The functional relationship between CstF64 and CstF64τ, two paralogous mammalian mRNA 3′ processing factors, is not well understood. Our comprehensive characterization revealed that these two factors are widely expressed in mammalian tissues, have highly similar RNA-binding specificities, but they have overlapping and distinct protein interactomes as well as roles in regulating mRNA alternative polyadenylation.
Keywords: 3′ end formation, alternative polyadenylation, mRNA processing, sequencing
Abstract
mRNA 3′ processing is dynamically regulated spatially and temporally. However, the underlying mechanisms remain poorly understood. CstF64τ is a paralog of the general mRNA 3′ processing factor, CstF64, and has been implicated in mediating testis-specific mRNA alternative polyadenylation (APA). However, the functions of CstF64τ in mRNA 3′ processing have not been systematically investigated. We carried out a comprehensive characterization of CstF64τ and compared its properties to those of CstF64. In contrast to previous reports, we found that both CstF64 and CstF64τ are widely expressed in mammalian tissues, and their protein levels display tissue-specific variations. We further demonstrated that CstF64 and CstF64τ have highly similar RNA-binding specificities both in vitro and in vivo. CstF64 and CstF64τ modulate one another's expression and play overlapping as well as distinct roles in regulating global APA profiles. Interestingly, protein interactome analyses revealed key differences between CstF64 and CstF64τ, including their interactions with another mRNA 3′ processing factor, symplekin. Together, our study of CstF64 and CstF64τ revealed both functional overlap and specificity of these two important mRNA 3′ processing factors and provided new insights into the regulatory mechanisms of mRNA 3′ processing.
INTRODUCTION
mRNA 3′ end formation is an essential step of eukaryotic gene expression, and it typically involves an endonucleolytic cleavage followed by polyadenylation (Colgan and Manley 1997; Zhao et al. 1999; Chan et al. 2011; Proudfoot 2011). mRNA 3′ processing not only significantly impacts many other steps in the mRNA maturation process, it also plays an important role in gene regulation. Approximately 70% of mammalian genes produce multiple mRNAs with distinct 3′ ends by selecting different cleavage/polyadenylation sites (PAS), a phenomenon known as APA (Di Giammartino et al. 2011; Proudfoot 2011; Shi 2012; Tian and Manley 2013). APA isoforms from the same genes may encode different proteins and/or have distinct 3′ untranslated regions (UTRs), which can differentially impact the stability and/or translation efficiency of the mRNAs. Recent studies have shown that APA is highly regulated in development and in a tissue-specific manner (Wang et al. 2008; Ji et al. 2009; Shepard et al. 2011; Derti et al. 2012; Smibert et al. 2012). Aberrant APA patterns have been linked to human diseases including cancer and neuromuscular disorders (Danckwardt et al. 2008; Mayr and Bartel 2009; Jenal et al. 2012). Given the recent progress, APA has emerged as a critical mechanism for eukaryotic gene regulation. However, it remains poorly understood how APA is regulated.
Recent studies have provided strong evidence that the core components of the mRNA 3′ processing machinery are also key regulators of the global APA profiles (Takagaki et al. 1996; Kubo et al. 2006; Martin et al. 2012; Yao et al. 2012). In mammals, the mRNA 3′ processing complex consists of the poly(A) polymerase (PAP) and four multisubunit protein complexes, the cleavage/polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factor I (CF Im), and cleavage factor II (CF IIm) complexes (Shi et al. 2009; Chan et al. 2011). CPSF and CstF bind cooperatively the AAUAAA hexamer and the U/GU-rich downstream element, respectively, two key cis-elements found in the majority of mammalian PASs. CstF consists of three subunits—CstF77, CstF64, and CstF50. CstF64 directly interacts with RNAs via its RNA recognition motif (RRM) and plays important roles in mRNA 3′ processing and APA regulation (Takagaki et al. 1996; Chan et al. 2011). Previous SELEX analysis using the CstF64 RRM selected GU-rich sequences, similar to those found in many downstream sequence elements (Takagaki and Manley 1997). Recently, we and others have mapped CstF64-RNA interactions in vivo at the transcriptome level (Martin et al. 2012; Yao et al. 2012), and our results showed that CstF64 binds to U/GU-rich downstream sequence elements at many PASs, but CstF64-DSE interactions are highly variable in affinity (Yao et al. 2012). In addition to the RRM at its N terminus, CstF64 also contains a hinge domain, a P/G-rich region that contains twelve pentapeptide repeats, and a C-terminal domain (CTD) (Chan et al. 2011). CstF64 hinge domain mediates its interactions with both CstF77 and the CPSF subunit symplekin (Takagaki and Manley 2000). The P/G-rich region and the CTD remain poorly characterized but may mediate additional interactions with other 3′ processing factors and/or regulators (Qu et al. 2007). These studies suggest that both protein-RNA and protein–protein interactions of CstF64 are critical for mRNA 3′ processing and its regulation.
CstF64τ is a conserved paralog of CstF64 in mammals (Wallace et al. 1999). Earlier studies suggested that CstF64τ is highly expressed in the testis and also present at low levels in the brain, but undetectable in other tissues (Wallace et al. 1999). In vitro binding assays with homopolymer RNAs indicate that CstF64τ has distinct RNA-binding specificity than that of CstF64 (Monarez et al. 2007). Based on these observations, it was proposed that CstF64τ might recognize a distinct set of poly(A) sites than those bound by CstF64 and mediate testis-specific APA regulation (MacDonald and McMahon 2010). However, the functions of CstF64τ in mRNA 3′ processing and APA regulation and its functional relationship with CstF64 are still poorly characterized. For examples, it has been shown that CstF64τ mRNAs are detected ubiquitously in mouse tissues, and it is unclear how testis-specific expression of CstF64τ proteins is achieved (Huber et al. 2005). In addition, we and others have shown recently that CstF64 depletion in human cells has relatively little effect on the global APA profile (Martin et al. 2012; Yao et al. 2012), but codepletion of both CstF64 and CstF64τ leads to greater APA changes. These observations suggest that CstF64 and CstF64τ functions may be more similar than previously thought.
To define the functions of CstF64τ in mRNA 3′ processing and in APA regulation, we carried out comprehensive functional characterizations of CstF64τ and compared its properties to those of CstF64. Our results demonstrate that both CstF64 and CstF64τ are widely expressed in mammalian tissues. Although highly similar in their RNA-binding specificities, these two factors have overlapping and distinct protein interactomes and functions in mRNA 3′ processing.
RESULTS
Cell line- and tissue-specific expression of CstF64 and CstF64τ
In order to monitor the expression of CstF64 and CstF64τ, we carried out Western blotting analyses of seven human and mouse cell lines as well as five divergent mouse tissues. To ensure the specificity of the CstF64 and CstF64τ antibodies, we performed dual color Western analysis in which CstF64 (recognized by a mouse monoclonal antibody) is detected as a green band(s) of ∼64 kDa and CstF64τ (recognized by a rabbit polyclonal antibody) a red band of ∼70 kDa. As shown in Figure 1A, our Western analyses detected CstF64 and CstF64τ as distinct bands at the expected sizes. It was noted that the CstF64τ band in the mouse cell lines migrated higher compared to that in the human cell lines (Fig. 1A, cf. lanes 1–4,6–7). This is consistent with the fact that the mouse CstF64τ has a greater predicted molecular weight than its human homolog. Furthermore, our Western analyses using these antibodies detected specific decreases in CstF64 or CstF64τ in HeLa cells transfected with specific shRNA constructs targeting the respective genes (Fig. 4A, below; Yao et al. 2012). Together these observations confirmed the specificity of the antibodies used in our analyses.
FIGURE 1.
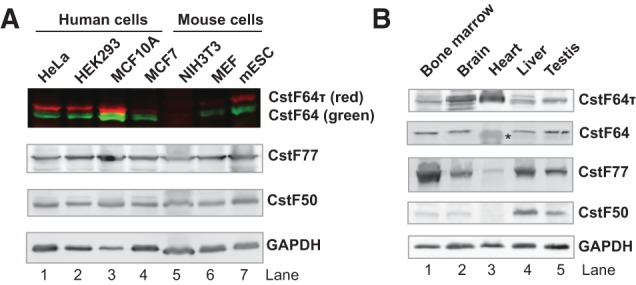
Expression profiles of CstF-64 and CstF-64τ. (A) Dual color Western blot analysis of CstF64 and CstF64t expression level in different cell lines as marked. The red bands correspond to CstF64τ and the green bands correspond to CstF64. (B) Western blotting analyses of CstF64, CstF64τ, CstF77, CstF50, and GAPDH expression in the mouse tissues as marked. For each lane, ∼20 μg total protein was loaded. (*) This band migrated faster than CstF64 in other tissues and is a potential nonspecific band.
FIGURE 4.
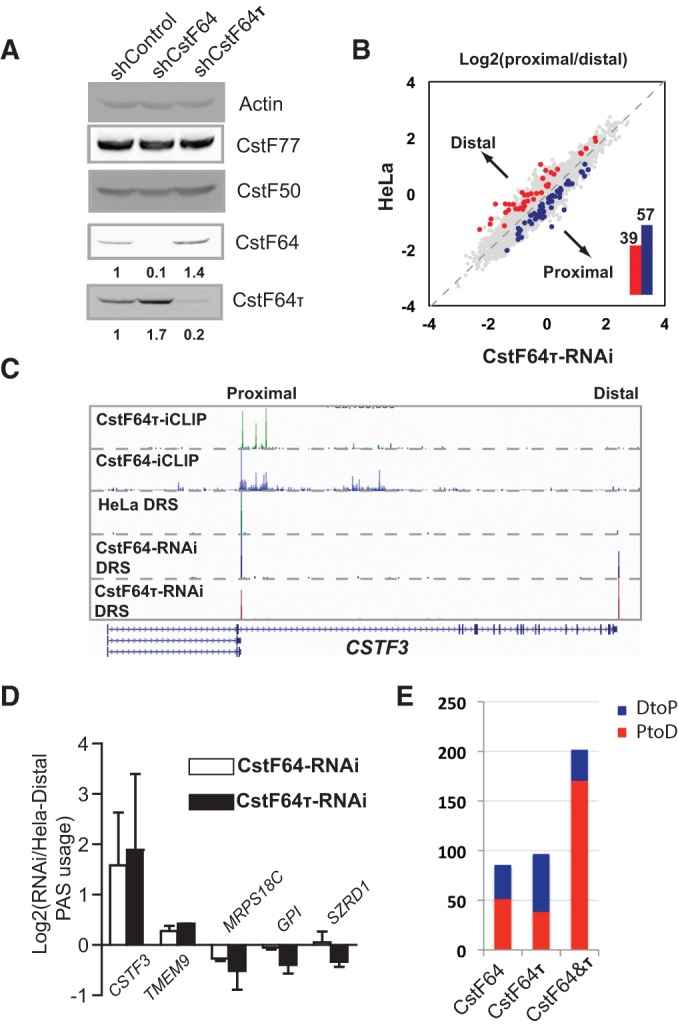
APA regulation by CstF64 and CstF64τ. (A) Western blotting analysis of control HeLa, CstF64-RNAi, and CstF64τ-RNAi cells. CstF64 and CstF64τ signals were quantified by using the ImageQuant program, and their levels in HeLa were set to 1. Relative levels in all samples are listed below. (B) Pairwise comparison of PAS usage in HeLa and CstF64τ-RNAi cells: (y-axis) log10(proximal/distal)–HeLa; (x-axis) log10(proximal/distal)–CstF64τ-RNAi. PAS pairs with statistically significant differences are highlighted in blue (DtoP shift) or red (PtoD shift). The numbers of PtoD and DtoP shifts are shown in the column graph in the inset. (C) UCSC Genome Browser tracks of the direct RNA sequencing results for CSTF3 in HeLa, CstF64-RNAi, and CstF64τ-RNAi cells. The proximal and distal PASs are marked on top. CstF64 and CstF64τ iCLIP data for the same region are also shown. (D) RT-qPCR validation of the APA changes in five selected genes: (y-axis) log2(extended 3′ UTR/common region)(CstF64τ-RNAi/HeLa). (E) A column graph showing the number of genes with significantly different APA profiles in CstF64-, CstF64τ-, or CstF64&τ-RNAi cells and the percentage of PtoD (red) and DtoP (blue) APA changes.
Our Western blotting results showed that both CstF64 and CstF64τ are expressed in most cell lines, but there are cell line-specific differences in their expression profiles (Fig. 1A). For example, the lowest levels of CstF64 or CstF64τ were observed in NIH3T3 cells (Fig. 1A, lane 5), and the highest levels of both proteins were detected in the breast epithelial cell line, MCF10A (Fig. 1A, lane 3). Compared to HeLa and HEK293 cells, the breast cancer cell line MCF7 expresses similar levels of CstF64 but significantly less CstF64τ (Fig. 1A, cf. lanes 1,2,4). Thus, significant variations exist in the ratio between CstF64 and CstF64τ as well as in the relative levels of both proteins compared to CstF77 and CstF50, the other two subunits of the CstF complex (Fig. 1A).
We also examined the expression of CstF64 and CstF64τ in five divergent mouse tissues, including the bone marrow, brain, heart, liver, and testis (Fig. 1B). CstF64 was detected at relatively constant levels in most tissues except for the heart, where its level seems significantly lower (Fig. 1B). Surprisingly, CstF64τ was detected in all five tissues, and the highest expression levels were observed in the heart and brain (Fig. 1B). Our results demonstrated that, similar to CstF64, CstF64τ is widely expressed in mammalian tissues. These results are in contrast with a previous report suggesting that CstF64τ is specifically expressed in the testis and the brain (Wallace et al. 1999). The possible reasons for this discrepancy are discussed below. These results reveal that both CstF64 and CstF64τ are widely expressed, but their protein levels display significant tissue-specific variations.
Characterization of the in vitro RNA-binding sequence specificity of CstF64 and CstF64τ by SELEX-seq
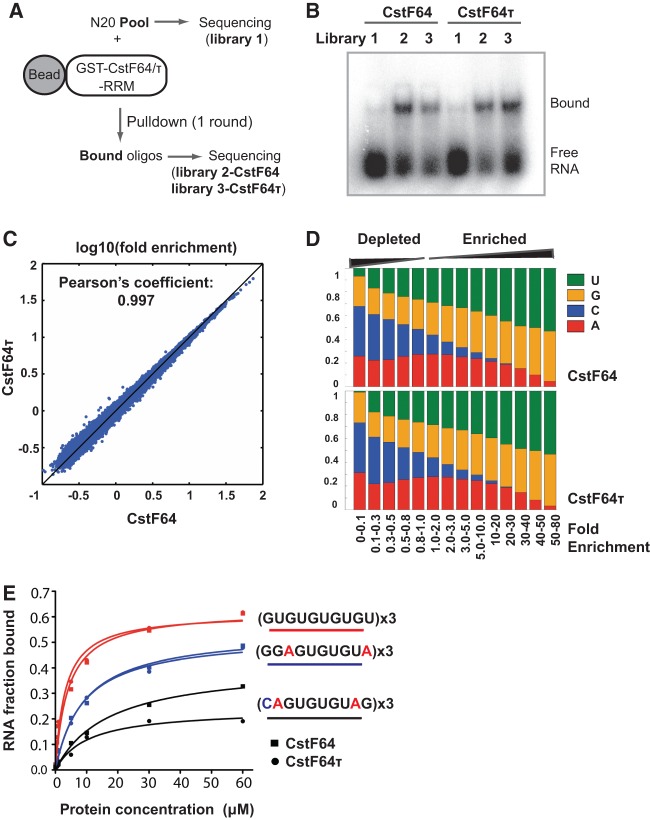
Previous studies have characterized CstF64 RNA-binding specificity in vitro using the conventional SELEX procedure (Takagaki and Manley 1997). However, the sequence specificity of CstF64τ remains unknown. To comprehensively characterize the RNA-binding specificity of both CstF64 and CstF64τ, we performed SELEX coupled with high-throughput sequencing (SELEX-seq) (Fig. 2A). Compared to the conventional SELEX, which only selects a small number of the most avid binders, SELEX-seq provides a more comprehensive characterization by identifying protein–RNA interactions with a wide range of affinities (Dittmar et al. 2012). To this end, we incubated the recombinant GST-CstF64-RRM or GST-CstF64τ-RRM fusion proteins with a pool of random 20-nucleotide (nt) RNAs. After GST fusion proteins were precipitated using glutathione beads and washed, bound RNAs were eluted and amplified by reverse transcription and PCR (RT-PCR) (Fig. 2A). After just one round of selection, CstF64 and CstF64τ cognate sequences were significantly enriched as shown by gel mobility shift assays (Fig. 2B). Additionally, CstF64-selected RNAs were bound by CstF64τ and vice versa (Fig. 2B), indicating that the two proteins have selected similar sequences. Next, we sequenced the CstF64- and CstF64τ-bound sequences along with the original RNA pool by using the Illumina HiSeq platform. Approximately 80 million reads were obtained for each library. Based on the sequencing data, we first compared the fold enrichment of all possible 10-mer sequences in CstF64- and CstF64τ-selected RNAs compared to the original RNA pool. Strikingly, the fold enrichment of all 10-mer sequences was almost identical for CstF64 and CstF64τ (Pearson's coefficient: 0.997) (Fig. 2C). We next compared the nucleotide composition of sequences that were enriched to different extents. For both CstF64 and CstF64τ, the most highly enriched sequences consisted almost entirely of Us and Gs (Fig. 2D). In sequences with intermediate enrichment levels, A was also present at a significant frequency. In contrast, C-containing sequences were depleted during selection (Fig. 2D). To verify our SELEX-seq results, we performed gel mobility shift assay with GST-CstF64-RRM or GST-CstF64τ-RRM and three selected 10-mer sequences. Consistent with our sequencing results, both CstF64 and CstF64τ bound to UG-repeats with the highest affinity (Fig. 2E; Supplemental Fig. S1). The affinities of both proteins were lower for the A-containing sequence and further decreased and became more divergent when C was present (Fig. 2E). Finally, we identified specific sequence motifs that are enriched at different levels (Supplemental Table S1). Long UG-repeats had the highest fold enrichment (Supplemental Table S1). Sequences containing shorter UG-rich regions were enriched to a lesser extent. CACGACG was among the most depleted motifs. These analyses revealed that CstF64 and CstF64τ have nearly identical RNA-binding sequence specificities in vitro, and both proteins preferentially bind to UG-rich motifs while avoiding C-rich sequences.
FIGURE 2.
Comparison of in vitro RNA binding specificity of CstF-64 and CstF-64τ using SELEX-seq. (A) A schematic representation of the SELEX-seq procedure (details described in the text and Materials and Methods). (B) Gel mobility shift assay using RNAs prepared from the random pool (library 1), CstF64-selected sequences (library 2), and CstF64τ-selected sequences (library 3), with GST-CstF64 (marked as CstF64) or GST-CstF64τ RRM (marked as CstF64τ). (C) Comparison of fold enrichment of all possible 10-mer RNA sequences in CstF64 and CstF64τ selected sequences: (x-axis) log10(frequency of each 10-mer sequence in library 2/its frequency in library 1); (y-axis) log10(frequency of each 10-mer sequence in library 3/its frequency in library 1). (D) Comparison of nucleotide composition of RNA sequences with fold enrichment in CstF64- and CstF64τ-selected sequences. Fold enrichment levels are marked on the bottom. The height of the boxes corresponds to the frequency of each nucleotide. (E) Quantification of the gel shift assays in Supplemental Figure S1. Three sequences were tested (different colored lines) with GST-CstF64-RRM (squares) or GST-CstF64τ-RRM (dots).
Mapping the in vivo RNA-binding sites of CstF64 and CstF64τ at the transcriptome levels by iCLIP-seq
We recently mapped CstF64-RNA interactions in vivo at the transcriptome level through iCLIP-seq analysis (Yao et al. 2012). To globally characterize CstF64τ-RNA interactions in vivo, we carried out iCLIP-seq of CstF64τ in HeLa cells. As shown in Figure 3A (lanes 1,2), CstF64τ was efficiently crosslinked to RNAs in vivo by UV irradiation and specifically purified by immunoprecipitation (IP) using our CstF64τ antibody described earlier. Following similar procedures that were used for CstF64 iCLIP-seq (Yao et al. 2012), we prepared CstF64τ iCLIP-seq libraries and subjected them to high-throughput sequencing using the Illumina HiSeq platform, and 6.1 million uniquely mapped reads were obtained for the CstF64τ iCLIP library. The distribution of CstF64τ iCLIP tags showed significant enrichment in the 3′ UTRs (Fig. 3B), consistent with its function in mRNA 3′ end processing. A global analysis of the distribution of CstF64τ-binding sites near actively used PASs (based on our RNA 3′ end mapping, see below) detected a peak at 30∼35 nt downstream from the AWTAAA hexamer (Fig. 3C, red line), suggesting that CstF64τ binds to RNA together with CPSF. The distribution of CstF64τ iCLIP tags is very similar to that of CstF64 (Fig. 3C, blue line); but overall, CstF64τ iCLIP signals are lower than those of CstF64 (Fig. 3C, cf. red and blue lines). Motif analysis of CstF64τ crosslinking sites downstream from AWTAAA identified an UG-rich motif (Fig. 3C, inset), which resembles the UG-repeat motif identified by our SELEX-seq. As shown in two specific examples of the CstF64τ iCLIP analyses results (Fig. 3D,E), CstF64τ and CstF64 bind to the same regions downstream from the cleavage sites in Rps12 and Basp1 transcripts. Together, these results demonstrated that CstF64τ-RNA interaction landscape in vivo is highly similar to that of CstF64.
FIGURE 3.
Mapping CstF64τ-RNA interactions in vivo at the transcriptome level by iCLIP-seq analysis. (A) HeLa cells were UV irradiated or mock treated, and cell lysates were treated with RNase I as labeled. CstF64τ was IPed. After the 3′ linker ligation and 5′ end labeling, the purified RNP complexes were resolved by PAGE and visualized by phosphorimaging. CstF64τ and β-actin levels in all cell lysates were monitored by Western blotting. (B) Distribution of CstF64τ iCLIP tags in different regions compared to their frequency in the genome. (C) Distribution of the cleavage sites (green line), CstF64- (blue line), and CstF64τ binding sites (red line) relative to the closest upstream AWTAAA. Position 0 represents the 5′ end of AWTAAA. Weblogo of the top 20 over-represented motifs at CstF64τ crosslinking sites near poly(A) sites are shown in the inset. (D,E) CstF64 and CstF64τ iCLIP-seq results for Rps12 and Basp1 visualized on the UCSC genome browser.
The role of CstF64τ in regulating mRNA alternative polyadenylation
We next characterized the role of CstF64τ in regulating global APA. To this end, we established HeLa cell lines that stably express a CstF64τ-targeting shRNA (CstF64τ-RNAi cell lines). As shown in Figure 4A (right lane), CstF64τ was significantly depleted in CstF64τ-RNAi cells. In these cells, an increase in CstF64 protein levels was observed. Similar reciprocal changes were also observed in a cell line in which CstF64 is stably silenced by RNAi (CstF64-RNAi) (Fig. 4A, middle lane). These results suggest that CstF64 and CstF64τ negatively regulate one another's expression. Quantitative RT-PCR (RT-qPCR) analyses did not detect significant changes at the mRNA levels (Supplemental Fig. S2), indicating that CstF64 and CstF64τ modulate one another's expression primarily at the post-transcriptional level. Next, we characterized the global APA profile in CstF64τ-RNAi cells by direct RNA sequencing (DRS) analysis using the Helicos platform as described before (Yao et al. 2012). By comparing the APA profiles between control HeLa cells and CstF64τ-RNAi cells, we identified 96 genes (of the 3800 genes that have at least two polyadenylation sites, each with 10 DRS reads or more) that displayed significantly different APA patterns in CstF64τ-RNAi cells (false discovery rate <0.05, Fisher's exact test) (Fig. 4B). Among them, 57 showed a relative increase in the mRNA isoforms polyadenylated at the proximal PAS (distal-to-proximal or DtoP shift), whereas the remainder showed APA changes in the opposite direction (proximal-to-distal or PtoD shift) (Fig. 4B). For example, the APA profiles of CSTF3 in control HeLa cells and CstF64τ-RNAi cells are shown in Figure 4C. The APA profile of the same gene in CstF64-RNAi cells was also shown for comparison (Fig. 4C). A significant increase in the mRNAs polyadenylated at the distal PAS in CSTF3 was observed in both CstF64- and CstF64τ-RNAi cells (Fig. 4C). This and a number of other APA changes identified by our DRS analyses were validated by RT-qPCR (Fig. 4D), confirming the accuracy of our sequencing analysis. To further characterize the CstF64τ-mediated APA regulation, we next compared the APA changes induced by CstF64τ depletion and the previously reported APA changes caused by CstF64 depletion or CstF64&τ codepletion (Fig. 4E). It is apparent that depletion of CstF64 or CstF64τ had a relatively small effect on the global APA profile compared to the codepletion of both factors (Fig. 4E; Yao et al. 2012), strongly suggesting functional redundancy between the two proteins. Additionally, APA changes induced by CstF64 or CstF64τ did not seem to have strong biases between PtoD and DtoP changes (Fig. 4E; Supplemental Fig. S3). In contrast, codepletion of CstF64 and CstF64τ led to PtoD shifts in 85% of the affected genes (Fig. 4E; Supplemental Fig. S3). These observations are consistent with a model in which CstF64 and CstF64τ play largely redundant roles in APA regulation. Depletion of either proteins induces the up-regulation of the other, resulting in a relatively stable general mRNA 3′processing activity and relatively few changes in APA. However, codepletion of both proteins leads to a decrease in mRNA 3′ processing activity and thus lower efficiency of cleavage/polyadenylation at the proximal PASs. This, in turn, results in higher transcription read-through and usage of the distal PASs and thus PtoD shifts.
Characterization of protein interactions of CstF64 and CstF64τ
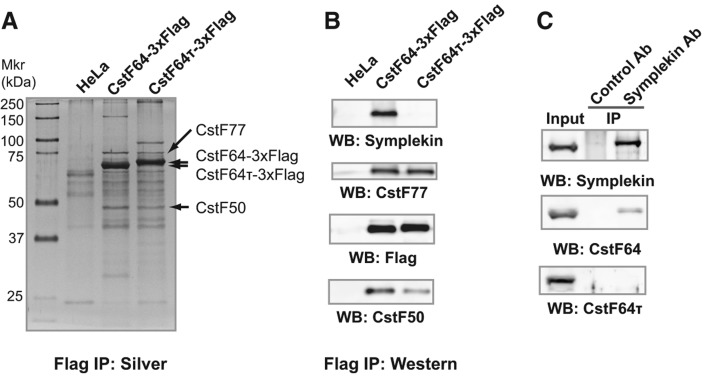
Finally, we compared the protein interactomes of CstF64 and CstF64τ in vivo. To this end, we established stable HeLa cell lines expressing Flag-tagged CstF64 or CstF64τ and carried out IPs using anti-Flag antibodies. As shown in Figure 5A,B, Flag-tagged CstF64 and CstF64τ were specifically IPed from the respective cell lines. Similar levels of CstF77 and CstF50 were co-IPed in both samples (Fig. 5A,B), suggesting that both CstF64 and CstF64τ were assembled into the CstF complex. Next, we analyzed CstF64- and CstF64τ-associated proteins by mass spectrometry analyses, and the identified proteins are listed in Supplemental Table S2. As expected, CstF77 and CstF50 were identified in both samples. Additionally CstF64τ was detected in CstF64-associated proteins and vice versa (Supplemental Table S2). This is consistent with previous studies showing that CstF dimerizes (Takagaki and Manley 2000; Bai et al. 2007) and further suggests that CstF64-containing and CstF64τ-containing CstF complexes form heterodimers. Symplekin, a subunit of the CPSF complex and a known binding partner of CstF64 (Takagaki and Manley 2000), was also co-IPed with CstF64. Surprisingly, however, symplekin was not detected in the Flag-CstF64τ IP sample by our mass spectrometry analysis (Supplemental Table S2). Consistently, our Western analysis detected symplekin in the CstF64 IP sample, but very little symplekin was detected in the CstF64τ IP sample (Fig. 5B). To further verify this result, we carried out IP using anti-symplekin antibodies and HEK293 cell lysates. As shown in Figure 5C, the endogenous symplekin was efficiently precipitated, and CstF64 was co-IPed. However, very little CstF64τ was detected in the symplekin IP sample (Fig. 5C). Since our IP experiments were performed under stringent conditions (in RIPA buffer), we next repeated the symplekin IP under milder conditions (see Materials and Methods for details) to determine whether CstF64τ can associate with symplekin. Under this condition, CstF64 and a small amount of CstF64τ were co-IPed with symplekin (Supplemental Fig. S3). Together these results suggest that CstF64 is more strongly associated with symplekin in vivo than CstF64τ.
FIGURE 5.
Proteomic analyses of the CstF64 and CstF64τ interactomes. (A) Flag IPs were carried out using cell lysates from control HeLa cells or CstF64-3×Flag or CstF64τ-3×Flag cell lines. IP samples were resolved by SDS-PAGE and visualized by silver staining. (B) Western analyses of the IP samples described in A. (WB) Western blotting. (C) IP was carried out using anti-symplekin antibodies and HEK293 cell lysates. IP samples were resolved by SDS-PAGE and analyzed by Western blotting (WB).
As mentioned earlier, CstF64 and CstF64τ are highly similar in their domain structures, and each contains an N-terminal RRM domain, a hinge domain, a P/G-rich domain, and a C-terminal domain (CTD). Since previous studies have shown that CstF64 directly binds to symplekin via its hinge domain (Takagaki and Manley 2000), we next determined whether CstF64τ can directly interact with symplekin. To this end, we synthesized the full-length symplekin, CstF64, and CstF64τ by in vitro translation using the rabbit reticulocyte system. Then symplekin was incubated with CstF64 or CstF64τ followed by IP with anti-symplekin antibodies. Interestingly, CstF64, but not CstF64τ, was co-IPed with symplekin (Fig. 6A). This result suggests that CstF64 has significantly higher affinity for symplekin than CstF64 in vitro.
FIGURE 6.
Characterization of the interactions between symplekin and CstF64/τ. (A) Symplekin and full-length CstF64/τ were prepared by in vitro translation and incubated in the specified combination. IP was carried out using control or anti-symplekin antibodies. The IP samples were resolved by SDS-PAGE and visualized by phosphorimaging: (*) an apparent truncated symplekin fragment. (B) GST pulldown assays with in vitro translated symplekin and GST or GST-CstF64-hinge or GST-CstF64τ-hinge. GST pulldown samples were resolved by SDS-PAGE and visualized by phosphorimaging. (C,D) IP experiments were carried out as described in A except that the P/G-rich domain (C) or the hinge-P/G-rich domain fragments (D) were used, respectively. (E) A summary of the in vitro binding assay results shown in A–D. (F) A schematic model for the interactions between symplekin and CstF64/τ. See text for details.
We next mapped the domains that mediate the differential binding of CstF64 and CstF64τ to symplekin. As mentioned earlier, the hinge domain of CstF64 has been shown to mediate its interaction with symplekin (Takagaki and Manley 2000; Ruepp et al. 2010). We next expressed and purified recombinant CstF64- or CstF64τ-hinge domain fused to a GST tag and used them in GST pulldown assays with in vitro translated symplekin. Consistent with previous studies, CstF64 hinge domain interacts with symplekin (Fig. 6B). Interestingly however, CstF64τ hinge domain also pulled down symplekin with similar efficiency (Fig. 6B), suggesting that the hinge domains of both CstF64 and CstF64τ are capable of binding to symplekin. Using the same co-IP assay as described in Figure 6A, we found that the P/G-rich domains of CstF64 or CstF64τ do not interact with symplekin (Fig. 6C). Since the hinge domains of both CstF64 and CstF64τ can bind to symplekin (Fig. 6B), we reasoned that the interaction between the CstF64τ hinge domain and symplekin might be inhibited by other domains in the full-length CstF64τ. To test this idea, we compared the interactions between symplekin and an internal fragment of CstF64 or CstF64τ consisting of the hinge domain and the P/G-rich domain. As shown in Figure 6D, this internal fragment of CstF64, but not that of CstF64τ, interacted with symplekin, recapitulating the differential binding of the full-length proteins. The results of all the in vitro binding assays are summarized in Figure 6E. Together, these results revealed a significant difference in the affinities of CstF64 and CstF64τ for symplekin. Although the hinge domains of both CstF64 and CstF64τ are capable of binding to symplekin, this interaction is inhibited by the P/G-rich domain in CstF64τ (Fig. 6F). The potential implications of this difference on the functions of CstF64 and CstF64τ in mRNA 3′ processing are discussed below.
DISCUSSION
In this report, we comprehensively compared the properties of CstF64τ and CstF64, including their expression patterns, protein-RNA and protein–protein interactions, and functions in global APA regulation. Our Western analyses of human and mouse cell lines as well as mouse tissues reveal that both CstF64 and CstF64τ are widely expressed. We demonstrated that the two proteins have highly similar RNA binding sequence specificity in vitro and in vivo. Additionally, CstF64 and CstF64τ modulate one another's expression, and they play partially redundant roles in regulating global APA. Finally, we demonstrate that CstF64 and CstF64τ have overlapping but distinct protein interactomes and differ significantly in their interactions with another core 3′ processing factor, symplekin. Below, we discuss the implications of these findings for mRNA 3′ processing and APA regulation.
CstF64τ has been proposed to function in mediating tissue-specific APA regulation (MacDonald and McMahon 2010). This is based on the observations that CstF64τ seems to be specifically expressed in the testis (hence, the name CstF64τ) and the brain (Wallace et al. 1999). Here, our study revealed that both CstF64 and CstF64τ are widely expressed in mouse tissues (Fig. 1B). This discrepancy is most likely due to the different sensitivity and/or specificity of the antibodies used for the CstF64τ Western analyses. In the previous report (Wallace et al. 1999), a mouse monoclonal antibody (6A9) was used to detect CstF64τ. Although this antibody detected a protein of ∼70 kDa that was interpreted as CstF64τ in the testis and brain, it only detected a ∼64-kDa band in HeLa cells that was believed to be CstF64 (Wallace et al. 1999). These observations suggest that this antibody has cross-reactivities and is not specific for CstF64τ. On the other hand, multiple lines of evidence demonstrated that the CstF64τ antibody used in our study is highly specific. First, it consistently detected the ∼70-kDa CstF64τ in cell lines and tissues of human or mouse origins. Second, our dual color Western analyses detected no cross-reactivity with CstF64 for this antibody (Fig. 1A). Finally, this antibody detected the specific depletion of CstF64τ by RNAi in our stable knockdown cell lines (Fig. 4A). These results suggest that our CstF64τ antibody is highly specific, and our Western analyses demonstrate that CstF64τ, similar to CstF64, is widely expressed. This is consistent with previous reports that CstF64τ mRNAs are detected ubiquitously in mouse tissues (Huber et al. 2005). However, given that the CstF64:CstF64τ ratio and the total levels of these two proteins are highly variable in different tissues (Fig. 1B), CstF64τ may still contribute to tissue-specific regulation of mRNA 3′ processing and APA.
Our SELEX-seq and iCLIP-seq results demonstrated that CstF64 ad CstF64τ have highly similar RNA-binding specificities in vitro and in vivo (Figs. 2, 3). This high degree of functional redundancy may be part of the reason why depletion of either CstF64 or CstF64τ alone had relatively small effect on the global APA profile. For comparison, we found that depletion of another mRNA 3′ processing factor, Fip1, led to at least five times more APA changes than CstF64 or CstF64τ depletion (data not shown). Additionally, the functional impact of CstF64τ depletion is mitigated by the compensatory increase in CstF64 and vice versa (Fig. 4A). Therefore, the observed APA changes in CstF64τ-RNAi cells are the combined effects of CstF64τ depletion and higher levels of CstF64. The specific APA changes observed in these cells are most likely determined by minor differences in CstF64 and CstF64τ binding at the regulated PASs. To test this, we compared the relative CstF64- and CstF64τ-RNA interactions at the proximal and distal PASs regulated by CstF64τ (Supplemental Fig. S5). Although CstF64 and CstF64τ bind to the same regions at these PASs, we detected differences in their iCLIP tag densities at the proximal and distal PASs, which may reflect their binding strength and/or frequencies (Supplemental Fig. S5). These results indicate that the similarities and differences in CstF64- and CstF64τ-RNA interactions may contribute to their functional redundancy and specificity in mRNA 3′ processing.
Although CstF64 and CstF64τ are highly similar in their interactions with RNAs, our study revealed key differences in their protein–protein interactions. Importantly, we showed that CstF64 interacts with symplekin with significantly higher affinity than CstF64τ (Fig. 5). Although the hinge domains of both CstF64 and CstF64τ can directly bind symplekin, this interaction is inhibited by the P/G-rich domain in CstF64τ (Fig. 6). In keeping with this, the P/G-rich domain is the most divergent region between CstF64 and CstF64τ. Although CstF64τ affinity for symplekin is low, it is possible that CstF64τ can indirectly associate with symplekin in vivo through dimerization with CstF64, which explains the co-IP of CstF64τ with symplekin under mild conditions (Supplemental Fig. S3). As symplekin links CstF to CPSF (Takagaki and Manley 2000), our results indicate that CstF64 may interact with CPSF more strongly; thus, the CstF64-containing CstF complexes may be recruited to the mRNA 3′ processing complex more efficiently than the CstF64τ-containing CstF complexes. This may lead to functional differences between CstF64 and CstF64τ in mRNA 3′ processing. As such, tissue-specific variations in the CstF64:CstF64τ ratio could provide a mechanism for fine-tuning the mRNA 3′ processing activities.
Given their functional similarities, it is surprising that both CstF64 and CstF64τ are well conserved during mammalian evolution. This may be explained by at least two possible scenarios. First, it has been suggested that, in mammalian testis, expression of the X-linked CstF64 is suppressed during spermatogenesis due to the formation of meiotic XY bodies (Wallace et al. 1999). During this period, CstF64τ, encoded by an autosomal gene, becomes essential for mRNA 3′ processing. Consistent with this model, the main phenotype of the CstF64τ knockout mice is a defect in spermatogenesis (Dass et al. 2007). Alternatively, although CstF64 and CstF64τ have highly similar RNA-binding specificities, they differ in protein–protein interactions (Figs. 5, 6) and therefore may have distinct functions in some aspects of mRNA 3′ processing or in other cellular processes. Indeed, a recent study reported differential expression of retrotransposon RNAs in the testis of CstF64τ knockout mice (Li et al. 2012). Interestingly, the functional relationship between CstF64 and CstF64τ is reminiscent of that of PTB (or Ptbp1) and nPTB (or Ptbp2), two related hnRNPs that function in splicing regulation (Boutz et al. 2007; Coutinho-Mansfield et al. 2007; Licatalosi et al. 2012). Although the two proteins have very similar RNA-binding specificities, they are expressed in different developmental stages and in different tissues. They negatively regulate one another's expression and have both overlapping and distinct functions in regulating global mRNA alternative splicing (Boutz et al. 2007; Coutinho-Mansfield et al. 2007; Licatalosi et al. 2012). Therefore, coordinated expression of functionally related RNA-binding proteins may be an important regulatory mechanism for alternative mRNA processing in development.
MATERIALS AND METHODS
Western blot
Mouse tissues were obtained from wild-type C57BL/6 mice (gifts from Dr. Paolo Casali's laboratory). Cell lines were obtained from ATCC and grown in recommended medium. Primary antibodies used include CstF64τ (Bethyl A301-487A), CstF64 (monoclonal antibody 3A7), CstF77 (Bethyl A301-096A), CstF50 (Bethyl A301-251A), symplekin (Bethyl A301-465A), GAPDH (Santa Cruz Biotechnology sc-32233), and β-actin (Santa Cruz Biotechnology sc-8432).
SELEX-seq
GST-CstF64/τ-RRM was expressed in E. coli and purified with glutathione-conjugated beads according to the manufacturer's instructions (GE Healthcare Life Sciences). SELEX was carried out as previously described with minor modifications (Takagaki and Manley 1997). Briefly, template DNA containing 20 nt random sequence was generated by annealing two oligos (T7 and N20), and the DNA ends were filled with Klenow. The RNA pool was prepared by in vitro transcription with T7 RNA polymerase and purified on a urea-polyacrylamide gel. For RNA pulldown, 7.4 µg GST fusion protein was conjugated with 20 µL glutathione sepharose 4B beads. After washing, the beads were mixed with 200 μL of binding buffer (8 mM HEPES, pH 7.9; 40 mM NaCl; 2 mM EDTA; 0.2 mM DTT; 0.2 mM PMSF; 8% glycerol) containing 16 µg of heparin and 32 µg random RNA pool (final concentration 7.5 µM), and incubated for 10 min at 30°C with occasional shaking. After washing, bound RNAs were released by proteinase K digestion and precipitated by ethanol. Recovered RNAs as well as the original RNA pool were amplified by RT-PCR to generate barcoded libraries for sequencing. Oligo sequences are provided in Supplemental Material.
iCLIP-seq
CstF64τ iCLIP-seq was carried out as described previously (Yao et al. 2012). For simplicity, one master iCLIP-seq library was generated and used for all analyses. For CstF64τ iCLIP motif analysis, we ranked all CstF64τ crosslinking sites according to their cDNA counts. We iteratively chose the crosslinking site with the highest cDNA count that did not overlap with the 21-nt region spanning a site that had been chosen previously. For all nonoverlapping crosslinking sites, we determined the enrichment score of 6-mer motifs in the 21-nt surrounding region. We further classified those CstF64τ crosslinking sites into two groups: those with A(A/U)UAAA within 40 nt upstream (AAUAAA+) and those without (AAUAAA−). We aligned the top 20 most enriched motifs in each group and generated sequence logos using WebLogo 3 (http://weblogo.threeplusone.com/) as previously described (Yao et al. 2012).
Gel shift assay
RNA oligos were prepared by in vitro transcription with T7 RNA polymerase and purified on denaturing polyacrylamide gels. For gel shift assays, 5′ radio-labeled RNAs (5 nM) were incubated with 0.5 µM to 60 µM GST-CstF64/t RRM protein in 10 µL binding buffer for 10 min at 30°C. The reaction mixtures were resolved on a 4% nondenaturing polyacrylamide gel and visualized by phosphorimaging.
Direct RNA sequencing analyses
Direct RNA sequencing (DRS) was performed by Helicos BioSciences, and DRS reads were aligned to hg19 using the index-DPgenomictool in Helisphere (Helicos BioSciences). Only uniquely mapped reads with a minimum mapped length of 25 and an alignment score of 4.0 were kept. We further filtered and clustered all individual PASs in the same way as described previously (Yao et al. 2012). To compare the alternative polyadenylation (APA) profiles in HeLa and CstF64τ-RNAi cells using DRS data, we first removed PASs that had zero or one read in both samples. For the remaining PASs to be mapped to RefSeq genes, we used the Fisher exact test to compare the ratio of the DRS read counts of one PAS to the sum of the read counts of all of the other PASs within the same gene. The P-values were adjusted by the Benjamini–Hochberg method for calculating the FDR. PASs with an FDR <0.05 were defined as significantly changed PASs. To create the scatter plot shown in Figure 4B, we selected two PASs with the smallest P-values for each gene with multiple PASs and calculated the corresponding proximal/distal ratio. In the figure, PAS pairs with an FDR <0.05 and log10(proximal/distal PAS read count-control/RNAi) >0.2 are highlighted in red for proximal-to-distal switches and in blue for distal-to-proximal switches.
Protein–protein interaction studies
For proteomic analyses of CstF64- and CstF64τ-associated proteins, retroviral vectors for expressing CstF64- or CstF64τ-3×Flag proteins were used to transduce HeLa cells. Transduced cells were selected with puromycin and expanded. Cells were lysed in RIPA buffer, and the cell lysates were used for immunoprecipitation using anti-Flag antibodies (M2, Sigma). Bound proteins were eluted using 3×Flag peptide, precipitated with TCA, and subjected to MudPIT mass spectrometry analyses (details for MudPIT analyses are provided in Supplemental Material). Common contaminants from Flag IP were filtered out as previously described (Shi et al. 2009). For the interaction studies shown in Figure 6, CstF64, CstF64τ, or symplekin cDNAs were cloned into pcDNA3.1-Flag vector. 35S methionine-labeled proteins were prepared by using the TNT Quick Coupled Transcription/Translation System (Promega). CstF64 or CstF64τ were mixed with symplekin in five volumes of buffer D100 (20 mM HEPES, pH 7.9, 100 mM NaCl, 1 mM MgCl2, 0.2 mM EDTA, 10% Glycerol, 10 mM β-ME, 0.5 mM PMSF). Immunoprecipitation was performed overnight at 4°C using anti-symplekin antibodies (Bethyl A301-465A) or control antibodies. After washing, the bound proteins were eluted by boiling in SDS sample buffer, resolved on SDS-PAGE, and visualized by phosphorimaging. For IPs in Supplemental Figure S4, IPs were carried out in Buffer D300 (similar to Buffer D100 described above except that 300 mM NaCl was used).
DATA DEPOSITION
All sequencing data have been deposited into the Gene Expression Omnibus (GEO) database (accession number: GSE51156).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Paolo Casali's laboratory (University of California, Irvine) for the generous gifts of mouse tissues and Dr. Charles Limoli's lab (University of California, Irvine) for sharing equipment. This study was supported by grants from NIH (R01GM090056) and ACS (RSG-12-186) (Y.S); an NIH grant (R01GM088342) and a junior faculty grant from the Edward Mallinckrodt Jr. Foundation (Y.X.); grants from the National Center for Research Resources (5P41RR011823-17), the National Institute of General Medical Sciences (8 P41 GM103533-17), and the National Institute on Aging (R01AG027463-04) (J.R.Y.).
REFERENCES
- Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L. 2007. Crystal structure of murine CstF-77: Dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell 25: 863–875. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M Jr, Black DL. 2007. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21: 1636–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S, Choi EA, Shi Y. 2011. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip Rev RNA 2: 321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766. [DOI] [PubMed] [Google Scholar]
- Coutinho-Mansfield GC, Xue Y, Zhang Y, Fu XD. 2007. PTB/nPTB switch: A post-transcriptional mechanism for programming neuronal differentiation. Genes Dev 21: 1573–1577. [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE. 2008. 3′ end mRNA processing: Molecular mechanisms and implications for health and disease. EMBO J 27: 482–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dass B, Tardif S, Park JY, Tian B, Weitlauf HM, Hess RA, Carnes K, Griswold MD, Small CL, Macdonald CC. 2007. Loss of polyadenylation protein τCstF-64 causes spermatogenic defects and male infertility. Proc Natl Acad Sci 104: 20374–20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. 2012. A quantitative atlas of polyadenylation in five mammals. Genome Res 22: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Jiang P, Park JW, Amirikian K, Wan J, Shen S, Xing Y, Carstens RP. 2012. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol Cell Biol 32: 1468–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber Z, Monarez RR, Dass B, MacDonald CC. 2005. The mRNA encoding τCstF-64 is expressed ubiquitously in mouse tissues. Ann N Y Acad Sci 1061: 163–172. [DOI] [PubMed] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kühn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, et al. 2012. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149: 538–553. [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. 2006. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res 34: 6264–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yeh HJ, Shankarling GS, Ji Z, Tian B, MacDonald CC. 2012. The τCstF-64 polyadenylation protein controls genome expression in testis. PLoS One 7: e48373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. 2012. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev 26: 1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald CC, McMahon KW. 2010. Tissue-specific mechanisms of alternative polyadenylation: Testis, brain, and beyond. Wiley Interdiscip Rev RNA 1: 494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Gruber AR, Keller W, Zavolan M. 2012. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep 1: 753–763. [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monarez RR, MacDonald CC, Dass B. 2007. Polyadenylation proteins CstF-64 and τCstF-64 exhibit differential binding affinities for RNA polymers. Biochem J 401: 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. 2011. Ending the message: Poly(A) signals then and now. Genes Dev 25: 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Perez-Canadillas JM, Agrawal S, De Baecke J, Cheng H, Varani G, Moore C. 2007. The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3′-end processing. J Biol Chem 282: 2101–2115. [DOI] [PubMed] [Google Scholar]
- Ruepp MD, Schweingruber C, Kleinschmidt N, Schümperli D. 2010. Interactions of CstF-64, CstF-77, and symplekin: Implications on localisation and function. Mol Biol Cell 22: 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi E, Lu J, Flanagan LA, Hertel KJ, Shi Y. 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. 2012. Alternative polyadenylation: New insights from global analyses. RNA 18: 2105–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR III, Frank J, Manley JL. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, et al. 2012. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep 1: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. 1997. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol 17: 3907–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 20: 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952. [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2013. Alternative cleavage and polyadenylation: The long and short of it. Trends Biochem Sci 38: 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AM, Dass B, Ravnik SE, Tonk V, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. 1999. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc Natl Acad Sci 96: 6763–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Biesinger J, Wan J, Weng L, Xing Y, Xie X, Shi Y. 2012. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci 109: 18773–18778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.