This paper examines A-to-I RNA-editing conservation among infant and mature humans, chimpanzees, and macaques in two brain regions. While RNA-editing patterns are largely conserved within and among species, as well as between brain regions, >15% of sites display human-specific editing patterns. Further, in all three species, RNA-editing levels tend to increase with age.
Keywords: RNA editing, human, brain, evolution, chimpanzee, aging
Abstract
Adenosine-to-inosine (A-to-I) substitutions are the most common type of RNA editing in mammals. A-to-I RNA editing is particularly widespread in the brain and is known to play important roles in neuronal functions. In this study we investigated RNA-editing changes during human brain development and maturation, as well as evolutionary conservation of RNA-editing patterns across primates. We used high-throughput transcriptome sequencing (RNA-seq) to quantify the RNA-editing levels and assess ontogenetic dynamics of RNA editing at more than 8000 previously annotated exonic A-to-I RNA-editing sites in two brain regions—prefrontal cortex and cerebellum—of humans, chimpanzees, and rhesus macaques. We observed substantial conservation of RNA-editing levels between the brain regions, as well as among the three primate species. Evolutionary changes in RNA editing were nonetheless evident, with 40% of the annotated editing sites studied showing divergent editing levels among the three species and 16.5% of sites displaying statistically significant human-specific editing patterns. Across lifespan, we observed an increase of the RNA-editing level with advanced age in both brain regions of all three primate species.
INTRODUCTION
RNA editing is a co-/post-transcriptional RNA modification process that affects various types of RNA and is known to contribute to transcriptome diversity (for reviews, see Gott and Emeson 2000; Schaub and Keller 2002; Bogdanov et al. 2011). RNA editing is the umbrella term for several mechanisms that generate nucleotide insertions or deletions as well as base conversions in RNA (Simpson and Emeson 1996). An evolutionary advantage of RNA editing in comparison to DNA level changes is flexibility regarding spatial and temporal regulation, as well as a capacity for quick adaptation to environmental changes (Gommans et al. 2009; Nishikura 2010). The most widespread type of RNA editing in higher eukaryotes is known as adenosine-to-inosine (A-to-I) RNA editing. A-to-I RNA editing is catalyzed by a group of enzymes called adenosine deaminases acting on RNA (ADARs), which use hydrolytic deamination to convert adenosine (A) to inosine (I) on dsRNA substrates (Nishikura et al. 1991; Bass 2002). As inosine base pairs with cytidine, it is recognized as guanosine (G) in sequencing experiments. Thus, the term A-to-G RNA editing is also commonly used to describe A-to-I RNA editing.
Specifically, A-to-I RNA editing is important for determining the functional amino acid sequence of several proteins expressed in the central nervous system. Known examples include glutamate receptor subunits (Sommer et al. 1991) and serotonin receptors (Burns et al. 1997) in mouse, potassium channels in squid (Patton et al. 1997), and ion channel proteins in the fruit fly (Peixoto et al. 1997; Semenov and Pak 1999; Hanrahan et al. 2000). In comparison to other tissues, inosine levels, as well as expression levels of ADAR enzymes, are higher in the mammalian brain (O'Connell et al. 1995; Paul and Bass 1998). This hints at higher RNA-editing activity in the brain tissue. The mis-regulation of A-to-I RNA editing has been shown to affect neural functions in various organisms from humans to worms (Higuchi et al. 2000; Palladino et al. 2000; Tonkin et al. 2002; Miyamura et al. 2003; Maas et al. 2006).
A-to-I RNA editing affects not only the protein sequences themselves, but also RNA stability, cellular localization, splicing, and translation efficiency, especially when localized in non-coding parts of the transcript (for review, see Nishikura 2010). As ADARs’ activity requires double-stranded RNA as a substrate, most of the RNA-editing sites found in the brain or in other tissues are located within repetitive elements (Morse and Bass 1999; Morse et al. 2002). Recent computational screens have revealed tens of thousands of A-to-I RNA-editing sites in transcripts originating from Alu-repeat elements and located outside of protein-coding regions (Athanasiadis et al. 2004; Blow et al. 2004; Kim et al. 2004; Levanon et al. 2004). Several studies have further reported only little overlap of A-to-I RNA-editing sites between human individuals or between experiments (Park et al. 2012; Peng et al. 2012). These observations challenge the overall functional significance of A-to-I RNA editing and highlight difficulties in identification of functionally relevant RNA-editing events.
Here, we used a cross-species comparison of the A-to-I RNA-editing patterns in the brains of humans, chimpanzees, and rhesus macaques to study the evolutionary conservation and the possible functional significance of this editing. Given that a previous study investigating A-to-I RNA editing at several functionally relevant sites in the mouse brain reported that the editing level changes with age (Wahlstedt et al. 2009; Dillman et al. 2013), we included a temporal dimension by measuring RNA-editing levels in the brains of newborn, young adults, and aged individuals. Further, we investigated the RNA-editing patterns in two functionally distinct brain regions: prefrontal cortex (PFC) and cerebellar cortex (CBC). The two brain regions were chosen to represent evolutionary and functionally different brain areas. PFC is one of the most evolutionarily recent brain regions and is associated with cognitive functions such as abstract thinking, executive planning, decision making, and social behavior (Yang and Raine 2009). In contrast, CBC is an evolutionary ancient brain region that is mainly responsible for movement coordination and other aspects of motor activity (Wolf et al. 2009).
RESULTS
Transcriptome data and RNA-editing sites
To study RNA-editing changes with age in the human brain and to estimate the evolutionary conservation of RNA-editing patterns among primate species, we utilized published RNA-seq data from 30 newborn, young adult, and aged humans collected from two brain regions, PFC and CBC (Mazin et al. 2013). In addition, we sequenced PFC and CBC transcriptomes of five newborn and five adult chimpanzees, as well as five newborn and five adult rhesus macaques, following the same procedure as the one used for the human samples. In order to reduce the influence of individual variation, RNA samples from five individuals of a similar age were pooled prior to sequencing (Supplemental Table S1).
All samples were sequenced using a paired-end sequencing protocol with read lengths of 75 nucleotides (nt). An average of 18 million paired-end reads per sample were mapped uniquely to the corresponding genomes allowing a maximum of three mismatches and no gaps, with a range of 17–23 million mapped reads per sample (Supplemental Table S2; Materials and Methods). Previous studies have indicated that de-novo identification of RNA-editing sites based on high-throughput RNA-seq data, including data obtained using the Illumina platform, requires high sequencing read coverage to ensure confident RNA-editing site identification (Li et al. 2009b). Accordingly, fewer than 500 de-novo RNA-editing sites could be identified in humans based on our RNA-seq data (Materials and Methods; Supplemental Table S3). Application of the same procedure to chimpanzee and macaque RNA-seq data failed to identify de-novo RNA-editing sites, mainly due to the lack of detailed genomic sequence polymorphism data for these species (Supplemental Fig. S1). We, therefore, did not rely on RNA-editing sites identified based on our RNA-seq data, but instead focused on the set of RNA-editing sites previously identified in the human transcriptome from a meticulous analysis of human EST (expressed sequence tags), available from the Database of RNA Editing (DARNED, http://darned.ucc.ie/) (Kiran and Baranov 2010). Among the more than 333,000 RNA-editing sites provided by this database, we selected 8041 A-to-I editing sites located within exons of protein-coding genes, including coding and untranslated regions, overlapping with no human SNPs (single nucleotide polymorphisms) reported in NCBI dbSNP (Sherry et al. 2001).
Of these 8041 human A-to-I editing sites, 7449 and 5488 could be mapped uniquely to orthologous sites in the chimpanzee and macaque genomes, respectively. Of them, 5257 sites were mapped unambiguously in all three species (Materials and Methods). For simplicity we denote the 8041 human editing sites downloaded from DARNED and the orthologous sites in chimpanzee and macaque as DARNED sites. Only the DARNED sites present in all three species and with sufficient sequence read coverage (more than four sequence reads) were considered in the species comparison study. On average, 2909 DARNED sites had sufficient sequence read coverage in humans, 2446 in chimpanzees, and 1802 in macaques (Supplemental Table S2).
In accordance with the A-to-I editing mechanism, the DARNED sites displayed a substantially and significantly higher rate of A-to-G substitutions in our RNA-seq data, in all three species, when compared with neighboring non-DARNED sites with a similar read coverage (on average 7.1% vs. 0.4%, one-sided binomial test, P< 0.0001) (Fig. 1A). Further, in agreement with the reported tandem modification of A-sites by ADAR enzymes (Bass and Weintraub 1988; Bass 2002; Nishikura 2010), A-sites located in the vicinity of DARNED sites (within an ∼50-bp region) also showed an elevated rate of A-to-G substitutions (Supplemental Fig. S2). Substitution count data also show that the excess of A-to-G substitutions at the DARNED sites cannot be caused by sequencing errors, as no enrichment for non-A-to-G (A-to-T and A-to-C) substitutions was observed at DARNED sites when compared with the neighboring non-DARNED sites (Fig. 1A; Supplemental Fig. S2).
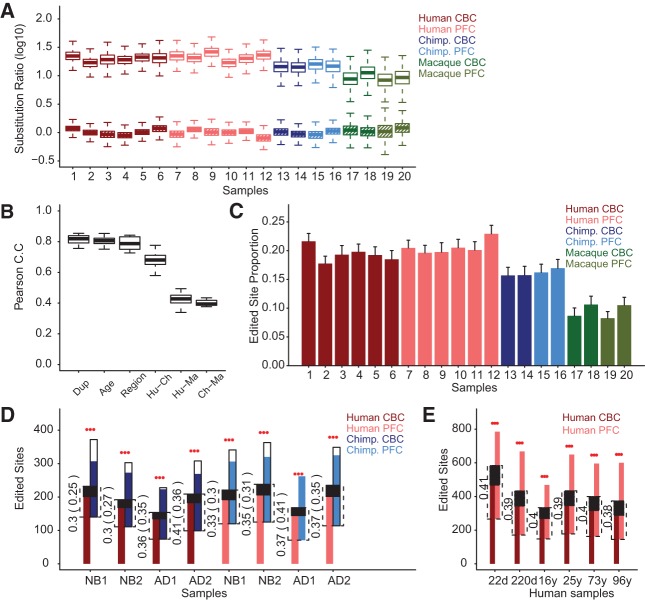
FIGURE 1.
Characteristics of A-to-I RNA editing at DARNED sites in the PFC and CBC of humans, chimpanzees, and rhesus macaques. (A) Comparison of the A-to-G and non-A-to-G (A-to-C and A-to-T) substitution frequencies between DARNED sites and neighboring non-DARNED sites located within 1000 bp on the same genome strand. Shown are nucleotide substitution ratios between DARNED and neighboring non-DARNED sites for A-to-G substitutions (empty boxes) and non-A-to-G substitutions hatched boxes). The colors represent species, and shades of colors represent brain regions. The variance of nucleotide substitution ratio estimates was obtained by bootstrapping over genomic sites 1000 times. (B) Editing level correlation between samples. Shown are Pearson correlation coefficients (Pearson C.C) of RNA-editing level between samples representing biological replicates sharing the same species, age, and brain region (Dup), samples representing different age groups for the same brain region of the same species (Age), samples representing different brain regions for the same age group of the same species (Region), and human-chimpanzee (Hu-Ch), human-rhesus macaque (Hu-Ma), and chimpanzee-rhesus macaque (Ch-Ma) comparisons conducted within the same brain region and the same age group. This analysis is based on 162 DARNED sites with sufficient sequence read coverage (greater or equal to five reads) in all samples and showing detectable A-to-I RNA editing in at least one sample. (C) Proportion of DARNED sites with detectable A-to-I RNA editing. The bar represents the mean proportion of A-to-I edited sites among all DARNED sites with sufficient sequence read coverage. The error bars show 95% confidence intervals obtained by bootstrapping over genomic sites 1000 times. The colors represent species, and shades of colors represent brain regions. (D) Overlap of DARNED sites with detectable A-to-I RNA editing between humans and chimpanzees in CBC and PFC. The height of colored bars represents numbers of edited sites identified in each sample. The colors represent species, and shades of colors the brain regions. Comparisons were conducted between all human and chimpanzee individuals within the same brain region and age group (NB, newborns; AD, adults). Comparisons involving younger and older human samples within the same age group are distinguished by numbers (NB1 or AD1, younger human samples; NB2 or AD2, older human samples). Extended white parts of chimpanzee bars represent edited sites specific to chimpanzees that might have been lost in the analysis, assuming that chimpanzees and humans have equal numbers of species-specific edited sites. The dashed lines show the overlap of detected edited sites between a human and a chimpanzee sample. The numbers show overlap proportions. The proportions calculated, including assumed chimpanzee-specific editing (white part of the bar), are shown in parentheses. The black part shows the overlap expected by chance, calculated in 1000 permutations of editing status labels (edited or non-edited) among detected DARNED sites, and assuming that chimpanzees have as many edited sites as humans (colored and white part of the bar combined). The red stars above bars indicate overlap significance; (***) permutation P< 0.001. (E) Overlap of DARNED sites with detectable A-to-I RNA editing between CBC and PFC samples from the same human individual. Colors and bars are as in D.
Editing level variation among species
After removal of DARNED sites with poor sequencing quality or removal of those containing non-A/G variants, we determined A-to-I edited DARNED sites using Single Nucleotide Polymorphism (SNP) identification methods implemented in the SAMtools algorithm (Materials and Methods; Li et al. 2009a). Human samples contained an average of 584 DARNED sites that showed detectable A-to-I RNA editing (A-to-I edited DARNED sites) (Fig. 1C; Supplemental Table S2), based on the “AG” genotype probability determined by the SAMtools algorithm (error probability < 0.01). For more than a half of these sites, RNA editing was detected in at least two samples within each human brain region (54.2% and 56.4% in CBC and PFC, respectively), significantly more than would be expected by chance (permutation P< 0.001). Chimpanzee samples contained an average of 396 A-to-I edited DARNED sites and macaque samples had an average of 172 A-to-I edited DARNED sites (Fig. 1C; Supplemental Table S2). In agreement with previous studies, the majority of the A-to-I edited DARNED sites (90%) were located in Alu-repeats positioned within 3′ untranslated regions (3′ UTRs) (Supplemental Fig. S3). Since DARNED sites were originally identified based on human transcription data, lower levels of RNA editing observed in the chimpanzee and macaque samples may reflect editing level divergence among species. Expression levels of the two main A-to-I RNA-editing enzymes, ADAR and ADARB1, were not decreased in non-human brain samples (one-sided t-test, P> 0.1) (Supplemental Fig. S4). This indicates that overall RNA-editing levels might be similar in the three species.
Despite the substantial drop in RNA-editing levels in chimpanzee and macaque samples, we still observed a strong and significant positive correlation of A-to-I editing levels (proportion of high quality A-to-G substitutions, see Materials and Methods) among the three species, especially between humans and chimpanzees (average Pearson correlation r= 0.67, P< 0.0001) (Fig. 1B). Further, we observed a significant and substantial overlap of A-to-I edited DARNED sites between humans and two non-human primates in both brain regions: >30% of the sites overlapped for humans and chimpanzees and >17% for humans and macaques. This overlap was significantly greater than expected by chance, even assuming that the chimpanzee and macaque transcriptomes contain the same number of species-specific editing sites as the human transcriptome (permutation P< 0.001) (Fig. 1D; Supplemental Fig. S5). A good overlap of A-to-I edited sites was also observed in the two human brain regions: >38% of sites overlapped between PFC and CBC (permutation P< 0.001) (Fig. 1E).
Evolutionary and tissue conservation of RNA editing in the brain
To study the evolutionary divergence in A-to-I RNA editing among human, chimpanzee, and macaque brain transcriptomes in more detail, we visualized the editing level variation among samples using principle component analysis (PCA) and hierarchical clustering (HC). The analysis was based on the editing levels of 162 DARNED sites with sufficient sequence read coverage in all samples of the three species. Further, we required these sites to show detectable A-to-I RNA editing in at least one of the three species (Materials and Methods). Both the PCA and HC trees showed a clear separation of RNA-editing patterns among species (Fig. 2A,B). In contrast, brain regions and age had a limited effect on the editing pattern variation among samples when compared with the effect of species identity, according to PCA, HC, as well as principal variance component analysis (PVCA) (Supplemental Fig. S6; Materials and Methods).
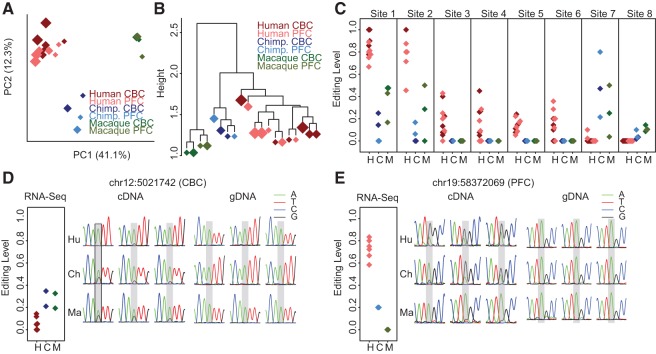
FIGURE 2.
Editing level variation among species and brain regions. (A) PCA and (B) the UPGMA tree based on A-to-I RNA-editing levels across all samples (refer to Materials and Methods for detail). Each diamond represents a sample. The diamond size is proportional to the median age of individuals within the sample. The colors represent species: brown, humans; blue, chimpanzees; and green, rhesus macaques. The shades of color represent brain regions: dark, CBC; light, PFC. This analysis is based on 162 DARNED sites with sufficient sequence read coverage in all samples and showing detectable A-to-I RNA editing in at least one sample. (C) Examples of DARNED sites showing significant human-specific A-to-I RNA editing-level changes. Each diamond represents a sample. The color scheme is the same as in panels A and B. Different species samples are located separately on the x-axis (H, human; C, chimpanzee; and M, rhesus macaque). The eight sites shown correspond to the following genomic locations based on the hg19 human genome assembly: chr2:201842479, chr2:128950497, chr1:204521712, chr1:6282587, chr5:115165632, chr9:136229572, chr5:156904785, and chr7:44917278 (sites 1 to 8, respectively). (D,E) Validation A-to-I RNA-editing level differences among species at two DARNED sites using conventional sequencing. Shown are A-to-I RNA-editing levels estimated using RNA-seq (left panels) and conventional sequencing (cDNA, central panels), as well as genomic DNA sequences determined using conventional sequencing (gDNA, right panels). RNA-seq data represented in the same way as in panel C. Conventional sequencing results are represented by the chromatogram traces from three individuals of each species—humans (Hu), chimpanzees (Ch), and rhesus macaques (Ma)—at two DARNED sites: chr12:5021742 (KCNA1 gene) showing editing level difference in the CBC and chr19:58372069 (ZNF587 gene) showing editing level difference in the PFC.
To identify the DARNED sites that showed species-specific editing level changes, we used the generalized linear model (GLM) to fit high-quality A-to-G substitution rate change among samples, with species and brain regions as predictors. We then used analysis of variance (ANOVA) to estimate the relative contribution to the total variance for each of these predictors and permuted sample identifiers to determine the false discovery rate (FDR) (Materials and Methods). We identified 96 sites with significant editing level differences among the three species (ANOVA P <0.05 and mean editing level difference [MELD]>20% for either species pair; permutation P< 0.0001) (Supplemental Table S4) among the 236 DARNED sites that had sufficient sequence coverage in both humans and non-human primates, as well as A-to-I RNA editing detected in at least four human or non-human samples (Materials and Methods). Among them, 39 sites showed human-specific A-to-I editing (ANOVA P< 0.05 and MELD between humans and non-human primates >20%; permutation P= 0.004) (examples shown in Fig. 2C). Using the same criteria, we found only five DARNED sites with significant editing level differences between the two brain regions (ANOVA P< 0.05 and MELD>20%; permutation P= 0.002) (Supplemental Table S4). Taken together, these results indicate rapid RNA-editing level divergence among primate species, but not between the brain regions studied, PFC and CBC.
To test the authenticity of RNA-editing divergence among primate species according to our RNA-seq data of pooled samples, we picked two DARNED sites with different editing change patterns between non-human primates and human for validation by more accurate conventional Sanger sequencing on three individuals from each species (Supplemental Table S5). Conventional cDNA sequencing results were consistent with the RNA-seq analysis results for editing change patterns among the species (Fig. 2D,E). Further, there was no detectable nucleotide polymorphism at the genome level for both sites according to genomic DNA (gDNA) sequencing results (Fig. 2D,E; Materials and Methods).
RNA editing changes with age in the human brain
Age-related changes in A-to-I RNA-editing levels were previously reported for 27 specific sites in the mouse brain (Wahlstedt et al. 2009) and for a larger number of sites in a more recent study (Dillman et al. 2013). To estimate the scope and the prevalence of age-related A-to-I RNA-editing changes in the human brain, we analyzed RNA-editing changes in our data using general linear models (GLM) at 388 DARNED sites that had sufficient sequencing coverage in all human samples, and with A-to-I RNA editing detected in at least two human samples. We then used F-test to estimate the significance of the age-related A-to-I RNA-editing level changes (age effect) (Supplemental Table S6). In agreement with the general analysis of RNA-editing variation, age effects on RNA-editing level changes were substantially lower than species effects (Fig. 2A,B; Supplemental Fig. S6); out of the 388 sites, only 44 (11.3%) showed significant RNA-editing level changes with age (GLM P-value < 0.05; permutation P= 0.043) (Fig. 3A; Supplemental Table S6; Supplemental Fig. S7). Twenty-nine of the 44 sites showed editing level changes with age of >10% with an average of 21.1%. At the DNA sequence level, these 29 age-related DARNED sites tended to be more conserved than the other DARNED sites: mean primate lineage phastCons scores 0.137 vs. 0.081, respectively. Moreover, there was a general trend of increased DNA sequence conservation for DARNED sites showing more significant age-related editing level changes (Fig. 3B; Supplemental Fig. S8).
FIGURE 3.
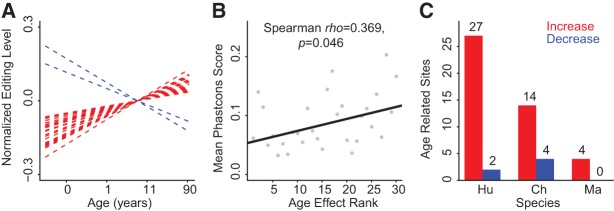
Age-related A-to-I RNA-editing level changes. (A) Linear regression trajectories based on A-to-I RNA-editing levels in human brain samples at 29 DARNED sites showing significant editing level changes with age with amplitude >10% (GLM, P< 0.05). Editing level increases with age are shown in red and decreases in blue. (B) Correlation between DNA sequence conservation and significance of the age-related A-to-I RNA-editing change calculated across 388 DARNED sites with sufficient sequence read coverage across human samples. The significance estimates were binned into 30 intervals based on their ANOVA F-statistics values. Higher age effect rank corresponds to higher F-statistics values. Higher phastCons scores represent higher DNA sequence conservation (refer to Materials and Methods for detail). (C) Numbers of DARNED sites showing significant age-related editing level changes with amplitude >10% in each species (GLM, P< 0.05). Editing level increases with age are shown in red and decreases in blue.
Among the 29 human age-related RNA-editing changes, almost all (N= 27, one-sided binomial test, P< 0.0001) represented a gradual increase with advanced age (Fig. 3A,C). This observation was further confirmed by investigation of the age effect on RNA-editing levels in the other two non-human primate species using the same method (Fig. 3C; Supplemental Tables S7, S8; Materials and Methods). Age-related A-to-I editing level increases cannot be caused by mismatches, as there was no general age-related increase of A-to-G nucleotide substitution rates on the neighboring genomic “A” sites within 1000 bp (Supplemental Fig. S9) (Spearman correlation test, P> 0.1 for both human brain regions). Intriguingly, a gradual increase of RNA-editing levels over the course of brain development was previously reported in mice (Wahlstedt et al. 2009; Dillman et al. 2013). Thus, the observed age-related increase in RNA-editing levels might be characteristic of all mammals. In further agreement with observations made in the mouse brain, age-related increases in RNA editing could not be directly linked to changes in the expression of ADAR genes. As in mice, expression of the two ADAR genes did not show consistent statistically significant age-related increase in our human data across the two brain regions (Supplemental Fig. S4).
DISCUSSION
Our results show that A-to-I RNA editing is widespread in the brain transcriptomes of humans and non-human primates. In agreement with previous reports (Athanasiadis et al. 2004; Blow et al. 2004; Kim et al. 2004; Levanon et al. 2004; Kiran and Baranov 2010), the majority of the A-to-I edited DARNED sites were located in Alu-repeat elements inside the 3′UTRs of the transcripts (90.4%), with only a small fraction of editing affecting coding regions (2.1%) (Supplemental Fig. S3). Our results indicate substantial conservation of RNA editing among the three primate species (average Pearson correlation between human and chimpanzee: r= 0.67, between human and macaque: r= 0.45, P< 0.001), within species (average Pearson correlation r= 0.81, P< 0.001), as well as between the two brain regions (average Pearson correlation r= 0.78, P< 0.001) (Fig. 1B).
Despite the conservation of RNA-editing sites, there are multiple differences in RNA-editing levels among species. Even though our study is limited with respect to sequence coverage and the number of samples examined, significant editing level differences separating humans from both chimpanzees and macaques were detected at 16% of RNA-editing sites with sufficient sequence read coverage. Even though our analysis was based on annotation of human RNA-editing sites (DARNED sites), a large fraction of RNA-editing level differences (12.5%) showed a higher editing level in the brains of chimpanzees and macaques compared with the human brain (Supplemental Table S4). The fact that we do not consider RNA-editing sites specific to the chimpanzee and/or the macaque brain constitutes an obvious limitation of our study. As a consequence, we observe a substantial drop in the transcriptome editing levels from humans to chimpanzees to macaques. Not all of this difference, however, may be due to annotation bias, as higher RNA-editing levels in the human brain compared with the brains of non-human primates were previously reported, based on an analysis of six transcribed Alu-repeats (Paz-Yaacov et al. 2010).
In contrast to the evident RNA-editing level divergence between the human brain and the brains of chimpanzees and macaques, we detect few significant RNA-editing level differences between the two brain regions analyzed in our study: PFC and CBC. This is unexpected, as differences in gene expression levels (Khaitovich 2004), histological organization, and functional roles (Kandel et al. 2000) of these two brain regions are well documented. As our study is limited with respect to transcriptome coverage and the number of samples analyzed, more detailed studies of editing level differences among human brain regions may reveal region-specific editing events. It is, however, clear that RNA-editing divergence within the human brain is far smaller than editing level differences between humans and closely related non-human primates.
The most intriguing finding of our study is a general increase in RNA-editing levels in the brains of humans and non-human primates with advanced age (Fig. 3C; Supplemental Tables S6–S8). These results match the editing level increase with age reported in the mouse brain based on a study of 28 sites (Wahlstedt et al. 2009), as well as in a more recent genome-wide mouse cerebral cortex transcriptome study (Dillman et al. 2013), and suggest the presence of a general age-related RNA-editing trend in the mammalian brain transcriptome. Since there was no consistent age-related increase in the expression of ADAR genes across the three primate species, the increase in RNA-editing levels with age may be caused by other yet unknown factors. Gene ontology analysis (Ashburner et al. 2000) of age-related editing genes produced no significant enrichment after multiple testing correction (Supplemental Table S9), even though most of the top-scoring functional groups were involved in biological processes related to cell fate determination.
Overall, substantial conservation of RNA-editing patterns among species and brain regions, the presence of a common trend for RNA-editing increase with advanced age, as well as greater sequence conservation of sites showing age-related increase at the genome sequence level indicate that RNA editing may play substantial functional roles in the primate and mammalian brains. Further studies are clearly needed to investigate the possible functional significance of this phenomenon.
MATERIALS AND METHODS
Sample collection and RNA sequencing
Cerebellar cortex (CBC) and prefrontal cortex (PFC) samples were collected postmortem from 10 chimpanzees (aged 0 d–44 yr) and 10 rhesus macaques (aged 0.5 d–14 yr). Chimpanzee samples were obtained from the Yerkes Primate Center (Georgia, USA), the Anthropological Institute & Museum of the University of Zürich-Irchel (Switzerland), and the Biomedical Primate Research Centre (Netherlands). Macaque samples came from the Suzhou Experimental Animal Center (China). All subjects were defined as normal on the basis of gross morphology. All subjects suffered sudden death with no prolonged agonal state and for reasons other than their participation in this study, and without any relation to the tissue used.
CBC dissections were dissected from the lateral CBC, and PFC dissections were made from the frontal part of the superior frontal gyrus, for each individual. RNA was isolated using Trizol reagent (Invitrogen). Oligo(dT) selection was performed twice using Oligotex mRNA Midi Kit (Qiagen). After selection, 100 ng mRNA was first fragmented by addition of 5× fragmentation buffer (200 mM Tris acetate, pH 8.2, 500 mM potassium acetate, and 150 mM magnesium acetate) with heating at 94°C for 2 min 30 sec in a thermo-cycler. The fragments were then transferred to ice and run over a Sephadex-G50 column (USA Scientific) to remove the fragmentation ions. Equal quantities of total RNA from five individuals of similar age were pooled to obtain two chimpanzee and two macaque samples for the two separate brain regions. All samples had good and comparable RNA quality. Detailed sample information can be found in Supplemental Table S1.
We used random hexamer primers (Invitrogen, cat. no. 48190-011) for reverse transcription of fragmented mRNA to double-stranded cDNA. Sequencing libraries were prepared according to the paired-end non-strand-specific sample preparation protocol of Illumina (http://www.illumina.com). Each sample was sequenced in a separate lane in the Illumina Genome Analyzer II system, using the 75-bp paired-end sequencing protocol.
The 75-bp paired-end RNA-seq reads generated from 12 human pooled samples (six for CBC and six for PFC) (sample information included in Supplemental Table S1) were downloaded from the Short Read Archive.
Sequence data information for samples of all three species is summarized in Supplemental Table S2.
Read mapping
Read pairs containing low-quality nucleotides, as defined by the Illumina sequencing procedure, were filtered out. The remaining reads were mapped to the reference genomes of human, chimpanzee, and rhesus macaque (hg19, panTro2, and rheMac2 genome assemblies were used for these three species throughout this work) using the RNA-seq read mapping software TopHat (v1.0.10, http://tophat.cbcb.umd.edu/) (Trapnell et al. 2009) in the paired-end-mode allowing up to three mismatches and no gaps. We removed reads that mapped to multiple genomic locations using an in-house PERL script. For more accurate and reliable editing quantification and detection, we removed potential PCR artifacts by collapsing multiple paired reads that mapped to the same genomic location. After these filtering steps we obtained 14–23 million reliably mapped reads per sample (Supplemental Table S2).
De-novo identification of A-to-I editing sites
For the de-novo identification of editing sites, we used the following criteria:
More than 10 HQ (high sequencing quality, Phred quality score >20 or base-calling error probability <1%) bases.
No more than two nucleotide variants (≤2).
Greater than or equal to three HQ variants that accord to 10%–90% of all HQ bases.
No significant strand bias of variant bases (30%–70% of plus strand reads).
Non-SNP probability <1% (estimated by genotyping algorithm in MAQ).
Not annotated as SNPs (only in human).
Identified in ≥2 samples for each brain region and species.
The human de-novo identified A-to-I RNA-editing sites are listed in Supplemental Table S3. The numbers of de-novo identified A-to-I RNA-editing sites in each brain region of each of the three species are shown in Supplemental Figure S1.
DARNED sites and A-to-I editing
Genomic locations of annotated human A-to-I RNA-editing sites (genome assembly version: hg19) were downloaded from the DARNED (http://darned.ucc.ie/) (Kiran and Baranov 2010). Among all DARNED sites we selected 8041 sites located in annotated exonic regions and with no single nucleotide polymorphisms as listed in the NCBI RefSeq database: SNP131 (Sherry et al. 2001). To determine the orthologous genomic sites in chimpanzee and macaque for these editing sites, we converted their coordinates between each pair of the three species genomes using the standalone UCSC tool LiftOver (http://genome.ucsc.edu). Among the 8041 DARNED sites, 7449 and 5488 could be unambiguously and reciprocally converted between the genomes of human and chimpanzee as well as between human and macaque respectively, with 5257 unambiguously converted among the three species’ genomes.
Due to the loss of genomic strand information in the sequencing process, we considered sequences that mapped within exonic regions as coming from the sense strand. The sequence alignment manipulation software SAMtools (http://samtools.sourceforge.net/, v.0.1.18) was used to extract base calling information and predict consensus genotypes for each genomic site. Only sites covered by more than four sequence reads (later defined as “sufficient sequence read coverage”) were used in the analysis.
In each sample, A-to-I RNA-editing level at each DARNED site was estimated as the proportion of “G” base nucleotides among all HQ “A” and “G” nucleotides.
To detect DARNED sites that were A-to-I edited in our RNA-seq data, we applied the following requirements to remove sequencing errors and potential DNA polymorphism sites:
Only “A” and/or “G” base nucleotides should be detected in RNA-seq data at these sites.
At least two HQ “G” base nucleotides had to be detected in RNA-seq data at these sites, constituting 10%–90% of all nucleotides detected at the site.
The probability of a genotype not identical to the “AG” type estimated by SAMtools (see below) had to be <1%.
As a result, among 1774–3387 DARNED sites with sufficient sequence read coverage in each sample, we obtained 146–734 DARNED sites that showed detectable A-to-I RNA editing (A-to-I edited DARNED sites) (Supplemental Table S2).
We used SAMtools for genotyping based on RNA-seq. SAMtools uses a consensus genotype-calling model implemented in MAQ (http://maq.sourceforge.net/). Briefly, it estimates the probability of observing a certain set of base-calling events along with the base-calling error probabilities assuming each genotype. It then calculates the posterior probability for each genotype by applying Bayesian statistic methods with the assumption of prior genotype probability in the population. For details of the method please refer to Li et al. (2008).
Conservation of A-to-I editing levels among species and brain regions
To estimate the significance of the edited DARNED sites overlap between humans and non-human primates, chimpanzees and macaques, for each pair of species we calculated edified sites overlap between each pair of individuals from the corresponding age interval. In each comparison, we then removed bias toward excess of RNA editing detected in humans due to use of human edited sites annotation by assigning the same number of edited sites, as it was detected in its human counterpart, to a non-human individual. Note that this step was not performed when more A-to-I RNA edited sites were detected in a non-human individual (AD1 comparison in Fig. 1D). We next estimated significance of the overlap by randomly permuting 1000 times edited status labels (edited or non-edited) among all DARNED sites with sufficient read coverage in two compared species, including additional edited sites assigned to non-human individuals in the previous step of this procedure. The average overlap values obtained in these simulations are shown as black areas within human and non-human edited values overlap region in Figure 1D,E and Supplemental Figure S5, while P-values were calculated as a proportion of permutations where the number of overlapping edited sites was equal to or greater than the number of overlapping sites observed in the actual data. The significance of edited site overlap between two brain regions, PFC and CBC, of the same individual was calculated the same way, but skipping bias-removing step, as all comparisons were conducted between human samples.
Editing level variation analysis
PCA of the A-to-I RNA-editing level data was performed by singular value decomposition, using the “prcomp” function in R. The first two principal components are shown in Figure 2A. The UPGMA tree based on A-to-I RNA-editing levels across all samples was constructed using the “hclust” function in R with the Euclidean distance and average agglomeration method, as shown in Figure 2B.
PVCA of the A-to-I RNA-editing level data was performed in two steps. First, we determined principal components based on the editing level data across all samples using the “eigen” function in R. Second, we used the variance Component analysis to calculate the effect of species, age, and brain region on editing level variation based on the first nine principal components generated in the previous step (explaining 91% of the total variance) using the “lmer” function in R. Averages of the standardized (to same sum) and weighted (proportional to the explained variance) variance component for each effect are shown in Supplemental Figure S6.
The PVCA, PCA, and UPGMA tree construction analyses were all based on the A-to-I RNA-editing levels of 162 DARNED sites with sufficient sequence read coverage in all samples and showing detectable A-to-I RNA editing in at least one sample. The R script for the above analysis can be provided upon request.
Statistical analysis of editing level variation
To estimate the significance of A-to-I editing level changes among species, between brain regions or during aging, we fit the changes of A-to-G substitution frequency using the generalized linear models (GLMs, implemented using “glm” function in R) with species (human, chimpanzee, and rhesus macaque), brain region (CBC and PFC), and age (log2 d) as predictors, and used “quasibinomial” as the error distribution so as to be flexible to over dispersion of sequencing data that cannot be accounted for solely by a “binomial” distribution (Baggerly et al. 2003).
To detect DARNED sites with significant age-related A-to-I RNA-editing level changes or brain region-specific A-to-I RNA-editing level changes in humans, we fit the following GLM model for 388 sites with sufficient sequence coverage in all human samples and with A-to-I RNA editing detected in at least two human samples:
Here (G, A) represents a sequence input with each row containing counts of HQ “G” and “A” base nucleotides sequenced for each sample. The term “Brain Regions” is a categorical variable of a sample's brain region, “CBC” or “PFC”, and the term “Ages” is a continuous variable of the median of the individual ages of each sample pool measured as log2 transformed days postnatal.
To detect DARNED sites with significant species-specific changes or brain region-specific A-to-I RNA-editing level changes, we fitted the following GLM model for 236 sites with sufficient sequence read coverage in at least six samples of both human and non-human primates (chimpanzee and rhesus macaque combined), which also showed detectable A-to-I RNA editing in at least four samples in either group:
 |
Here the term “Species” represents a categorical variable of each sample's species. And for detection of human-specific editing sites, we combined “chimpanzee” and “macaque” as “non-human primates” for comparison with “human.” Other terms are the same as in the age test model described above.
The significance of the variation contribution of each predictor in the model was estimated by ANOVA using the F-test (“anova” function in R). All the sequencing data and test results are shown in Supplemental Tables S4, S6–S8. To estimate the false discovery rate (FDR) and discovery significance (permutation P-value) at different ANOVA significance level cutoffs, we conducted all possible permutations of samples’ species (210 times), brain regions (924 times), or age labels (720 times), depending on the factor tested, and repeated ANOVA. The A-to-I RNA-editing level change between human and non-human primates at each DARNED site was calculated as the mean editing level difference between human samples and non-human primate samples. The A-to-I RNA-editing level change with age was calculated as the difference between the minimal and maximum fitted values from linear regression.
To investigate the relationship between DNA sequence conservation and the significance of the age-related A-to-I RNA-editing level changes, the 388 DARNED sites described above were binned into 30 intervals according to their ANOVA F-statistics values (which can represent the age effect on editing level). The 30 intervals were ordered according to increasing F-statistics values. The relationship between the binned ANOVA significance rank (increasing from left to right along figures’ x-axis) and the mean phastCons score (primates) for the binned intervals is shown in Figure 3B. Results using other numbers of binned intervals (10, 20, 40, and 50) are shown in Supplemental Figure S8.
Experimental verification by Sanger sequencing
First-strand cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen) using CBC and PFC RNA samples from three individuals of each species (prepared in the same way as for RNA-seq data, refer to Sample Collection and RNA Sequencing). The CBC and PFC samples were dissected as described elsewhere (Mazin et al. 2013). To check the editing level change of two DARNED sites (chr12:5,021,742 from the KCNA1 gene and chr19:58,372,069 from the Zinc Finger gene ZNF587) among the three species in CBC and PFC respectively, six primer pairs were manually designed at the upstream and downstream flanks of the two DARNED sites for the three species. PCR reactions were performed at 95°C for 2 min followed by 30 cycles of 94°C for 30 sec, 50°C/58°C for 30 sec, 72°C for 50 sec, and 72°C for 5 min with rTaq DNA polymerase (TAKARA). Then 5 µL of each PCR product was used for electrophoresis in a 2% (w/v) agarose gel. The size of the fragments was appraised using visual estimation based on the DL 2000 DNA marker (TAKARA). The PCR products of appropriate sizes were dissected and purified from the electrophoresis gel and sequenced using the ABI3730XL sequencing platform (http://www.appliedbiosystems.com/). Sequencing chromatogram traces of cDNA and genomic DNA are shown in Figure 2D,E. Details about primer sequence and sample information are listed in Supplemental Table S5.
DATA DEPOSITION
All sequence data are available at the Gene Expression Omnibus (GEO) database, accession number GSE45244. Human RNA-seq data were downloaded from the Short Read Archive. The study accession number is SRP005169.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Yerkes Primate Center, the Biomedical Primate Research Centre, the Anthropological Institute, the Museum of the University of Zurich, and Dr. R. Martin and Dr. W. Scheffran, in particular, for providing chimpanzee samples; and the Suzhou Drug Safety Evaluation and Research Center and C. Lian, H. Cai, and X. Zheng, in particular, for providing the macaque samples; P. Mazin and J. Xiong for help in sharing the human RNA-seq data; and J. Boyd-Kirkup for help in improving the manuscript. We also thank J. Jing for assistance; M. Somel, K. Weng, X. Liu, and all members of the Comparative Biology Group in Shanghai for helpful discussions and suggestions. This work was supported by the National Natural Science Foundation of China (grant number 31171232); Ministry of Science and Technology of China (grant number 2012DFG31940); and the Foreign Expert 1000 Talents Plan program.
Footnotes
Freely available online through the RNA Open Access option.
REFERENCES
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S. 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggerly KA, Deng L, Morris JS, Aldaz CM. 2003. Differential expression in SAGE: Accounting for normal between-library variation. Bioinformatics 19: 1477–1483. [DOI] [PubMed] [Google Scholar]
- Bass BL. 2002. RNA Editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71: 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL, Weintraub H. 1988. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55: 1089–1098. [DOI] [PubMed] [Google Scholar]
- Blow M, Futreal PA, Wooster R, Stratton MR. 2004. A survey of RNA editing in human brain. Genome Res 14: 2379–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov AA, Zinovkin RA, Zamyatnin AA Jr. 2011. RNA editing: Breaking the dogma. Biochemistry (Mosc) 76: 867–868. [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308. [DOI] [PubMed] [Google Scholar]
- Dillman AA, Hauser DN, Gibbs JR, Nalls MA, McCoy MK, Rudenko IN, Galter D, Cookson MR. 2013. mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat Neurosci 16: 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommans WM, Mullen SP, Maas S. 2009. RNA editing: A driving force for adaptive evolution? Bioessays 31: 1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. 2000. Functions and mechanisms of RNA editing. Annu Rev Genet 34: 499–531. [DOI] [PubMed] [Google Scholar]
- Hanrahan CJ, Palladino MJ, Ganetzky B, Reenan RA. 2000. RNA editing of the Drosophila para Na+ channel transcript. Evolutionary conservation and developmental regulation. Genetics 155: 1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81. [DOI] [PubMed] [Google Scholar]
- Kandel ER, Schwartz JH, Jessell TM. 2000. Principles of neural science. McGraw-Hill, Health Professions Division, New York. [Google Scholar]
- Khaitovich P. 2004. Regional patterns of gene expression in human and chimpanzee brains. Genome Res 14: 1462–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. 2004. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res 14: 1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran A, Baranov PV. 2010. DARNED: A DAtabase of RNa EDiting in humans. Bioinformatics 26: 1772–1776. [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005. [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009a. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. 2009b. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324: 1210–1213. [DOI] [PubMed] [Google Scholar]
- Maas S, Kawahara Y, Tamburro KM, Nishikura K. 2006. A-to-I RNA editing and human disease. RNA Biol 3: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin P, Xiong J, Liu X, Yan Z, Zhang X, Li M, He L, Somel M, Yuan Y, Phoebe Chen YP, et al. 2013. Widespread splicing changes in human brain development and aging. Mol Syst Biol 9: 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamura Y, Suzuki T, Kono M, Inagaki K, Ito S, Suzuki N, Tomita Y. 2003. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am J Hum Genet 73: 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Bass BL. 1999. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc Natl Acad Sci 96: 6048–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Aruscavage PJ, Bass BL. 2002. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci 99: 7906–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem 79: 321–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K, Yoo C, Kim U, Murray JM, Estes PA, Cash FE, Liebhaber SA. 1991. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J 10: 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A, Keller W. 1995. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol 15: 1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. 2000. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 102: 437–449. [DOI] [PubMed] [Google Scholar]
- Park E, Williams B, Wold BJ, Mortazavi A. 2012. RNA editing in the human ENCODE RNA-seq data. Genome Res 22: 1626–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton DE, Silva T, Bezanilla F. 1997. RNA editing generates a diverse array of transcripts encoding squid Kv2 K+ channels with altered functional properties. Neuron 19: 711–722. [DOI] [PubMed] [Google Scholar]
- Paul MS, Bass BL. 1998. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J 17: 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, Amariglio N, Eisenberg E, Rechavi G. 2010. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci 107: 12174–12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto AA, Smith LA, Hall JC. 1997. Genomic organization and evolution of alternative exons in a Drosophila calcium channel gene. Genetics 145: 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. 2012. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol 30: 253–260. [DOI] [PubMed] [Google Scholar]
- Schaub M, Keller W. 2002. RNA editing by adenosine deaminases generates RNA and protein diversity. Biochimie 84: 791–803. [DOI] [PubMed] [Google Scholar]
- Semenov EP, Pak WL. 1999. Diversification of Drosophila chloride channel gene by multiple posttranscriptional mRNA modifications. J Neurochem 72: 66–72. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. 2001. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Emeson RB. 1996. RNA editing. Annu Rev Neurosci 19: 27–52. [DOI] [PubMed] [Google Scholar]
- Sommer B, Kohler M, Sprengel R, Seeburg PH. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19. [DOI] [PubMed] [Google Scholar]
- Tonkin LA, Saccomanno L, Morse DP, Brodigan T, Krause M, Bass BL. 2002. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J 21: 6025–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlstedt H, Daniel C, Enstero M, Ohman M. 2009. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res 19: 978–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf U, Rapoport MJ, Schweizer TA. 2009. Evaluating the affective component of the cerebellar cognitive affective syndrome. J Neuropsychiatry Clin Neurosci 21: 245–253. [DOI] [PubMed] [Google Scholar]
- Yang Y, Raine A. 2009. Prefrontal structural and functional brain imaging findings in antisocial, violent, and psychopathic individuals: A meta-analysis. Psychiatry Res 174: 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.