Eukaryotic ribosome biogenesis requires a large number (more than 200) assembly factors, one of which is Rlp7. This article shows that Rlp7 binding to pre-rRNA induces conformational changes in ITS2 that may regulate pre-rRNA processing. These findings do not support the hypothesis that Rlp7 serves as a placeholder for ribosomal protein L7.
Keywords: ribosome biogenesis, Rlp7, RNA binding protein, crosslinking and analysis of cDNAs (CRAC), ribosomal protein L7
Abstract
Eukaryotic ribosome assembly requires over 200 assembly factors that facilitate rRNA folding, ribosomal protein binding, and pre-rRNA processing. One such factor is Rlp7, an essential RNA binding protein required for consecutive pre-rRNA processing steps for assembly of yeast 60S ribosomal subunits: exonucleolytic processing of 27SA3 pre-rRNA to generate the 5′ end of 5.8S rRNA and endonucleolytic cleavage of the 27SB pre-rRNA to initiate removal of internal transcribed spacer 2 (ITS2). To better understand the functions of Rlp7 in 27S pre-rRNA processing steps, we identified where it crosslinks to pre-rRNA. We found that Rlp7 binds at the junction of ITS2 and the ITS2-proximal stem, between the 3′ end of 5.8S rRNA and the 5′ end of 25S rRNA. Consistent with Rlp7 binding to this neighborhood during assembly, two-hybrid and affinity copurification assays showed that Rlp7 interacts with other assembly factors that bind to or near ITS2 and the proximal stem. We used in vivo RNA structure probing to demonstrate that the proximal stem forms prior to Rlp7 binding and that Rlp7 binding induces RNA conformational changes in ITS2 that may chaperone rRNA folding and regulate 27S pre-rRNA processing. Our findings contradict the hypothesis that Rlp7 functions as a placeholder for ribosomal protein L7, from which Rlp7 is thought to have evolved in yeast. The binding site of Rlp7 is within eukaryotic-specific RNA elements, which are not found in bacteria. Thus, we propose that Rlp7 coevolved with these RNA elements to facilitate eukaryotic-specific functions in ribosome assembly and pre-rRNA processing.
INTRODUCTION
Eukaryotic ribosome assembly requires precise folding of ribosomal RNA (rRNA), binding of ribosomal proteins (r-proteins) to rRNA, and processing of pre-rRNA to generate mature 40S and 60S ribosomal subunits (Henras et al. 2008). Processing of pre-rRNA occurs through a series of consecutive steps that involves endo- and exonucleolytic removal of internal and external transcribed spacer (ITS and ETS) sequences (Fig. 1A). These irreversible steps are coupled with rRNA folding, r-protein binding, and transit of preribosomes from the nucleolus to the nucleus and cytoplasm and may therefore serve as checkpoints for proper assembly.
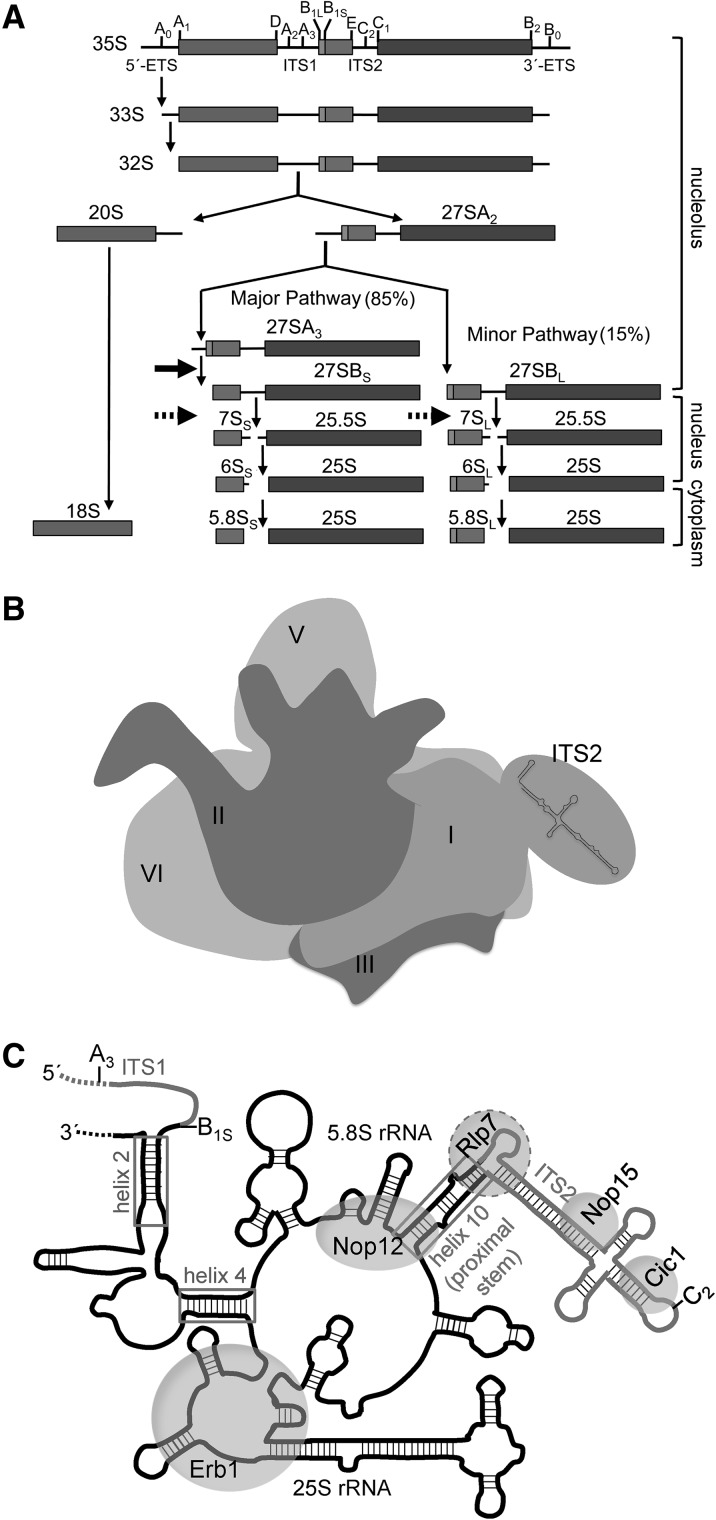
FIGURE 1.
Yeast ribosome assembly. (A) Pre-rRNA processing pathway. Eukaryotic pre-rRNA processing occurs through a series of consecutive steps. The locations of the ETS and ITS sequences (lines), pre-rRNA processing sites (vertical ticks), and mature rRNA sequences (boxes) are shown. The solid arrow marks the 27SA3 pre-rRNA processing step, which is blocked in the absence of Rlp7 and the other A3 factors. The dashed arrows indicate the 27SB pre-rRNA processing step, which also is blocked in rlp7 mutants. The 40S subunit contains 18S rRNA and the 60S subunit contains 5.8S, 25S, and 5S rRNAs. 5S rRNA is transcribed and processed separately and is not shown. (B) Cartoon of the solvent-accessible interface of pre-60S ribosomes containing mature rRNA domains I–VI and ITS2 in the hairpin conformation. Domain IV is on the back in this view (subunit interface) and thus is not labeled. (C) Cartoon representation of base-pairing between 5.8S and 25S rRNAs in domain I. Sequences of mature rRNA are black, and ITS1 and ITS2 are gray. Boxes indicate regions where 5.8S rRNA base-pairs with 25S rRNA. Helix numbers are listed for 25S rRNA. Helix 10 is also known as the ITS2-proximal stem. Gray circles indicate the binding sites of Rlp7, Erb1, Nop12, Nop15, and Cic1 on preribosomes. The A3, B1S, and C2 processing sites are indicated.
In Saccharomyces cerevisiae, ribosome biogenesis involves about 200 assembly factors that are present in preribosomal intermediates but not in mature ribosomal subunits. These factors include endo- and exonucleases, RNA helicases, AAA-ATPases, GTPases, protein and RNA modifying enzymes, and nonenzymatic scaffolding and RNA binding proteins. Scaffolding proteins contain predicted protein–protein interaction domains and are sometimes found in subcomplexes with other assembly factors (Miles et al. 2005). RNA binding proteins are thought to stabilize or chaperone pre-rRNA folding (Granneman et al. 2011; Young and Karbstein 2011), target other proteins to preribosomes (Woolls et al. 2011; Young et al. 2013), or act as placeholders for r-proteins (Rodriguez-Mateos et al. 2009).
Currently, we have an almost complete catalog of assembly factors and r-proteins that are required for each of the pre-rRNA processing steps to occur. However, why specific steps are blocked in the absence of individual assembly factors or how rRNA folding and r-protein binding are coupled to pre-rRNA processing is not known. Recently, crosslinking and cryo-EM assays have been adapted to locate the binding sites of assembly factors on preribosomes (Bohnsack et al. 2009; Granneman et al. 2010, 2011; Strunk et al. 2011; Bradatsch et al. 2012; Greber et al. 2012; Segerstolpe et al. 2013). Identification of binding sites of assembly factors is necessary to understand the specific functions of these proteins during ribosome assembly and to understand why pre-rRNA processing steps are blocked in their absence.
The A3 factors are a group of assembly factors required for exonucleolytic trimming of 27SA3 pre-rRNA to generate 27SBS pre-rRNA (Fig. 1A, solid horizontal arrow). This step removes the remaining fragment of ITS1 after cleavage at the A3 site and generates the 5′ end of the more abundant form of 5.8S rRNA, 5.8SS. In addition to the exonucleases Rat1 and Rrp17 (Henry et al. 1994; Oeffinger et al. 2009), RNA helicase Has1 (Emery et al. 2004), and r-proteins L7 and L8 (Jakovljevic et al. 2012), the known A3 factors include the scaffolding proteins Pwp1, Nop7, Ytm1, and Erb1 and RNA binding proteins Nop15, Cic1, Nop12, and Rlp7 (Dunbar et al. 2000; Pestov et al. 2001; Adams et al. 2002; Gadal et al. 2002; Oeffinger et al. 2002; Fatica et al. 2003; Oeffinger and Tollervey 2003; Miles et al. 2005; Granneman et al. 2011; Sahasranaman et al. 2011). Remaining questions in the study of A3 factors include (1) what are the functions of these nonenzymatic proteins in ribosome assembly and (2) why is the 27SA3 pre-rRNA processing step blocked in their absence.
25S rRNA is separated into six secondary structure domains, which are brought together to form one globular domain in the mature 60S subunit (Fig. 1B). In domain I, 5.8S rRNA base-pairs with 25S rRNA in three places: helix 2 at the 5′ end of 5.8S rRNA, helix 4, and the ITS2 proximal stem (helix 10) between the 3′ end of 5.8S rRNA and 5′ end of 25S rRNA (Fig. 1C, gray boxes). In mature ribosomes, r-proteins L8, L15, L17, L25, L26, L35, and L37 contact this rRNA domain. Of this group, L17, L26, L35, and L37 are missing from preribosomes in the absence of A3 factors (Sahasranaman et al. 2011; Jakovljevic et al. 2012; Dembowski et al. 2013). Because these four r-proteins make the most contacts with domain I, it is possible that the A3 factors play a role in stabilizing this rRNA–r-protein neighborhood.
The rRNA binding sites of nonenzymatic A3 factors Nop7, Erb1, Nop15, Cic1, and Nop12 have been previously identified by CRAC (crosslinking and analysis of cDNAs) (Fig. 1C, gray circles; Granneman et al. 2011). Nop7 binds to domain III and Erb1 binds to domain I. Because these two proteins interact in a stable trimeric subcomplex with Ytm1 (Miles et al. 2005), they may form an interdomain bridge to bring together domains I and III during assembly. Ytm1 and Pwp1 do not crosslink to RNA, suggesting that they do not directly interact with pre-rRNA. Nop15 and Cic1 crosslink to ITS2 and are thought to hold ITS2 in an open ring conformation as opposed to a closed hairpin structure (Granneman et al. 2011). Transitions between these two conformations may regulate the timing of 27S pre-rRNA processing (Cote et al. 2002). Nop12 binds to sequences in 5.8S and 25S rRNAs, suggesting that it may promote folding of domain I by stabilizing interactions between these two rRNAs. Because the A3 factors bind to domain I of 5.8S/25S rRNA or to ITS2 and are required for stable association of r-proteins to domain I, folding or stabilization of this rRNA-r-protein neighborhood may be coupled to the 27SA3 pre-rRNA processing step.
Rlp7 has been implicated not only in 27SA3 pre-rRNA processing but also in the downstream step, cleavage of 27SB pre-rRNA to generate 7S and 25.5S pre-rRNAs (Fig. 1A, dashed horizontal arrows; Dunbar et al. 2000; Gadal et al. 2002). This step involving cleavage by an unknown endonuclease initiates removal of ITS2 from pre-60S ribosomes. Therefore, we predict that Rlp7 might bind near the other A3 factors in ITS2 or domain I to promote consecutive processing of 27SA3 and 27SB pre-rRNAs. However, because Rlp7 shares 32% sequence identity to r-protein L7 (Lalo et al. 1993; Mizuta et al. 1995), it has been proposed to bind to domain II of 25S rRNA to act as a placeholder for L7 during assembly (Gadal et al. 2002).
Here we sought to identify the binding site of Rlp7 on preribosomes, with the goal to better understand its function in 27S pre-rRNA processing steps. We found that Rlp7 binds to the ITS2-proximal stem of rRNA domain I and to adjacent sequences in ITS2, placing it in close proximity to the previously identified RNA binding sites for A3 factors Erb1, Nop15, Cic1, and Nop12 (Fig. 1C, gray circles). We identified an interaction network among Erb1, Nop15, Cic1, and Rlp7, consistent with these A3 factors clustering to a neighborhood within assembling ribosomes. We also carried out in vivo RNA structure probing of ITS2 in the presence and absence of Rlp7. Our results are consistent with Rlp7, Cic1 and Nop15 being interdependent for association with ITS2 and inducing an open ring conformation of ITS2 as opposed to a closed hairpin structure. Taken together, we conclude that Rlp7 binds at the junction of the ITS2-proximal stem and ITS2 and may act together with other assembly factors to promote the formation of domain I and ITS2 conformations that are conducive to consecutive 27S pre-rRNA processing steps.
RESULTS AND DISCUSSION
Rlp7 binds to ITS2 and the ITS2-proximal stem
In order to identify the binding sites and RNA substrates of Rlp7, we carried out CRAC (Granneman et al. 2009). We treated yeast cells expressing Rlp7-HTP with UV light to crosslink RNA to proteins in vivo. Rlp7 was purified, and crosslinked RNAs were detected (Fig. 2A). Consistent with Rlp7 acting as an RNA binding protein and directly contacting its RNA substrate, Rlp7 crosslinked to RNA in a UV-dose dependent manner. Crosslinked RNA was purified, amplified, and cloned. A TapeStation-generated gel image of representative cDNA libraries is shown (Fig. 2B). The negative control cDNA was not detectable and yielded very few transformants when cloned.
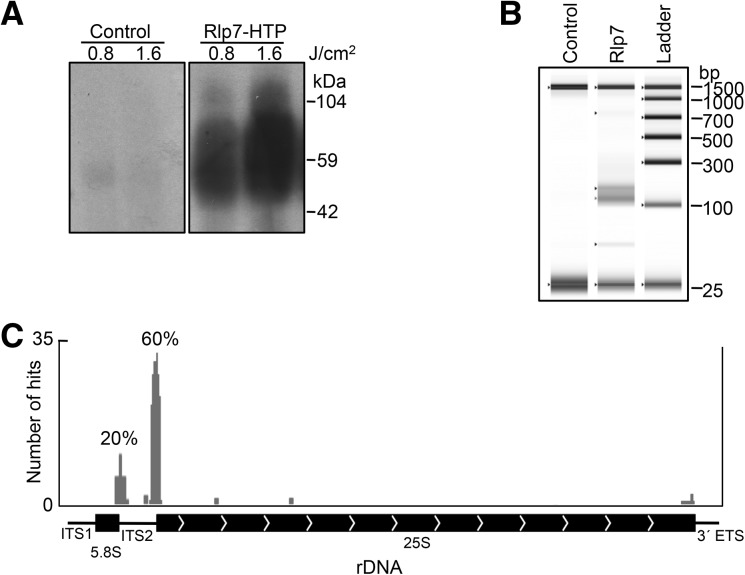
FIGURE 2.
Rlp7 crosslinks to ITS2 and the ITS2-proximal stem of domain I. (A) Purification of RNA crosslinked to Rlp7. An autoradiogram of affinity purified Rlp7 and crosslinked RNA is shown. His-tagged Rlp7 (∼38 kDa) with crosslinked RNA and linkers (∼21–26 kDa) is expected to migrate around 59–64 kDa in an SDS–polyacrylamide gel. Results from the untagged parent strain, BY4741, are shown as a negative control. A faint band ∼55 kDa is sometimes seen in the negative control, consistent with published observations (Granneman et al. 2009). (B) TapeStation-generated gel image of cDNA libraries. The expected length of cDNA libraries derived from 15- to 30-nucleotide RNA fragments is 130–145 bp. For reference the 25- and 1500-bp markers are shown in all lanes. The negative control consistently yielded no detectable cDNA. (C) Graphical view of Rlp7 CRAC sites on 5.8S and 25S rRNAs and ITS1, ITS2, and 3′ETS. The number of hits to each site is indicated on the y-axis, and the frequency is indicated above (percentage of time site was observed).
For Rlp7, 59 clones containing a cDNA insert were sequenced. We identified crosslinked RNAs that localized to the 3′ end of 5.8S rRNA or the 5′ end of 25S rRNA, as well as adjacent sequences in ITS2 (80% of total hits) (Fig. 2C; see Fig. 4B, below; Supplemental Fig. S1). We also identified a less common sequence in ITS2 (5% of hits) and background sequences in 18S and 25S rRNAs that were observed one or two times (Fig. 2C; Supplemental Fig. S2). Sites where Rlp7 crosslinked to RNA could be identified as skipped or changed nucleotides in the sequencing reaction (see Fig. 4B, yellow; Supplemental Fig. S1, red). Nop7 CRAC was carried out as a positive control. Consistent with published observations, we identified a site in domain III of 25S rRNA that crosslinked to Nop7 (see Fig. 4C; Supplemental Fig. S2; Granneman et al. 2011). For the negative control, we sequenced 33 clones. Of these clones, only three contained a cDNA insert (9%). These cDNAs mapped to different sites than Rlp7 cDNA on 25S rRNA and did not contain crosslinked nucleotides.
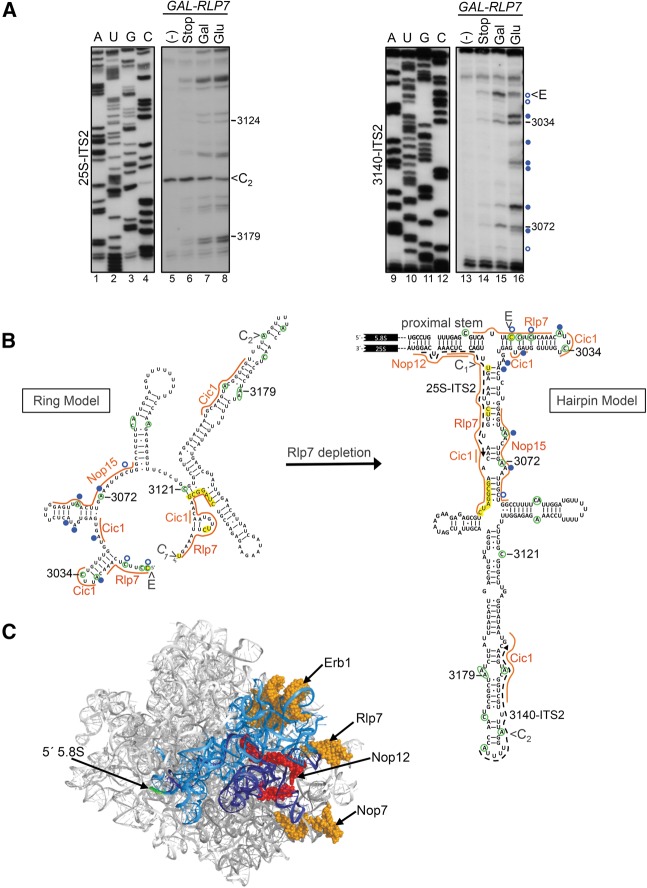
FIGURE 4.
The accessibility of nucleotides in ITS2 to DMS chemical modification is altered after Rlp7 depletion. (A) In vivo DMS probing of the GAL-RLP7 strain grown in galactose (Gal)-containing or glucose (Glu)-containing medium. Controls include no DMS control (−) and stop control (stop), in which β-mercaptoethanol was added to quench the reaction before addition of DMS. The corresponding sequencing ladders are shown (A, U, G, C). The 25S-ITS2 (lanes 1–8) and 3140-ITS2 (lanes 9–16) oligonucleotides were used for primer extension of extracted RNA. Nucleotides that become more modified in the absence of Rlp7 are indicated by closed blue circles, and nucleotides that became less modified are indicated by open blue circles. Nucleotide positions are designated for 35S pre-rRNA. The same RNA samples and loading were used for both sets of primer extensions. (B) Locations of nucleotides that are more (closed blue circles) or less (open blue circles) modified in the absence of Rlp7 are displayed on the predicted ring (Joseph et al. 1999) or hairpin (Yeh and Lee 1990) structure of ITS2. ITS2 contains nucleotides from sites E to C1. Nucleotides that are modified in wild-type cells are circled in green. Orange lines represent the binding sites of Rlp7, Nop15, Cic1, and Nop12 determined by CRAC (Granneman et al. 2011). Crosslinked nucleotides are highlighted in yellow. On the hairpin structure, locations of primers used for primer extension are shown as dashed arrows (25S-ITS2, 3140-ITS2), and the location of the proximal stem is indicated. (C) PyMOL representation of the solvent accessible surface of the yeast 60S rRNA (PDB file 3U5H) (Ben-Shem et al. 2011). Domain I is blue (5.8S rRNA dark blue, 25S rRNA light blue), the 5′ end of 5.8S rRNA is green, and protein binding sites are shown in space-fill view. The binding site of Nop12 is red, and the binding sites of Erb1, Rlp7, and Nop7 are orange.
The structure of the ITS2-proximal stem is essential for ribosome assembly (Peculis and Greer 1998). However, the sequence of the helix and 3′ and 5′ terminal nucleotides of 5.8S and 25S rRNA, respectively, can be changed with little effect on cell growth or 25S rRNA levels (Peculis and Greer 1998; Cote and Peculis 2001). This suggests that Rlp7 recognizes secondary or tertiary structure elements in the pre-rRNA rather than primary sequence. Because we identified both strands of the ITS2-proximal stem in our assays, we predict that the proximal stem forms upon or before Rlp7 binding.
The Rlp7 binding site places it in a close proximity to ITS2, supporting a function in 27S pre-rRNA processing. Because Rlp7 exhibits significant amino acid sequence homology to r-protein L7, it was previously hypothesized that Rlp7 might bind to the L7 binding site to function as a “placeholder” for L7 in preribosomes (Gadal et al. 2002). However, our data are not consistent with this hypothesis, since L7 binds to domain II of 25S rRNA in mature ribosomes. Furthermore, we previously found that Rlp7 and L7 both associate with 35S and 27S pre-rRNAs (Sahasranaman et al. 2011; Jakovljevic et al. 2012), suggesting that L7 binds to preribosomes before Rlp7 dissociates from them. In addition, we and others have shown that Rlp7 and L7 coimmunoprecipitate and thus are likely to be associated with the same preribosomes (Babiano et al. 2013; J Jakovljevic, unpubl.). Rather, it is more likely that Rlp7 evolved from L7 to use a similar RNA recognition motif to bind and function at a different RNA element during ribosome assembly.
The binding site of Rlp7 is within eukaryotic-specific RNA elements, including ribosomal expansion segment 4 of the ITS2-proximal stem and ITS2. These two segments are not conserved in bacteria, suggesting that they evolved to facilitate eukaryotic-specific functions. It is clear from this and other studies that at least one role of these eukaryotic-specific RNA elements is to provide a binding site for assembly factors (Granneman et al. 2011). It is therefore possible that Rlp7 coevolved with the proximal stem and ITS2 to carry out functions unique to eukaryotic ribosome assembly.
Rlp7 interacts with proteins that bind within and near ITS2 during 60S ribosome assembly
To verify the localization of Rlp7 within preribosomes, we carried out a two-hybrid assay to identify proteins that interact with Rlp7. We tested for potential interactions between the three A3 factors that bind to ITS2 (Rlp7, Nop15, and Cic1) and other assembly factors and r-proteins implicated in 27SA2 (Brx1, Ebp2), 27SA3 (Pwp1, Nop7, Ytm1, Erb1, Rrp1, Has1, Rrp17, Rat1, Rai1, L7, L8), or 27SB (L17, L26, L35, L37, Nsa2, Rrs1, Rpf2, Nog2, Nop2, Nip7, L11, Spb4, Dbp10, Tif6, Rlp24, Mak11, Nog1) pre-rRNA processing, as well as other r-proteins that contact domain I (L15, L25) (Henry et al. 1994; Emery et al. 2004; Zhang et al. 2004; Oeffinger et al. 2009; Ben-Shem et al. 2011; Granneman et al. 2011; Sahasranaman et al. 2011; Babiano et al. 2012; Jakovljevic et al. 2012; Shimoji et al. 2012; Talkish et al. 2012; Gamalinda et al. 2013). Only 11 of the 210 potential pair-wise two-hybrid interactions tested were positive, confirming the specificity of the assay (Fig. 3A; Supplemental Table S1).
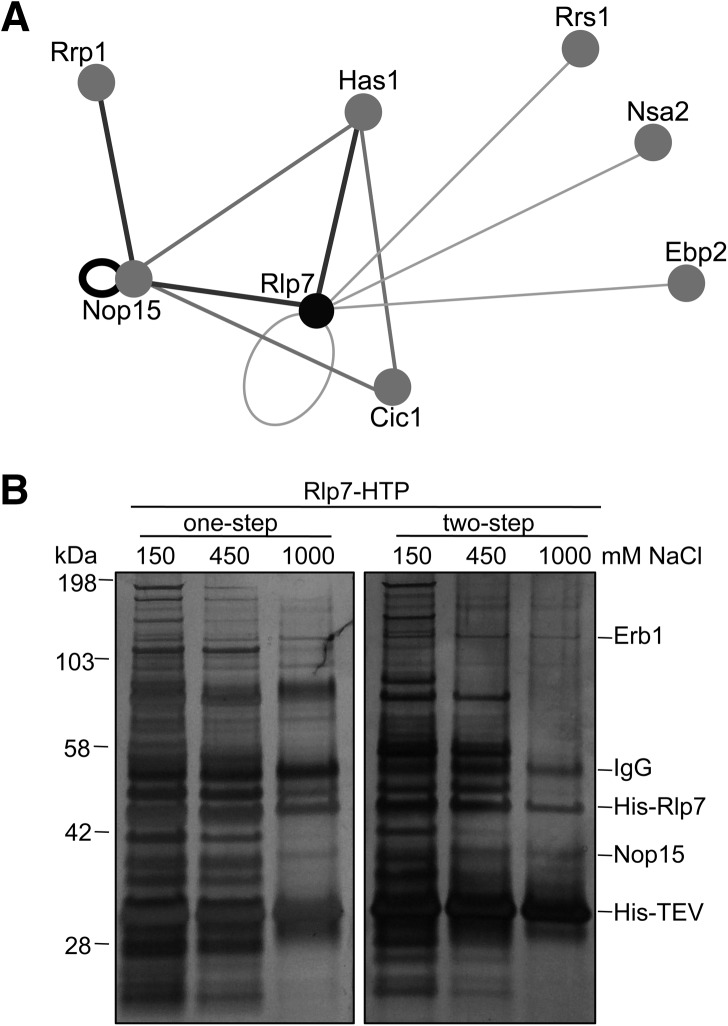
FIGURE 3.
Rlp7 interacts with a neighborhood of proteins that bind to or near ITS2 and the proximal stem. (A) Two-hybrid interaction map. Two-hybrid assays were carried out to test for interactions between the ITS2 neighborhood of proteins (Rlp7, Nop15, Cic1) and several other factors required for 27S pre-rRNA processing. Relative strengths of interactions are shown with dark gray, thick lines representing strong interactions and light gray, thin lines representing weak interactions as determined by growth of two-hybrid strains on media containing increasing concentrations (1, 2.5, 5, and 50 mM) of 3-aminotriazole (3-AT). (B) High-salt wash of Rlp7-associated proteins. Rlp7-HTP was affinity purified from yeast cell lysates, and complexes were washed with increasing concentrations of NaCl (150, 450, 1000 mM) to remove weakly associated proteins. One-step indicates that proteins were resolved after one-step purification on IgG-coated beads, and two-step indicates that proteins were resolved after two-step purification on IgG- and nickel-coated beads.
Consistent with the Rlp7 binding site in ITS2 and the proximal stem, our two-hybrid results show that Rlp7 interacts with proteins that bind to ITS2 or adjacent to ITS2 (Zhang et al. 2004; Granneman et al. 2011). Rlp7 interacted with Nop15, which also interacted with Cic1, consistent with Rlp7, Nop15, and Cic1 all contacting ITS2 during ribosome assembly. Rlp7 and Nop15 were positive for interactions with themselves and therefore may have the potential to form dimers on or off preribosomes. Rlp7, Nop15, and Cic1 all tested positive for interactions with the RNA helicase Has1. These three proteins are required for Has1 to enter pre-60S ribosomes (Sahasranaman et al. 2011) and may therefore recruit Has1. Has1 helps to stabilize the native conformation of domain I during ribosome assembly and is required for 27SA3 and 27SB pre-rRNA processing (Dembowski et al. 2013). We also observed interactions between Rlp7 and Ebp2. Ebp2 most likely binds near the ITS2-proximal stem since it copurifies with Brx1, Pwp1, Nop12, L8, and L15 in a subcomplex (Zhang et al. 2004). R-proteins L8 and L15 contact the proximal stem (Ben-Shem et al. 2011), and Nop12 contacts the proximal stem and nearby sequences in 5.8S rRNA (Granneman et al. 2011). In support of two-hybrid data, interactions between Rlp7 and Ebp2, as well as Cic1 and Nop15, have previously been identified by PCA (protein-fragment complementation assay) (Tarassov et al. 2008). We observed relatively weak two-hybrid interactions between Rlp7 and Rrs1 and Nsa2. These factors were previously implicated in 27SB pre-rRNA processing and may therefore bind near ITS2. However, the binding sites of Rrs1 and Nsa2 in preribosomes have yet to be identified.
As an orthogonal approach to identify factors that interact with Rlp7, we affinity purified Rlp7-associated preribosomes from yeast cell lysates and identified proteins by mass spectrometry that remained associated with Rlp7 after washing with high salt (Fig. 3B; Supplemental Table S2). We identified Nop15 and Erb1 by this assay. The observed interaction of Rlp7 with Nop15 is consistent with the two-hybrid data. Likewise, copurification of Erb1 with Rlp7 makes sense because Erb1 crosslinks to 25S rRNA of domain I (Granneman et al. 2011), which is <30 Å away from the Rlp7 CRAC site on the mature 60S ribosomal subunit (Fig. 4C). Whether copurification of Erb1 and Nop15 with Rlp7 reflects RNA- or protein-mediated interactions remains to be tested.
Taken together, these protein interaction data support the identified binding site of Rlp7 at ITS2 and the ITS2-proximal stem of domain I near assembly factors Cic1, Nop15, Nop12, and Erb1 (Fig. 4B,C). These proteins cluster near the 3′ end of 5.8S rRNA, yet they trigger processing of ITS1 to generate the 5′ end of 5.8S rRNA. Thus, a remaining question is why do these A3 factors bind at a distance from the A3 processing site? Perhaps they recruit the RNA helicase Has1 to preribosomes, which may couple domain I folding to 27SA3 pre-rRNA processing (Dembowski et al. 2013). Additional possibilities are that they directly chaperone domain I folding or regulate conformational changes within ITS2 that are propagated from the 3′ to 5′ end of 5.8S rRNA.
Rlp7 maintains an open ring conformation of ITS2
ITS2 has been proposed to form two different structures referred to as the closed “hairpin” and open “ring” conformations (Fig. 4B; Yeh and Lee 1990; Joseph et al. 1999). These two conformations differ in the regions closest to the ITS2-proximal stem. The ability to form both conformations is required for efficient 60S subunit assembly (Cote et al. 2002). In order to determine whether Rlp7 binding to the proximal stem and to ITS2 affects folding of ITS2, we carried out in vivo chemical probing of RNA using dimethyl sulfate (DMS) in the presence and absence of Rlp7. DMS modifies adenines and cytosines that are not involved in Watson-Crick base-pairing or that are not protected by proteins in a manner that causes pausing of reverse transcription. Modified RNAs were extracted and reverse transcribed using primers that bind within ITS2 to assay for changes in ITS2 and 5.8S rRNA.
In wild-type cells, the pattern of chemical modification within ITS2 is consistent with both the ring or hairpin structure of ITS2, suggesting that a mixed population of both structures exist in wild-type cells (Fig. 4A, lanes 7,15, B, green circles). For example, three cytosines adjacent to the E site were modified supporting the presence of the ring structure, while C3121 was modified supporting the presence of the hairpin structure. After Rlp7 depletion, three nucleotides became less modified (Fig. 4A,B, open blue circles; Supplemental Fig. S3), consistent with a shift from the ring to hairpin conformation. Nucleotide A3031 became more modified, which could indicate a shift to the hairpin structure or the loss of binding by Cic1. Because Rlp7 binds to the region of ITS2 that is most different between the two structural models, Rlp7 is a likely candidate to regulate transitions between the ring and hairpin conformations.
Because Cic1, Rlp7, and Nop15 are interdependent for their association with preribosomes (Sahasranaman et al. 2011), we also expected to observe increased modification of the binding sites of Cic1 and Nop15 in the absence of Rlp7. Six nucleotides become more modified in the absence of Rlp7 in the regions of the Cic1 and Nop15 binding sites (Fig. 4A,B, closed blue circles; Supplemental Fig. S3). We did not observe increased modification of the binding site of Rlp7 extending from the 3′ end of 5.8S rRNA through the 5′ end of ITS2. Because this site was not observed as frequently as the ITS2/25S rRNA CRAC site (Fig. 2C), Rlp7 may make fewer direct contacts with this region or may bind in a geometry that does not cause protection from chemical modification. Taken together, our data are consistent with Rlp7 being interdependent with Cic1 and Nop15 for binding to ITS2 and support a model in which Rlp7 helps to maintain an open ring conformation of ITS2.
Because the proximal stem does not become more accessible to chemical modification after depletion of Rlp7 and A3 factors, Cic1, Nop15, Nop7, and Has1, its structure is likely maintained without these proteins (Fig. 4A,B; Supplemental Fig. S4). This is consistent with CRAC data because both strands of the proximal stem were found crosslinked to Rlp7 (Fig. 2C). Therefore, we suspect that the proximal stem forms before the A3 factors bind to the pre-rRNA. Because these proteins associate with the earliest pre-rRNAs, 35S or 27SA2 (Granneman et al. 2011; Sahasranaman et al. 2011), it raises the possibility that the proximal stem forms cotranscriptionally. This would enable the proximal stem, as well as ITS2, to hold 5.8S and 25S rRNAs in close proximity to chaperone rRNA folding within domain I.
In a previous study, the length of the proximal stem was shown to be important for 60S ribosomal subunit assembly (Cote and Peculis 2001). Lengthening the ITS2-proximal part by 2 bp had no effect on 25S rRNA production. However, lengthening this region by 5 bp prevented processing. One possibility is that the longer insertions inhibit Rlp7 binding to this region of the pre-rRNA. Another possibility is that Rlp7 can still bind but that the critical length reflects a loss in Rlp7 communication across the proximal stem to the rest of domain I and the 5′ end of 5.8S rRNA. If Rlp7 recruits Has1 to preribosomes to trigger rearrangements within domain I, there may be a critical distance for this to occur.
Cote and Peculis (2001) previously predicted that two assembly machineries associate with the proximal stem: the ITS2-distal and ITS2-proximal machineries. These predictions were based on mutations made to sequences and structural elements within and flanking the proximal stem. We and others have determined that Rlp7 and Nop12 bind to ITS2-proximal and ITS2-distal regions of the proximal stem, respectively, supporting these predictions (Granneman et al. 2011).
CONCLUSIONS
A longstanding question in ribosome assembly is why ITS2 separates 5.8S and 25S rRNAs in eukaryotic preribosomes while this sequence is missing from 23S rRNA of bacterial preribosomes. Here we propose that ITS2 forms a platform for the association of eukaryotic-specific assembly factors during ribosome assembly. These assembly factors may directly promote folding of rRNA and binding of r-proteins or recruit helicases such as Has1 to trigger rRNA remodeling.
Our results also suggest that structural transitions in ITS2 near the 3′ end of 5.8S rRNA promote 27SA3 pre-rRNA processing at the 5′ end of 5.8S rRNA. One explanation for this observation is that ITS2 conformational changes might be propagated through the proximal stem to chaperone domain I folding, which is coupled to 27SA3 pre-rRNA processing. Therefore, the 27SA3 pre-rRNA processing step may serve as a checkpoint for proper construction of domain I. Domain I r-proteins L17, L26, L35, and L37 line the polypeptide exit tunnel in mature ribosomes. Therefore, proper formation of this rRNA–r-protein neighborhood is critical for ribosome function.
If ITS2 evolved as a landing point for assembly factors to stabilize domain I and promote 27SA3 pre-rRNA processing, it follows that it would be removed after these two events. Perhaps the role of Rlp7 in the 27SB pre-rRNA processing step is to recruit a yet to be identified nuclease to preribosomes or create ITS2 conformations to initiate cleavage and removal of ITS2 after domain I folding and 27SA3 pre-rRNA processing are complete. Furthermore, because Rlp7 binds to the junctions of ITS2 and the 5′ and 3′ ends of 25S and 5.8S rRNAs, respectively, it might be involved in proper processing of either of these ends.
MATERIALS AND METHODS
Yeast strains
Yeast strains are listed in Supplemental Table S3. Strains conditional for expression from the GAL1 promoter or expressing C-terminally His6-TEV-protein A (HTP)-tagged proteins were generated according to the method described previously (Longtine et al. 1998; Rigaut et al. 1999). Unless otherwise indicated, yeast were grown at 30°C in YEPD (2% dextrose, 2% peptone, 1% yeast extract) or YEPGal (2% galactose, 2% peptone,1% yeast extract) media and were harvested during mid-log phase growth.
Crosslinking and analysis of cDNAs
In vivo UV crosslinking, extract preparation, affinity purification, RNase digestion, and cloning of crosslinked fragments were carried out according to the method described previously (Granneman et al. 2009) with the following modifications: Yeast were UV irradiated at 0.8 J/cm2 instead of 1.6 J/cm2 and 200 μL IgG-conjugated dynal beads (Life Technologies) (Oeffinger et al. 2007) were used for affinity purification of the protein A tag instead of 250 μL IgG Sepharose beads (GE Healthcare). Alignments were visualized with Integrated Genome Viewer (Broad Institute). The Rlp7 CRAC assay was repeated two times, and the sequences at the junctions of 5.8S rRNA and ITS2 and ITS2 and 25S rRNA were reproducibly observed.
Two-hybrid assays
Movable open reading frames (Open Biosystems) were transferred into either the pACTGW-attR or pASGW-attR two-hybrid vector (Nakayama et al. 2002) using the Gateway recombination-based cloning system (Life Technologies). Plasmids were transformed into PJ69-4a (James et al. 1996) or PJ69-4α (Uetz et al. 2000) yeast, which were mated to test for potential protein–protein interactions. Pair-wise interactions were tested for activation of the GAL-HIS3 reporter gene by initial screening for the ability to grow on media lacking histidine in the presence of 1 mM 3-AT according to the method described previously (James et al. 1996). Positive interactions were further tested for the strength of interaction by growth in the presence of increasing concentrations of 3-AT (1–50 mM). Results are summarized in Supplemental Table S1.
Affinity purification and high-salt wash of Rlp7 associated proteins
Whole-cell lysates were prepared from 350 mL cultures of the RLP7-HTP strain (JWY9262) according to the method previously described (Granneman et al. 2009) followed by affinity purification of Rlp7-HTP and associated proteins on 60 μL IgG-conjugated Dynal beads (Life Technologies) for 1 h at 4°C (Oeffinger et al. 2007). After binding, protein complexes were washed three times with TN buffer (50 mM Tris-HCl at pH7.8, 0.1% IGEPAL, 5 mM β-mercaptoethanol) containing 150, 450, or 1000 mM NaCl. Protein complexes were eluted from IgG beads by cleavage with 10U TEV Protease (Life Technologies) for 1 h at room temperature. Eluted proteins were either precipitated with 10% TCA or subjected to a second purification step on 100 μL Ni-NTA Agarose beads (Qiagen) for 2 h at 4°C followed by elution with 250 mM imidazole and precipitation with 10% TCA. All steps were carried out in TN150 buffer (containing 150 mM NaCl) unless otherwise indicated. Proteins were separated on 4%–12% NuPAGE gels (Life Technologies) followed by silver staining according to standard procedures.
For identification of proteins that associate with Rlp7 after high-salt wash, two-step purifications were carried out as described except that cell lysates were obtained from 2-liter cultures followed by purification on 400 μL IgG beads and 1 mL Ni-NTA Agarose beads. Proteins were separated on 4%–12% NuPAGE gels followed by silver staining with the SilverQuest Silver Staining Kit (Life Technologies). Gel slices were destained and dried and sent to Penn State Hershey Core Research Facilities for reduction, alkylation, trypsin digestion, and MALDI-TOF mass spectrometry. Peptides were analyzed by the ProteinPilot 4.0 program, and results are summarized in Supplemental Table S2.
Chemical probing
In vivo structure probing with DMS was carried out according to the method previously described (Babiano et al. 2012) except that Transcriptor Reverse Transcriptase (Roche) was used for primer extensions with oligonucleotides designed to bind to ITS2 (Fig. 4B, dashed lines).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Woolford laboratory for critically reading the manuscript, Michael Gamalinda and Jason Talkish for images of domain I and ITS2, Sander Granneman for suggestions with the CRAC protocol, and the Penn State Hershey Core Proteomics Facility for mass spectrometry and data analysis. This work was supported by grants from the National Institutes of Health to J.L.W. (GM028301) and J.A.D. (GM099225).
REFERENCES
- Adams CC, Jakovljevic J, Roman J, Harnpicharnchai P, Woolford JL Jr. 2002. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA 8: 150–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiano R, Gamalinda M, Woolford JL Jr, de la Cruz J. 2012. Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function. Mol Cell Biol 32: 3228–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiano R, Badis G, Saveanu C, Namane A, Doyen A, Díaz-Quintana A, Jacquier A, Fromont-Racine M, de la Cruz J. 2013. Yeast ribosomal protein L7 and its homologue Rlp7 are simultaneously present at distinct sites on pre-60S ribosomal particles. Nucleic Acids Res 10.1093/nar/gkt726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. 2009. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell 36: 583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradatsch B, Leidig C, Granneman S, Gnadig M, Tollervey D, Bottcher B, Beckmann R, Hurt E. 2012. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol 19: 1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote CA, Peculis BA. 2001. Role of the ITS2-proximal stem and evidence for indirect recognition of processing sites in pre-rRNA processing in yeast. Nucleic Acids Res 29: 2106–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote CA, Greer CL, Peculis BA. 2002. Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA 8: 786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembowski JA, Kuo B, Woolford JL Jr. 2013. Has1 regulates consecutive maturation and processing steps for assembly of 60S ribosomal subunits. Nucleic Acids Res 41: 7889–7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar DA, Dragon F, Lee SJ, Baserga SJ. 2000. A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc Natl Acad Sci 97: 13027–13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, de la Cruz J, Rocak S, Deloche O, Linder P. 2004. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol Microbiol 52: 141–158. [DOI] [PubMed] [Google Scholar]
- Fatica A, Oeffinger M, Tollervey D, Bozzoni I. 2003. Cic1p/Nsa3p is required for synthesis and nuclear export of 60S ribosomal subunits. RNA 9: 1431–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Petfalski E, Gleizes PE, Gas N, Tollervey D, Hurt E. 2002. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J Cell Biol 157: 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M, Jakovljevic J, Babiano R, Talkish J, de la Cruz J, Woolford JL Jr. 2013. Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing. Nucleic Acids Res 41: 1965–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Kudla G, Petfalski E, Tollervey D. 2009. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci 106: 9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Swiatkowska A, Tollervey D. 2010. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J 29: 2026–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Tollervey D. 2011. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J 30: 4006–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Montellese C, Ban N. 2012. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol 19: 1228–1233. [DOI] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J 13: 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic J, Ohmayer U, Gamalinda M, Talkish J, Alexander L, Linnemann J, Milkereit P, Woolford JL Jr. 2012. Ribosomal proteins L7 and L8 function in concert with six A3 assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA 18: 1805–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph N, Krauskopf E, Vera MI, Michot B. 1999. Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res 27: 4533–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalo D, Mariotte S, Thuriaux P. 1993. Two distinct yeast proteins are related to the mammalian ribosomal polypeptide L7. Yeast 9: 1085–1091. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford JL Jr. 2005. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol Cell Biol 25: 10419–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta K, Hashimoto T, Otaka E. 1995. The evolutionary relationships between homologs of ribosomal YL8 protein and YL8-like proteins. Curr Genet 28: 19–25. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Kikuno R, Ohara O. 2002. Protein–protein interactions between large proteins: Two-hybrid screening using a functionally classified library composed of long cDNAs. Genome Res 12: 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Tollervey D. 2003. Yeast Nop15p is an RNA-binding protein required for pre-rRNA processing and cytokinesis. EMBO J 22: 6573–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Leung A, Lamond A, Tollervey D. 2002. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA 8: 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP. 2007. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods 4: 951–956. [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Zenklusen D, Ferguson A, Wei KE, El Hage A, Tollervey D, Chait BT, Singer RH, Rout MP. 2009. Rrp17p is a eukaryotic exonuclease required for 5′ end processing of pre-60S ribosomal RNA. Mol Cell 36: 768–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis BA, Greer CL. 1998. The structure of the ITS2-proximal stem is required for pre-rRNA processing in yeast. RNA 4: 1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov DG, Stockelman MG, Strezoska Z, Lau LF. 2001. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res 29: 3621–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mateos M, Garcia-Gomez JJ, Francisco-Velilla R, Remacha M, de la Cruz J, Ballesta JP. 2009. Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res 37: 7519–7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL Jr. 2011. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: Role of factors required for 27S pre-rRNA processing. EMBO J 30: 4020–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerstolpe A, Granneman S, Bjork P, de Lima Alves F, Rappsilber J, Andersson C, Hogbom M, Tollervey D, Wieslander L. 2013. Multiple RNA interactions position Mrd1 at the site of the small subunit pseudoknot within the 90S pre-ribosome. Nucleic Acids Res 41: 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoji K, Jakovljevic J, Tsuchihashi K, Umeki Y, Wan K, Kawasaki S, Talkish J, Woolford JL Jr, Mizuta K. 2012. Ebp2 and Brx1 function cooperatively in 60S ribosomal subunit assembly in Saccharomyces cerevisiae. Nucleic Acids Res 40: 4574–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL III, Karbstein K, Skiniotis G. 2011. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333: 1449–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J, Zhang J, Jakovljevic J, Horsey EW, Woolford JL Jr. 2012. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 41: 1965–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. 2008. An in vivo map of the yeast protein interactome. Science 13: 1465–1470. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Woolls HA, Lamanna AC, Karbstein K. 2011. Roles of Dim2 in ribosome assembly. J Biol Chem 286: 2578–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh LC, Lee JC. 1990. Structural analysis of the internal transcribed spacer 2 of the precursor ribosomal RNA from Saccharomyces cerevisiae. J Mol Biol 211: 699–712. [DOI] [PubMed] [Google Scholar]
- Young CL, Karbstein K. 2011. The roles of S1 RNA-binding domains in Rrp5′s interactions with pre-rRNA. RNA 17: 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young CL, Khoshnevis S, Karbstein K. 2013. Cofactor-dependent specificity of a DEAD-box protein. Proc Natl Acad Sci 110: E2668–E2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Morris QD, Chang R, Shai O, Bakowski MA, Mitsakakis N, Mohammad N, Robinson MD, Zirngibl R, Somogyi E, et al. 2004. The functional landscape of mouse gene expression. J Biol 3: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.