Google Scholar]
- Wasmuth EV, Lima CD. 2012. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol Cell 48: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT. 2011. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. WIREs RNA 3: 397–414. [DOI] [PubMed] [Google Scholar]
- Wolin SL, Sim S, Chen X. 2012. Nuclear noncoding RNA surveillance: Is the end in sight? Trends Genet 28: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737. [DOI] [PubMed] [Google Scholar]