This article ascribes novel functions to the precursor form of the liver-specific miR-122 molecule during hepatitis C virus replication, besides being merely an intermediate in the miR-122 pathway.
Keywords: hepatitis C virus, microRNAs, Dicer, Argonaute 2
Abstract
The hepatitis C viral RNA genome forms a complex with liver-specific microRNA (miR-122) at the extreme 5′ end of the viral RNA. This complex is essential to stabilize the viral RNA in infected cultured cells and in the liver of humans. The abundances of primary and precursor forms of miR-122, but not the abundance of mature miR-122, are regulated in a circadian rhythm in the liver of animals, suggesting a possible independent function of precursor molecules of miR-122 in regulating viral gene expression. Modified precursor molecules of miR-122 were synthesized that were refractory to cleavage by Dicer. These molecules were found to enhance the abundance of HCV RNA. Furthermore, they diminished the expression of mRNAs that contained binding sites for miR-122 in their 3′ noncoding regions. By use of duplex and precursor miR-122 mimetic molecules that carried mutations in the passenger strand of miR-122, the effects on viral and reporter gene expression could be pinpointed to the action of precursor miR-122 molecules. Targeting the circadian expression of precursor miR-122 by specific compounds likely provides novel therapeutic strategies.
INTRODUCTION
MicroRNAs (miRNAs) are 20- to 23-nucleotide (nt) RNA molecules that are processed from RNA polymerase II–transcribed long primary microRNAs (pri-miRNAs) (Bartel 2004, 2009). Briefly, pri-miRNAs are processed in the nucleus by the RNase III enzyme Drosha and its binding partner DGCR8 into ∼70-nt RNA hairpin structures, termed precursor miRNAs (pre-miRNAs) (Lee et al. 2002, 2003; Gregory et al. 2004; Han et al. 2004). Pre-miRNAs are exported to the cytoplasm, where they associate with the RNA-induced silencing complex (RISC) loading complex (Yi et al. 2003; Gregory et al. 2005; Maniataki and Mourelatos 2005; MacRae et al. 2008). The RISC component Dicer cleaves the pre-miRNA to create a 20- to 23-nt base-pair duplex. One RNA strand of the RNA duplex, termed the “guide strand,” is loaded into the RISC component Argonaute (Ago) (Bernstein et al. 2001; Park et al. 2011). The seed sequence (nucleotides 2–8) of the Ago-associated guide strand base pairs with complementary sequences in the 3′ noncoding region of a target mRNA (Lewis et al. 2003), which leads to inhibition of mRNA translation or degradation of the mRNA (for review, see Bartel 2009). Importantly, microRNA–mRNA interactions, which do not require seed-seed match base-pairing have been recently detected, using an unbiased Ago1–RNA crosslinking protocol (Helwak et al. 2013).
Accumulating evidence suggests that pre-miRNAs can also function in regulation of mRNA expression. In 2008, Liu et al. (2008) showed that mutations in the loop nucleotides in pre-miR-181 affected the activity of the miRNA in T-cell development by a mechanism that was independent of the processing to mature miRNA. Furthermore, unprocessed pre-miRNA–Ago2 complexes can cleave their target mRNAs in vitro in the absence of Dicer (Tan et al. 2009; Yoda et al. 2010). Recently, nucleotide changes in pre-miRNAs have been correlated with disease outcome. For example, a single nucleotide polymorphism in pre-miR-196a2 correlates with susceptibility to multiple cancers (Hu et al. 2008; Qi et al. 2010; Akkiz et al. 2011), and a mutation in the 3′ arm of the precursor of miR-96 correlates with deafness in humans (Solda et al. 2012). Therefore, it is of great interest to examine potential unique roles of precursor forms of miRNAs in gene regulation.
A hint that precursor forms of liver-specific miR-122 may have a regulatory role was provided by the observation that the abundances of pri-miRs and pre-miRs of miR-122, but not of mature miR-122, are regulated in a circadian rhythm (Gatfield et al. 2009). Specifically, the abundances of pri-miR-122 and pre-miR-122 are increased during the dark cycle in mouse liver (Gatfield et al. 2009). MiR-122 regulates cholesterol biosynthesis, and it is known that cholesterol abundance in the liver varies in a circadian manner (Edwards et al. 1972). Thus, we were curious as to whether precursor molecules of miR-122 regulate the expression of mRNAs that have binding sites for miR-122.
Hepatitis C virus (HCV) is among the RNAs that are targeted by miR-122. MiR-122 forms an oligomeric complex with 5′ terminal sequences of the RNA genome of HCV, thereby greatly enhancing viral RNA abundance (Jopling et al. 2005, 2008; Jangra et al. 2010; Norman and Sarnow 2010; Machlin et al. 2011; Shimakami et al. 2012a,b). This is an unusual example of an RNA whose function is enhanced by binding of microRNAs. HCV is a positive-stranded RNA virus that causes global health problems, with an estimated 170 million people infected worldwide (Hoofnagle 2002; Moradpour et al. 2007). In this study, we find that, like mature miR-122, pre-miR-122 both enhances HCV RNA abundance and diminishes the abundance of cellular mRNAs that contain binding sites for miR-122 in their 3′ noncoding regions. Therefore, precursor miR-122, like mature miR-122 (Janssen et al. 2013), is an excellent therapeutic target, whose increased size provides additional sites for antiviral intervention.
RESULTS
Processing and activities of synthetic pre-miR-122 in cultured cells
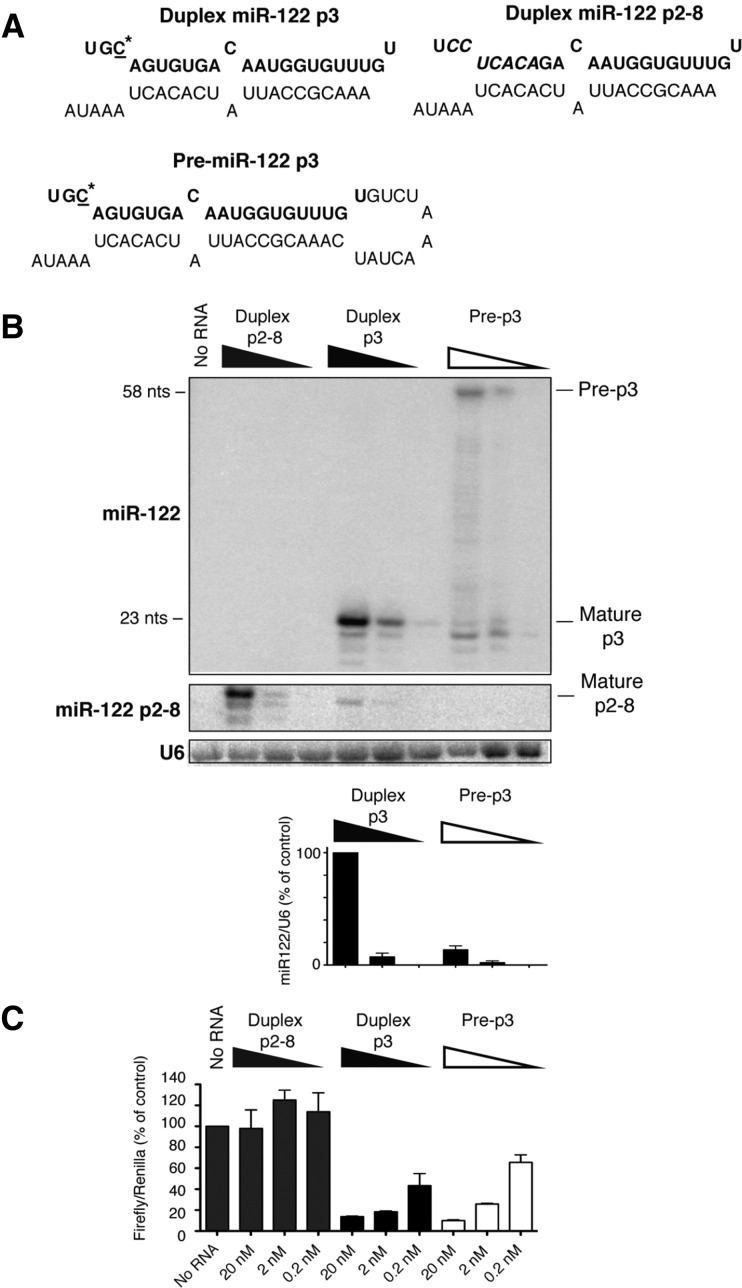
To test whether pre-miR-122 has a role in the regulation of gene expression, we measured the stability and function of transfected duplex miR-122 or pre-miR-122 RNA molecules (Fig. 1A). In HeLa cells that do not express endogenous miR-122 (Jopling et al. 2005), 22- to 23-nt miR-122s accumulated after transfection with duplex and precursor miR-122 RNAs, containing p3 seed-mutations (Fig. 1B). The 22-nt miR-122 isoform likely lacks the penultimate 3′ nucleotide. This led to a dose-dependent repression of luciferase expression in p3-miR-122 target site-containing luciferase reporter mRNAs (Fig. 1C). In cells transfected with pre-miR-122 RNAs, mature miR-122 was also produced, but ∼25% of precursor miR-122 remained unprocessed (Fig. 1B). Surprisingly, the repression of target mRNAs in cells transfected with duplex miR-122 and pre-miR-122 was very similar (Fig. 1C). These findings suggest that mature miR-122 that was processed from pre-miR-122 was sufficient to repress reporter mRNA expression or that full-length precursor miR-122 could directly down-regulate reporter gene expression.
FIGURE 1.
Processing and activities of synthetic miR-122 forms after transfection into HeLa cells. (A) Sequence and predicted structure of duplex p3 and pre-p3. The sequence of mature miR-122 is highlighted in bold. The mutated C-nucleotide at position 3 in miR-122 is underlined and marked with an asterisk. (B) Northern blot to visualize transfected miR-122 RNA species. Duplex p2-8 is a mutated miR-122 duplex with mutations in the seed sequence at positions 2–8. Duplex p3 is a phosphorylated miR-122 duplex with a single mutation at position 3. Pre-p3 is a phosphorylated pre-miR-122 with the same single mutation. Visualization of miR-122 p2-8 was accomplished using a miR-122 p2-8–specific hybridization probe (middle panel). Accumulation of 20- to 23-nt miR-122 bands was normalized to U6 snRNA, and abundance was measured using ImageQuant. (C) Luciferase-based microRNA assays. miR-122 species were transfected into HeLa cells concurrently with a DNA plasmid that expressed firefly luciferase mRNA, containing a miR-122 binding site, and a plasmid that expressed Renilla luciferase. Lysates were prepared and luciferase activities measured. Data from mock-transfected cells (no RNA) were set to 100. Data are representative of at least three independent replicates; error bars, SEM.
To test whether these effects involved base-pair interactions between the seed sequences in miR-122 and seed match sequences in target mRNAs, duplex p2-8 was synthesized. MiR-122 p2-8 molecules contain seven consecutive mutations in the entire seed sequence in miR-122 (positions 2–8). Transfected p2-8 accumulated in cells (Fig. 1B) but did not affect the expression of reporter mRNAs containing wild-type miR-122 seed match sequences (Fig. 1C). However, duplex p2-8 and pre-p2-8 were both functional in siRNA-based assays that measured the cleavage of p2-8 target site-containing green fluorescent protein (GFP) mRNAs (Supplemental Fig. S1; Machlin et al. 2011), arguing that the activities of duplex and pre-miR-122 in the RNAi pathway depended on canonical, seed–seed match interactions (Bartel 2009).
Stability and activity of Dicer-resistant pre-miR-122 molecules containing deoxynucleotides
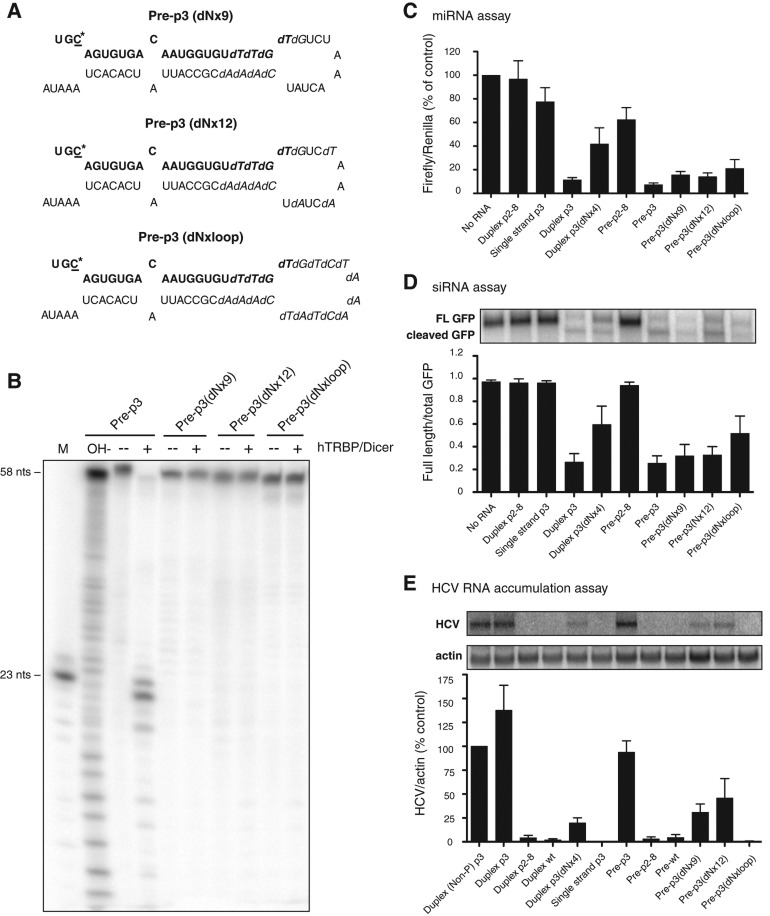
To examine whether pre-miR-122 molecules were active in miRNA, siRNA, and HCV RNA accumulation assays (see below), pre-miR-122s were synthesized that were predicted to be refractory to Dicer-mediated cleavage. Specifically, pre-miR-122 molecules were generated that contained substituted deoxynucleotides around the Dicer cleavage sites. Three versions of pre-miR-122 were generated: one with nine deoxynucleotides (pre-p3 dNx9), one with 12 (pre-p3 dNx12), and one in which the entire loop was composed of deoxynucleotides (pre-p3 dNxloop) (Fig. 2A). In vitro cleavage assays of radiolabeled pre-p3 showed that pre-miR-122 was processed to mature miR-122 in the presence of human Dicer and the Tar RNA binding protein (TRBP) (Fig. 2B). In contrast, all deoxynucleotide-containing pre-miR-122 molecules failed to accumulate detectable amounts of mature miR-122 (Fig. 2B), even after an extended incubation (Supplemental Fig. S2A). For example, ∼99% of precursor remained unprocessed in Pre-p3(dNx12). Some intermediate smaller bands were visible in both Dicer-treated and control-treated samples, indicating that they arose from Dicer-independent degradation (Supplemental Figs. S2A,B, S3).
FIGURE 2.
Stabilities and activities of precursor miR-122s containing deoxynucleotides. (A) Sequences and predicted structures of pre-p3 with nine (dNx9), 12 (dNx12), and 19 (dNxloop) deoxynucleotides. The sequence of mature miR-122 is highlighted in bold. The mutated C-nucleotide at position 3 in miR-122 is underlined and marked with an asterisk. Substituted deoxynucleotides are highlighted in italics. (B) hDicer/TRBP cleavage assays of deoxynucleotide-containing precursors. Precursors were labeled at their 5′ ends with 32P and incubated in the presence or absence of purified hDicer/TRBP. Products were separated by gel electrophoresis and exposed to X-ray film. A representative autoradiograph is shown. (C) Luciferase-based microRNA assays. Experiments were performed as in Figure 1C. Data are representative of at least three independent replicates; error bars, SEM. (D) GFP-based siRNA cleavage assays. Various miR-122 species were transfected into cells concurrently with a GFP-expressing plasmid that contains a perfectly complementary site to the mutant miR-122 molecules in its 3′ noncoding region. GFP mRNA was measured by Northern blot analysis. The ratio of full-length GFP mRNA to total (full-length plus cleaved) GFP mRNA was determined, and data from at least three independent replicates are shown. Error bars, SEM. (E) HCV RNA accumulation assays. Rescue of mutated HCV RNA by miR-122 species, carrying compensatory mutations, after transfection into Huh-7 cells. RNA accumulation was measured by Northern blot analysis. Autoradiographs were quantitated using ImageQuant, and HCV RNA amounts were normalized to that of γ-actin RNA. Data from cells transfected with p3 were set to 100% and used as a positive control. The data shown represent at least three independent replicates. Error bars, SEM.
Next, the deoxynucleotide-modified pre-miR-122 molecules were tested in miRNA (Fig. 2C), siRNA (Fig. 2D), and HCV RNA accumulation-based assays (Fig. 2E) to measure their functions as miRNAs, siRNAs, and HCV RNA stabilizers. The HCV RNA accumulation assay measures the rescue of a mutated HCV RNA (nucleotide 27 G-to-C change) by ectopic expression of miR-122 p3 (C-to-G change), which enables the interaction of mutated miR-122 with the mutated HCV genome in liver cells in the presence of endogenous miR-122 (Machlin et al. 2011).
Both duplex p2-8, which carries a mutated seed sequence motif, and single-stranded miR-122 p3 were inactive in all three assays (Fig. 2C–E). Curiously, transfection of precursors reduced translation of the luciferase reporter mRNA to a larger degree than microRNA duplexes, but this had no effect on siRNA and HCV RNA accumulation assays (see below), suggesting that precursor molecules may induce some innate immune response that affects translation. In contrast, duplex p3 functioned efficiently in all three assays. Importantly, effects on HCV RNA abundance were specific to the transfected p3 forms of miR-122 and, thus, functioned independently of endogenous miR-122 molecules, as expression of wild-type miR-122 or wild-type pre-miR-122 failed to enhance HCV RNA accumulation (Fig. 2E). Duplex p3 (dNx4), which contains four deoxynucleotides at positions 20–23 of miR-122, was able to function in all assays, although less efficiently than duplex p3 (Fig. 2C–E). The deoxynucleotide-containing precursor miR-122 molecules functioned as inhibitory miRNAs (Fig. 2C), as siRNAs (Fig. 2D), and as HCV RNA stabilizers (Fig. 2E), although with a slightly lower efficiency than pre-p3. The fully deoxynucleotide-substituted loop pre-miR-122, pre-p3 (dNxloop), functioned in miRNA and siRNA assays but not in the HCV RNA accumulation assay (Fig. 2C–E). It is possible that the additional deoxynucleotides in pre-p3 (dNxloop) perturbed an interaction important for HCV RNA accumulation, as has been suggested for deoxynucleotide-containing miR-122 duplexes (Machlin et al. 2011). Alternatively, it is possible that the Dicer-independent degradation products (Fig. 2B; Supplemental Fig. S3) or small amounts of miR-122 processed from pre-p3 (dNx9) and (dNx12) were responsible for the observed effects. To distinguish between these possibilities, an alternative approach was employed to investigate the contribution of full-length precursors to HCV RNA accumulation.
Target site recognition of bulge-containing duplex miR-122 and pre-miR-122 RNAs
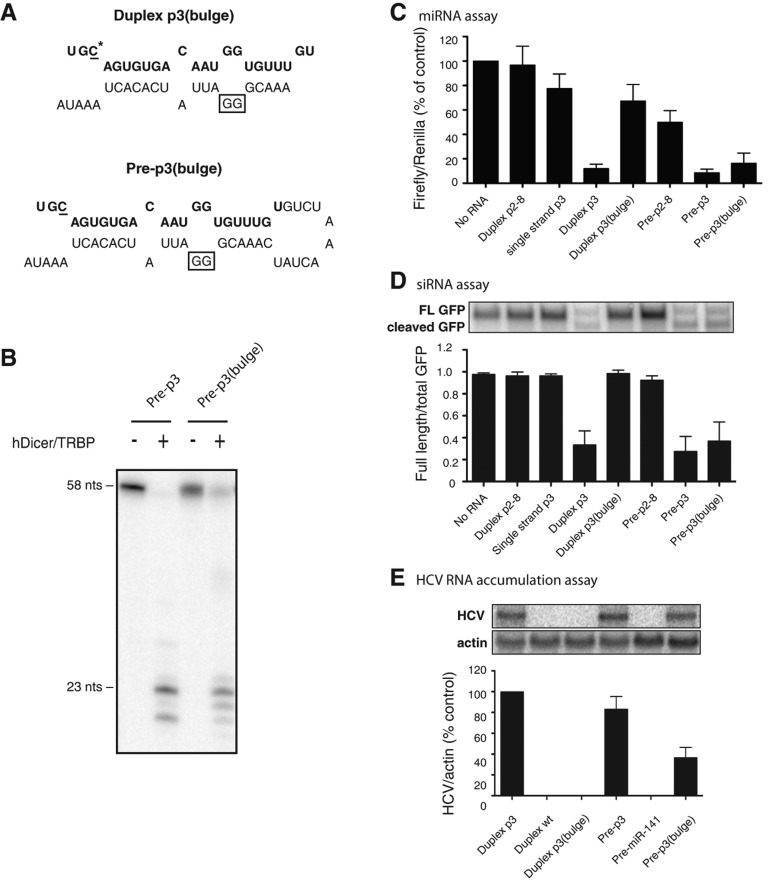
We reasoned that, if pre-miRNAs had independent functions, a sequence might be found that functioned only as a precursor molecule and not as a processed miRNA. One such mutation, p3 bulge, was found in which passenger strand nucleotides opposite of nucleotides 15 and 16 in the guide strand were changed from CC to GG (Fig. 3A). Both pre-p3 and pre-p3 (bulge) were processed in vitro by Dicer/TRBP to yield mature miR-122 molecules that were 20–23 nt in length; however while all of the pre-p3 was processed, ∼75% of pre-p3 (bulge) molecules remained unprocessed under these conditions (Fig. 3B). Duplex p3 (bulge) molecules functioned poorly in miRNA-based assays (Fig. 3C) and were completely nonfunctional in siRNA-based assays (Fig. 3D) and in HCV RNA accumulation assays (Fig. 3E). In contrast, pre-p3 (bulge) molecules functioned in all three assays (Fig. 3C–E). If the function of the pre-p3 (bulge) RNA was the result from the presence of any p3 (bulge) molecules, then duplex p3 (bulge) molecules, which are processed to yield the same miR-122 guide strand, should also have been functional in all three assays. Therefore, there must be a function of the pre-miRNA that is distinct from its processed form. Finally, we examined whether the 3′ end sequences in pre-miR-122 were important for function. Thus, we tested pre-p3 molecules containing either a 28-nt 3′ truncation or where the 28 nt of the 3′ end were exchanged with random sequences that are not predicted to base pair with the 5′ end sequences in pre-miR-122. These RNAs did not function in miRNA, siRNA, or HCV RNA accumulation assays (data not shown), suggesting that duplex structures are required for RISC loading.
FIGURE 3.
Differential activities of miR-122 RNA species containing an additional stem bulge. (A) Sequences and predicted structures of the duplex p3 (bulge) and the pre-p3 (bulge) mimetics. The sequence of mature miR-122 is highlighted in bold. The mutated C-nucleotide at position 3 in miR-122 is underlined and marked with an asterisk. The changed cytosines to guanosine nucleotides are boxed. (B) hDicer/TRBP cleavage assays were performed as described in Figure 2A. Samples were incubated with purified hDicer/TRBP for 1 h. (C) Luciferase-based microRNA assays were done as described in Figure 1C. Data include at least three independent replicates; error bars, SEM. (D) GFP-based cleavage assays were performed as described in Figure 2D. Quantitation is shown as the ratio of full-length GFP mRNA to total (full-length plus cleaved) GFP mRNA and represents at least three independent replicates. Error bars, SEM. (E) HCV RNA accumulation assays. Experiments were performed as in Figure 2E. Autoradiographs were quantitated using ImageQuant, and HCV RNA levels were normalized to the amount of γ-actin. Data from cells transfected with miR-122 p3 were set to 100% and used as a positive control. The data shown represent at least three independent replicates. Error bars, SEM.
A potential advantage for the pre-p3 (bulge) RNA over its “mature” p3 (bulge) RNA might be preferential loading into the RISC complex. To test this possibility, the ability of duplex and pre-miR-122, without and with bulged nucleotides, to form complexes with Ago 2 was examined in coimmunoprecipitation assays (Supplemental Fig. S4). No differences in the ability to form complexes with Ago 2 were observed (Supplemental Fig. S4).
Dependence of duplex and precursor miR-122 activities on the Ago 2 protein
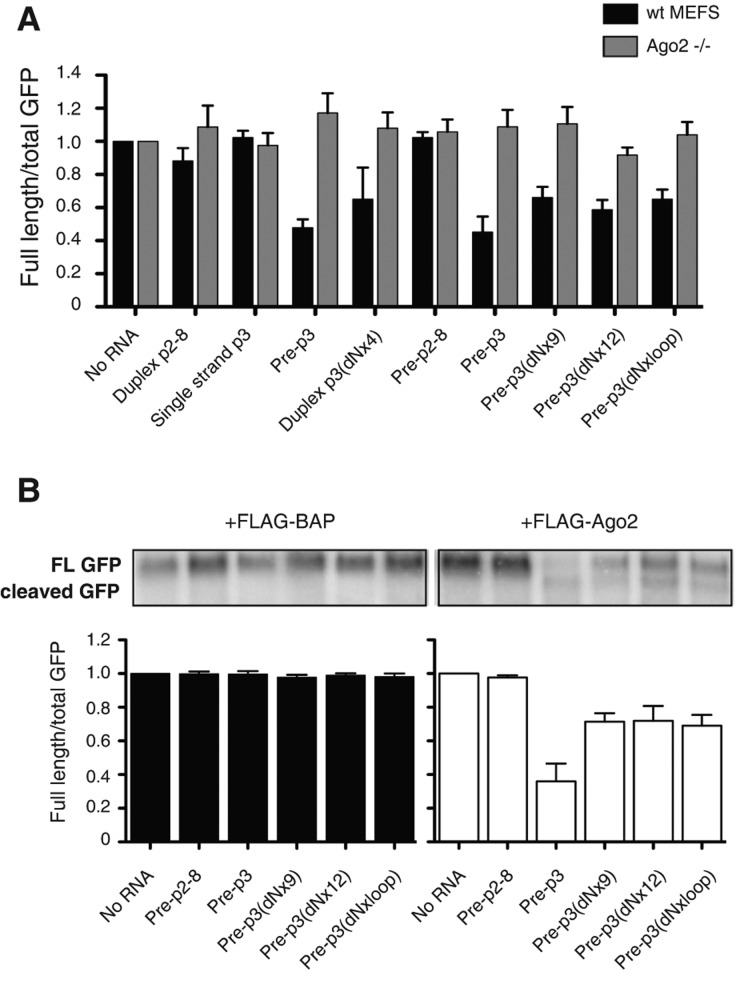
To test whether the function of pre-miR-122 was, like mature miR-122, dependent on Ago 2 protein, RNAs and reporter plasmids were transfected into wild-type and Ago 2−/− MEF cells. Figure 4A shows that the activities of all tested pre-miR-122s, regardless of whether they were susceptible (pre-p3) or resistant (pre-p3 [dNx9], pre-p3 [dNx12], p3 [dNxloop]) to Dicer cleavage, were dependent on Ago2 in siRNA-mediated cleavage assays. In each case, siRNA activities could at least be partially restored in Ago2−/− mouse embryo fibroblasts (MEFs) by the expression of Flag-tagged Ago2 proteins, but not by Flag-tagged BAP proteins (Fig. 4B; Supplemental Fig. S4). These findings argue that like mature miR-122 molecules, pre-miR-122 molecules utilize Ago 2 in target RNA cleavage.
FIGURE 4.
Argonaute (Ago) 2–dependent siRNA activities of precursor miR-122 molecules in wild-type mouse embryo fibroblasts (MEFs) and Ago2−/− MEFs. (A) GFP-based siRNA assays in the presence of pre-miR-122 molecules, as indicated. Experiments were done as described in Figure 2D. Quantitation is shown as the ratio of full-length GFP mRNA to total (full-length plus cleaved) GFP mRNA and represents at least three independent replicates. Error bars, SEM. (B) Rescue of siRNA activities by transfection of plasmids encoding chimeric Flag-Ago2 proteins. Experiments were performed as in A but with the addition of plasmids encoding either Flag-tagged Ago2 or Flag-tagged BAP proteins. Quantitation is shown as the ratio of full-length GFP mRNA to total (full-length plus cleaved) GFP mRNA and represents at least three independent replicates. Error bars, SEM.
DISCUSSION
Our findings show that pre-miR-122 molecules are able to function in siRNA, miRNA, and HCV RNA accumulation assays in cultured cells. We studied the activities of pre-miR-122 molecules in which ribonucleotides surrounding the predicted Dicer cleavage sites were replaced with deoxynucleotides, thus preventing processing by Dicer. The precursors that contained nine, 12, and 19 (dNxloop) deoxynucleotides still functioned in miRNA- and siRNA-based assays despite being resistant to processing by Dicer (Fig. 2; Supplemental Figs. S2, S3). These findings further the vitro observation that Ago2-associated pre-let-7 can cleave target mRNAs in vitro in the absence of Dicer (Tan et al. 2009; Yoda et al. 2010).
MiR-122 is essential to augment HCV RNA accumulation in infected cells (Jopling et al. 2005) and in the human liver (Janssen et al. 2013). Precursor miR-122 and Dicer-resistant pre-miR-122 was able to function in HCV RNA accumulation, despite undetectable Dicer processing (Fig. 2). However, pre-miR-122 molecules, in which the ribonucleotides in the loop were replaced with deoxynucleotides (dNxloop), were not able to function in the HCV RNA accumulation assay (Fig. 2).
Precursor miR-122 RNAs can function even when “mature” miR-122 RNAs cannot. Serendipitously, duplex miR-122 and pre-miR-122 molecules that contained two nucleotide changes in the miR-122 passenger strand illustrated this point. The introduction of the nucleotide changes did not change the primary sequence of the mature miR-122 (Fig. 3A). Surprisingly, while the duplex miR-122 bulge was completely nonfunctional, the pre-miR-122 (bulge) was functional in miRNA, siRNA, and HCV RNA accumulation assays (Fig. 3C–E). Because small amounts of mature miR-122 processed from pre-miR-122 (bulge) should have produced a miR-122 molecule that is the same as the mature miR-122, it is most likely that full-length pre-miR-122 (bulge) enhanced HCV RNA accumulation. Finally, we discovered that all pre-miR-122 molecules, like mature miR-122, require Ago2 to function in siRNA assays (Fig. 4). Whether pre-miR-122-loaded Ago2 is stably associated during HCV RNA accumulation remains to be investigated.
Overall, our data argue that full-length miR-122 precursors are able to function in canonical RNAi pathways and in enhancing HCV RNA accumulation. Recently, Zhang et al. (2012) reporter that Dicer-depletion decreased HCV RNA accumulation and that add-back of duplex miR-122 molecules restored HCV RNA abundance. While this finding seems to indicate that pre-miR-122 molecules are not sufficient to maintain HCV RNA abundance, it should be noted that the Huh-7 liver cells that are routinely used to study HCV gene expression do not express detectable amounts of pre-miR-122. Thus, studies on the functions of pre-miR-122 on viral and cellular gene expression rely on the ectopic expression of pre-miR-122, at least in Huh-7 cells. However, pre-miR-122 is abundantly expressed in mouse liver and, interestingly, in a circadian manner (Gatfield et al. 2009). This observation suggests that pre-miR-122 may control HCV expression in response to changes in light or feed in the liver of animals. While endogenous pre-miR-122 is undetectable in Huh-7 cells, the steady-state ratio of miR-122/pre-miR-122 is 400:1 (Gatfield et al. 2009). Consequently, there are about 150–200 pre-miR-122 molecules per liver cell (Chang et al. 2004). Unless they are dedicated to other distinct unknown functions, they could regulate the expression of HCV in the persistently infected liver. Thus, targeting the precursor form of miR-122 at particular times may offer a novel antiviral therapeutic approach.
MATERIALS AND METHODS
Oligonucleotides and DNA constructs
RNA oligonucleotides were synthesized by the Stanford PAN facility and Dharmacon. miRNAs ordered from PAN were as follows: p2-8 (UAAUCACAGACAAUGGUGUUUGU), p3 (UGCAGUGUGACAAUGGUGUUUGU), p3 (dNx4) (UGCAGUGUGACAAUGGUGUdTdTdGdT), pre-p3 (UGCAGUGUGACAAUGGUGUUUGUGUCUAAACUAUCAAACGCCAUUAUCACACUAAAUA), pre-p3 (bulge) (UGCAGUGUGACAAUGGUGUUUGUGUCUAAACUAUCAAACGGGAUUAUCACACUAAAUA). All RNAs ordered from Dharmacon contained a 5′ phosphate and were polyacrylamide gel electrophoresis (PAGE) purified. RNAs ordered from Dharmacon were as follows: p3 (5′P-UGCAGUGUGACAAUGGUGUUUGU), pre-p3 (5′P-UGCAGUGUGACAAUGGUGUUUGUGUCUAAACUAUCAAACGCCAUUAUCACACUAAAUA), pre-p3 (dNx9) (5′P- UGCAGUGUGACAAUGGUGUdTdTdGdTdGUCUAAACUAUdCdAdAdACGCCAUUAUCACACUAAAUA), pre-p3 (dNx12) (5′P-UGCAGUGUGACAAUGGUGUdTdTdGdTdGUCdTAAdACUdAUdCdAdAdACGCCAUUAUCACACUAAAUA), and pre-p3 (dNxloop) (5′P-UGCAGUGUGACAAUGGUGUdTdTdGdTdGdTdCdTdAdAdAdCdTdAdTdCdAdAdACGCCAUUAUCACACUAAAUA). All passenger strands were ordered from PAN. p2-8 was duplexed with p2-8 passenger (AAACGCCAUUAUCUGUGAGGAUA). p3 and p3 (dNx4) were duplexed to the wild-type passenger strand (AAACGCCAUUAUCACACUAAAUA). p3 (bulge) was produced by duplexing p3 with p3 (bulge) passenger (AAACGGGAUUAUCACACUAAAUA).
Plasmids pLuc-122 × 2p3, eGFP-122- G3, and pH77ΔE1/p7 HCV site 1 (G27) (here referred to as HCV S1:p3) have been previously described (Jopling et al. 2005; Machlin et al. 2011). The pFlag-Ago2 vector was a generous gift from Mark Kay's laboratory at Stanford University (available from Addgene) (Meister et al. 2004). p3xFlag-CMV™-7-BAP control vector (referred to in text as Flag-BAP) was ordered from Sigma-Aldrich.
Cell culture and transfection
Huh-7 cells were cultured in DMEM supplemented with 10% FBS, 1% nonessential amino acids, and 200 μM L-glutamate (Life Technologies), and HeLa cells were cultured in DMEM supplemented with 10% FBS and 200 μM L-glutamate. Wild-type MEFs and Ago2−/− MEFs were a generous gift from Mark Kay's laboratory at Stanford University (Liu et al. 2004) and were cultured in DMEM supplemented with 10% FBS, 1% nonessential amino acids, and 200 μM L-glutamate.
For luciferase-based microRNA assays, DNA constructs and oligonucleotides were transfected into HeLa, wild-type MEF, and Ago2−/− MEF cells in six-well plates using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Of course, it should be noted that the stability of Lipofectamine-delivered oligonucleotides is likely to be different from the stabilities of endogenously expressed small RNAs. Firefly luciferase (pLuc-122 × 2p3) and Renilla luciferase (pRL-SV40) plasmids were transfected at 1 μg and 0.1 μg per well, respectively. miR-122 duplexes, single strands, and precursors were transfected at a concentration of 20 nM unless otherwise specified. The Renilla luciferase plasmid was used as a control for transfection efficiency, and samples were normalized to Renilla luciferase expression. Transfected cells were cultured for 24 h and harvested in 1× Passive Lysis Buffer (Promega). Luciferase assays were performed according to the manufacturer's instructions. For small RNA Northern analysis, cells were split 8 h after transfection and were harvested in TRIzol and processed as described below.
For GFP-based siRNA assays, DNA constructs and oligonucleotides were transfected into HeLa, wild-type MEF, or Ago2−/− MEF cells in six-well plates using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. GFP plasmids were transfected at 100 ng per well, Flag-BAP and Flag-Ago2 at 1 μg per well, and miRNAs at 20 nM per well. Twenty-four hours after transfection, cells were lysed, and total RNA was extracted in TRIzol reagent (Invitrogen). HCV RNA electroporation assays were carried out in Huh-7 cells according to the method previously described (Jopling et al. 2005). For HCV assays, all miRNAs were transfected into cells at a concentration of 50 nM.
RNA isolation and Northern analysis
Total RNA was extracted using TRIzol reagent, according to the manufacturer's instructions. Ten micrograms of total RNA from Huh-7 cells and 12 μg of total RNA from wild-type MEFs and Ago2−/− MEFs were separated in 1% Agarose gels containing 1× MOPS buffer and 2.2 M formaldehyde and transferred to Zeta-probe membranes (Bio-Rad). Membranes were hybridized in Express Hyb Hybridization Buffer (ClonTech) to random-primed 32P-labeled DNA probes (RadPrime DNA Labeling System, Invitrogen) complementary to HCV (nucleotides 84–374), GFP (nucleotides 40–814), or γ-actin (nucleotides 685–1171). Autoradiographs were quantitated using ImageQuant (GE Healthcare).
Small RNA Northern analysis
Total RNA was extracted using TRIzol reagent, according to the manufacturer's instructions. Ten micrograms of total RNA from HeLa cells was separated on a 15% acrylamide/7 M urea gel. After transfer onto Hybond-N1 membrane (Amersham Pharmacia Biotech), small RNAs were detected with 32P-end-labeled DNA probes complementary to miR-122 (ACAAACACCAUUGUCACACUCCA), miR-122 p2-8 (ACAAACACCAUUGUCUGUGAUUA), and U6 (CACGAATTTGCGTGTCATCCTTGC). Autoradiographs were quantitated using ImageQuant (GE Healthcare).
Cleavage of pre-miR-122 substrates with human Dicer/TRBP complex
All pre-miR122 RNAs used in the cleavage assays except for pre-miR-122 p3 bulge included a 5′ phosphate. RNAs were dephosphorylated prior to radio-labeling using calf intestinal alkaline phosphatase (CIP; NEB) for 1 h at 37°C in a total volume of 50 µL containing 500 pmol RNA, 1× NEB buffer #3, and 10 units CIP. Following incubation, the CIP was removed by extraction with phenol/chloroform/isoamyl alcohol, and the RNA was recovered by ethanol precipitation of the aqueous layer and resuspended in 50 µL nuclease-free water. RNAs were subsequently 5′ end 32P-labeled using T4 polynucleotide kinase (PNK; NEB) for 1 h at 37°C in a total volume of 20 µL containing 50 pmol RNA, 1× PNK buffer, 50 µCi γ-32P-ATP, and 10 units PNK. Following incubation, 30 µL of nuclease-free water was added, and the excess label was removed using a G25 spin column (GE Healthcare). The 32P-labeled RNAs were used without further purification. RNA (2000 cpm, ∼0.2 pmol) was folded by heating for 5 min to 65°C followed by incubation for 5 min at 4°C in the presence of 2.5 mM MgCl2, 100 mM KCl, and 100 mM HEPES (pH 7.5) in a 4 µL reaction. Cleavage was initiated by adding 1 µL human Dcr/TRBP complex (250 nM), or 1 µL nuclease-free water for control reactions, followed by incubating for 1 or 5 h at 37°C. Reactions were stopped by adding an equal volume of RNA loading dye (95% [v/v] deionized formamide, 5 mM EDTA at pH 8.0, 0.025% [w/v] SDS, 0.025% bromophenol blue, and 0.025% xylene cyanol). The resulting cleavage products were resolved by PAGE (20% 29:1 acrylamide:bis-acrylamide, 7 M urea). The lengths of cleavage products were assigned by an adjacent hydrolysis ladder as well as a 23-nt mature miR-122 RNA.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karla Kirkegaard for many helpful comments during these studies. We also thank Chang-Zheng Chen for many discussions. We thank Cameron Noland (UC Berkeley) for the gift of purified human Dcr/TRBP complex, and Mark Kay (Stanford University) for the pFlag-Ago2 vector and 293-Flag-Ago2 cells. This study was supported by grants from the National Institutes of Health (AI47365, AI069000). E.M.C. received support from the Paul and Mildred Berg Stanford Graduate Fellowship. S.M.S. is an Amgen Fellow of the Life Sciences Research Foundation (LSRF). S.M.S. also thanks the National CIHR Research Training Program in Hepatitis C Virus (NCRTP-HepC) for post-doctoral funding. S.A.W.M. is a fellow of the Leukemia and Lymphoma Society.
REFERENCES
- Akkiz H, Bayram S, Bekar A, Akgollu E, Ulger Y. 2011. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: A case-control study. J Viral Hepat 18: E399–E407. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol 1: 106–113. [DOI] [PubMed] [Google Scholar]
- Edwards PA, Muroya H, Gould RG. 1972. In vivo demonstration of the circadian rhythm of cholesterol biosynthesis in the liver and intestine of the rat. J Lipid Res 13: 396–401. [PubMed] [Google Scholar]
- Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, et al. 2009. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev 23: 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123: 631–640. [DOI] [PubMed] [Google Scholar]
- Han JJ, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. 2013. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoofnagle JH. 2002. Course and outcome of hepatitis C. Hepatology 36: S21–S29. [DOI] [PubMed] [Google Scholar]
- Hu ZB, Chen JP, Tian T, Zhou XY, Gu HY, Xu L, Zeng Y, Miao RF, Jin GF, Ma HX, et al. 2008. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest 118: 2600–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra RK, Yi M, Lemon SM. 2010. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol 84: 6615–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. 2013. Treatment of HCV infection by targeting microRNA. N Engl J Med 368: 1685–1694. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309: 1577–1581. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Schuetz S, Sarnow P. 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J 21: 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han JJ, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- Liu G, Min H, Yue S, Chen CZ. 2008. Pre-miRNA loop nucleotides control the distinct activities of mir-181a-1 and mir-181c in early T cell development. PLoS One 3: e3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin ES, Sarnow P, Sagan SM. 2011. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci 108: 3193–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. 2008. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci 105: 512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. 2005. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev 19: 2979–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197. [DOI] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat Rev Microbiol 5: 453–463. [DOI] [PubMed] [Google Scholar]
- Norman KL, Sarnow P. 2010. Modulation of hepatitis C virus RNA abundance and the isoprenoid biosynthesis pathway by microRNA miR-122 involves distinct mechanisms. J Virol 84: 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. 2011. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475: 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi P, Dou TH, Geng L, Zhou FG, Gu X, Wang H, Gao CF. 2010. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum Immunol 71: 621–626. [DOI] [PubMed] [Google Scholar]
- Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. 2012a. Stabilization of hepatitis C virus RNA by an Ago2–miR-122 complex. Proc Natl Acad Sci 109: 941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakami T, Yamane D, Welsch C, Hensley L, Jangra RK, Lemon SM. 2012b. Base pairing between hepatitis C virus RNA and microRNA 122 3′ of its seed sequence is essential for genome stabilization and production of infectious virus. J Virol 86: 7372–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solda G, Robusto M, Primignani P, Castorina P, Benzoni E, Cesarani A, Ambrosetti U, Asselta R, Duga S. 2012. A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing. Hum Mol Genet 21: 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GS, Garchow BG, Liu XH, Yeung J, Morris JP, Cuellar TL, McManus MT, Kiriakidou M. 2009. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res 37: 7533–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye XC, Iwasaki S, Liu QH, Tomari Y. 2010. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol 17: 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Huys A, Thibault PA, Wilson JA. 2012. Requirements for human Dicer and TRBP in microRNA-122 regulation of HCV translation and RNA abundance. Virology 433: 479–488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.