It has been known that Lin28 negatively regulates let-7 production by recruiting 3′-terminal uridylyl transferases. As a result, the processing of uridylated pre-let-7 by Dicer is blocked. The identity of the RNase that acts to degrade uridylated pre-let-7 was not known until recently. In this paper, the authors report the identification of mammalian DIS3L2 as an oligo(U)-binding exonuclease that specifically targets uridylated let-7 miRNA precursors in vivo. These findings complement a recent report which identified Dis3l2 as the relevant mouse exonuclease. DIS3L2 has 3′–5′ exonuclease activity, is predominantly cytoplasmic and, unlike other mammalian DIS3 orthologs, is not stably associated with the RNA exosome complex.
Keywords: DIS3L2, RNA degradation, RNA uridylation, let-7 miRNA
Abstract
The mechanisms of gene expression regulation by miRNAs have been extensively studied. However, the regulation of miRNA function and decay has long remained enigmatic. Only recently, 3′ uridylation via LIN28A-TUT4/7 has been recognized as an essential component controlling the biogenesis of let-7 miRNAs in stem cells. Although uridylation has been generally implicated in miRNA degradation, the nuclease responsible has remained unknown. Here, we identify the Perlman syndrome-associated protein DIS3L2 as an oligo(U)-binding and processing exoribonuclease that specifically targets uridylated pre-let-7 in vivo. This study establishes DIS3L2 as the missing component of the LIN28-TUT4/7-DIS3L2 pathway required for the repression of let-7 in pluripotent cells.
INTRODUCTION
RNA processing and stability play key roles in the regulation of gene expression and have impacts on complex cellular processes including cell growth, proliferation, and differentiation. RNA stability is regulated by a dynamic interplay of post-transcriptional modifications and trans-acting protein or RNA molecules that can be stabilizing as well as destabilizing factors. MicroRNAs (miRNAs) were identified as post-transcriptional regulators of gene expression more than two decades ago. MicroRNAs are essential for normal development and overall cellular physiology, among other functions. Dysregulation of miRNA expression can be detrimental and is often associated with human disease (Croce 2009). Although miRNA transcription, processing, and function have been studied in great detail, the mechanisms of regulation and turnover of miRNAs remain unknown.
Recently, the post-transcriptional control of let-7 biogenesis in early embryogenesis was revealed. Let-7 miRNA is one of the key regulators of embryonic cell differentiation, and its expression must be tightly controlled because it targets mRNAs encoding factors required for the maintenance of pluripotency (Bussing et al. 2008). This regulation appears to take place predominantly on the post-transcriptional level: The RNA binding protein LIN28A specifically binds the pre-let-7 family of miRNAs and, in an RNA-dependent manner, recruits the terminal uridyltransferases TUT4 and TUT7, which add a stretch of uridines (Heo et al. 2008, 2009; Piskounova et al. 2008; Hagan et al. 2009; Thornton et al. 2012). In turn, this modification inhibits Dicer-mediated processing of pre-let-7 miRNAs. In contrast to its role in embryonic stem cells, the monouridylation of pre-let-7 catalyzed by TUT2/4/7 proteins enhances Dicer processing of certain types of miRNAs in somatic cells (Heo et al. 2012). Single UMP addition repairs the 3′ ends of some pre-miRNAs, forming a dinucleotide overhang that is preferred by Dicer (Zhang et al. 2004).
In mammals, nontemplated uridylation also plays a role in the processing of other small RNAs, such as U6 small nuclear RNA (snRNA) (Trippe et al. 2006), and in the degradation of replication-dependent histone mRNAs (Schmidt et al. 2011; Minasaki and Eckmann 2012; Su et al. 2013). Intriguingly, recent transcriptome-wide studies revealed extensive uridylation of a wide spectrum of RNAs (Rissland et al. 2007; Schmidt and Norbury 2010; Choi et al. 2012), suggesting a general role for (U)-tailing in RNA metabolism and stability.
Although the molecular mechanism of LIN28A-TUT-mediated oligouridylation has been studied in great detail (Yeom et al. 2011), the fate of the uridylated molecules remains enigmatic. Oligouridylation of pre-let-7 has been generally assumed to trigger its degradation (Hagan et al. 2009; Heo et al. 2009); however, the downstream-acting factors, such as the specific nucleases responsible for this activity, remain unknown. In this work, we have searched for factors involved in the downstream processing or degradation of uridylated RNAs, and we have identified mammalian DIS3L2 as an oligo(U)-binding exonuclease that specifically targets uridylated let-7 miRNA precursors in vivo. This finding establishes DIS3L2 as the missing component of the LIN28-TUT4/7-DIS3L2 pathway, which is required for the regulation of let-7 expression in pluripotent cells.
RESULTS AND DISCUSSION
Identification of Dis3l2 as an oligo(U)-binding nuclease in mouse embryonic stem cells
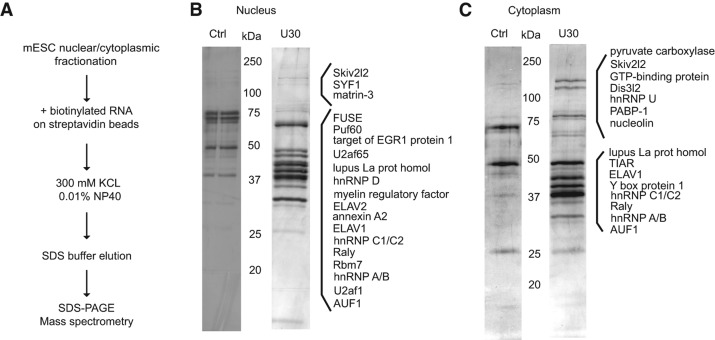
To identify factors involved in the regulation of uridylated RNAs, we carried out affinity purifications of nuclear and cytoplasmic fractions from mouse embryonic stem cells (mESC) with biotin-labeled U30 RNA and a nonspecific 30-nt RNA control (Fig. 1A). Analyses of both the nuclear and cytoplasmic U30-precipitated proteins revealed several proteins that were previously reported to specifically interact with (mostly internal) (U)-rich motifs, such as ELAV1 (Kim et al. 2012), TIAR (Kim et al. 2012), hnRNP C (Soltaninassab et al. 1998), and La protein (Stefano 1984; Fig. 1B,C; Supplemental Tables S1, S2). When searching for factors involved in RNA degradation, we found that the Skiv2l2/mMtr4 RNA helicase, which is a component of the yeast and human exosome targeting complexes TRAMP and NEXT (LaCava et al. 2005; Vanacova et al. 2005; Lubas et al. 2011), specifically coprecipitated with U30 RNA from both cellular compartments (Fig. 1B,C; Supplemental Tables S1, S2). However, the most intriguing finding was the identification of a putative exoribonuclease, Dis3l2, in the U30 sample from the cytoplasmic fraction (Fig. 1C; Supplemental Table S1). To further test the oligo(U)-specific binding of DIS3L2, we have performed filter-aided sample preparation of IP eluates obtained with random and U30 RNAs, and we analyzed the resulting peptide mixtures using LC-MS/MS. The quantification of label-free DIS3L2 showed more than sevenfold enrichment of DIS3L2 (two biological replicates) over the random RNA sample (Supplemental Fig. S1A,B; Supplemental Table S2). DIS3L2 protein has gained attention only recently due to its association with the Perlman syndrome in humans (Astuti et al. 2012). Although it has been reported that disruption of the DIS3L2 gene can lead to aneuploidy, mitotic errors, and expression changes in several mitosis-related proteins (Astuti et al. 2012), the molecular mechanism of DIS3L2 function has remained elusive.
FIGURE 1.
Identification of Dis3l2 as an oligo(U)-binding nuclease in mouse embryonic stem cells (mESCs). (A) Schematic overview of the protocol. (B) Proteins identified in control and U30 RNA-bound nuclear extracts. (C) Proteins identified in control and U30 RNA-bound cytoplasmic extracts. Equal amounts of eluates from control RNA (Ctrl) and U30 RNA bait fractions were separated on 12% SDS-PAGE gels and silver stained. Proteins identified only in the U30 RNA sample are indicated on the right.
Human DIS3L2 is an oligo(U)-binding, cytoplasmic, Mg++-dependent exoribonuclease that does not associate with exosomes
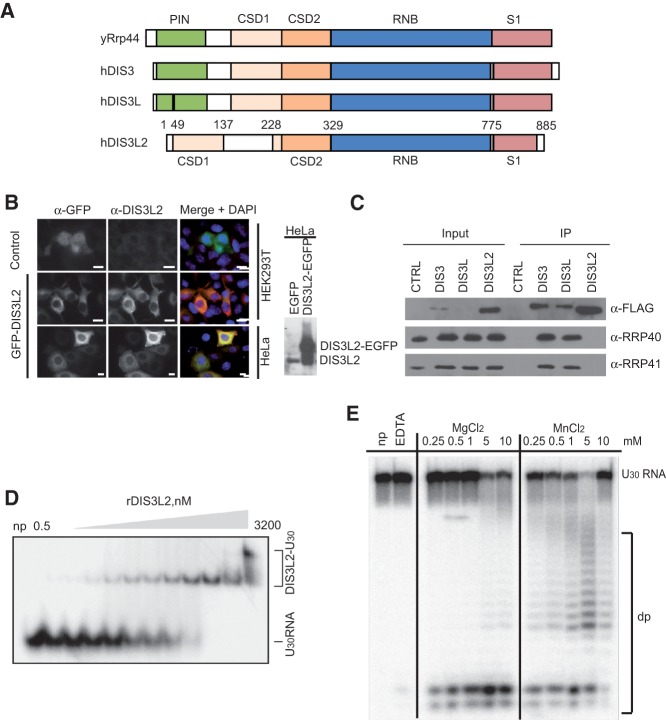
DIS3L2 is a member of the RNase II family of enzymes and is a sequence homolog of the main catalytic subunit of yeast and mammalian exosomes (Rrp44p in yeast, DIS3 and DIS3L in humans) (Fig. 2A; Dziembowski et al. 2007; Schneider et al. 2007; Tomecki et al. 2010). In mice, DIS3L2 is expressed in at least two isoforms. We have identified isoform 2 in our U30 RNA samples (Supplemental Tables S1, S2). Interestingly, isoform 2 was previously identified in the mouse embryo transcriptome (Diez-Roux et al. 2011). To further study the biochemistry and function of this protein, we have subcloned the human DIS3L2 isoform 1, which is the closest homolog of mouse DIS3L2 isoform 2.
FIGURE 2.
DIS3L2 is a cytoplasmic, oligo(U)-binding, Mg++-dependent exoribonuclease that does not associate with exosomes. (A) Schematic representation of the domain organization of DIS3L2 homologs from Saccharomyces cerevisiae and Homo sapiens. The PIN domain is shown in green, the CSD1 and CSD2 RNA binding domains are in orange, the RNB ribonuclease domain is in blue, and the S1 domain is in pink. (B) Immunofluorescence staining of HEK293T and HeLa cells transfected with either DIS3L2 C-terminally fused to an EGFP (DIS3L2-EGFP) or an empty EGFP-expressing plasmid (EGFP). DAPI was used to visualize nuclei. The scale bar corresponds to 10 μm. The Western blot on the right shows the relative abundance of endogenous and EGFP-tagged DIS3L2 in transfected HeLa cells as detected with a DIS3L2-specific antibody. (C) DIS3L2 does not interact with core exosome components. FLAG-tagged DIS3, DIS3L, and DIS3L2 were immunoprecipitated from stable cell lines inducibly overexpressing the individual proteins. The composition of the IP samples was analyzed by Western blot with the indicated antibodies. (D) DIS3L2 binds U30 RNA with nanomolar affinity. Electromobility shift assay with recombinant DIS3L2 and 5′ end-32P-labeled U30 RNA. The migration patterns of free RNA and protein-RNA complexes are indicated. (E) The catalytic activity of DIS3L2 requires divalent metal cations. A degradation assay using recombinant DIS3L2 with U30 RNA as a substrate was performed in the presence of different divalent metal ions as indicated. (EDTA) Reaction mixture containing 5 mM EDTA; (np) control reaction without the addition of any protein.
Human DIS3 and DIS3L show distinct subcellular localization patterns; DIS3 is primarily nuclear, whereas DIS3L is cytoplasmic (Staals et al. 2010; Tomecki et al. 2010). Here, we demonstrate that DIS3L2 is localized in the cytoplasm of mouse ESCs (Fig. 1C) as well as HeLa and HEK293 cells (Fig. 2B; Supplemental Fig. S2A). This finding is in agreement with recent reports indicating the cytoplasmic localization of human and fission yeast DIS3L2 (Astuti et al. 2012; Lubas et al. 2013; Malecki et al. 2013). In contrast to DIS3 and DIS3L, DIS3L2 lacks the PIN domain (Fig. 2A), which is essential for interaction with the exosome core (Dziembowski et al. 2007; Lebreton et al. 2008; Schneider et al. 2009). Here, we showed that DIS3L2 did not coprecipitate with exosome subunits from HEK293T cells, whereas these subunits were detected in purified control samples of DIS3 and DIS3L (Fig. 2C). This finding indicates the functional independence of DIS3L2 from the exosome core complex. To further explore the role of DIS3L2 in RNA uridylation, we evaluated the binding and activity of wild-type (WT) and catalytically inactive (D391N) recombinant DIS3L2 (rDIS3L2) (Supplemental Fig. S2B) toward U30 RNA. The recombinant protein showed nanomolar affinity for U30 RNA in vitro (Fig. 2D), required divalent metal ions for exonuclease activity (Fig. 2E), and degraded U30 RNA to 2- to 4-nt end-products (Fig. 2E; Supplemental Fig. S2C,D). No degradation was observed with the D391N mutant (Supplemental Fig. S2C).
Human DIS3L2 targets 3′-uridylated precursors of let-7 miRNA
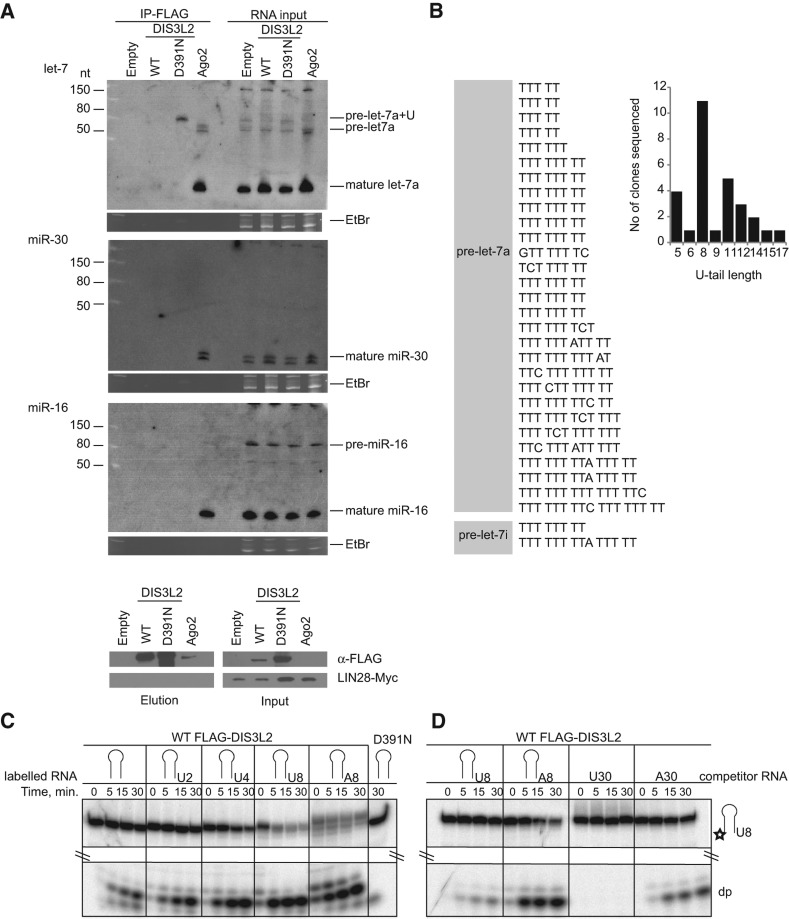
Because we initially pulled down DIS3L2 from embryonic stem cells, we next asked whether DIS3L2 could be the missing component of the pre-let-7 uridylation/degradation pathway. To test the ability of DIS3L2 to bind pre-let-7 miRNA, we ectopically expressed pri-let-7 miRNA and LIN28A in HEK293T-Rex cell lines stably expressing either the WT or D391N forms of FLAG-DIS3L2 to promote pre-let-7 uridylation according to Heo et al. 2008. The in vivo association between pre-let-7 and DIS3L2 was monitored by RNA immunoprecipitation (RIP) followed by Northern blot analyses. Mature let-7 miRNA coprecipitated only with AGO2, which was used as a positive control for let-7 interaction (Fig. 3A). Intriguingly, the D391N mutant of DIS3L2 coprecipitated with slower migrating forms of pre-let-7 miRNA (pre-let-7+U) (Fig. 3A). This binding was specific to pre-let-7 because no signal was observed with probes specific for miR-30 or miR-16 (Fig. 3A). The migration pattern of the extended form of pre-let-7 miRNA resembled that of the uridylated pre-let7 miRNAs induced in HEK293 cells upon LIN28A overexpression (Heo et al. 2008). To uncover the identity of D391N-bound fragments, we modified the immunoprecipitated RNAs with 3′ RNA linkers to allow for cDNA synthesis using linker-specific primers. The subsequent PCR amplification with pre-let-7-specific forward and linker-specific reverse primers revealed a product of ∼70 nt in length in D391N-bound RNAs but not in background controls from untransfected HEK293T-Rex cells (Supplemental Fig. S3A). The PCR products were subcloned, and 35 clones were sequenced. Sequencing analysis confirmed that the bound fragments corresponded to pre-let-7 miRNAs (Supplemental Fig. S3B). Most importantly, 31 out of the 35 sequenced clones displayed nontemplate uridine stretches at their 3′ ends (Fig. 3B). These oligo(U) extensions ranged between five and 17 nt in length (Fig. 3B), which strongly correlates with the length distribution of oligo(U) tails previously identified on pre-let-7a-1 in HEK293T cells ectopically expressing LIN28A and pri-let-7a-1 (Heo et al. 2008). To test the importance of oligouridylation for pre-let-7 degradation, we performed in vitro degradation assays with purified FLAG-DIS3L2. We observed that 4- to 8-nt oligo(U) extensions significantly enhanced pre-let-7 miRNA degradation by DIS3L2 in vitro (Fig. 3C; Supplemental Fig. S3C). Because the oligo(A8) modification also activated degradation by DIS3L2, we examined DIS3L2 nucleotide preference by using unlabeled RNA competitors. We demonstrated that DIS3L2 prefers oligo(U) over oligo(A), as only uridylated pre-let-7 and oligo(U) RNAs were able to inhibit the degradation of labeled uridylated pre-let-7 RNA substrates (Fig. 3D; Supplemental Fig. S3D).
FIGURE 3.
DIS3L2 targets 3′ end-uridylated precursors of let-7 miRNA. (A) Mutant DIS3L2 (D391N) specifically coprecipitates with extended forms of pre-let-7a miRNA. Northern blot analyses of RNAs coimmunoprecipitated with FLAG-tagged proteins as indicated on the upper side of the autoradiograph (IP-FLAG). The RNA input was total RNA isolated from whole cell lysates. The ethidium bromide (EtBr)-stained gels represent input loading controls. (Empty) Control RIP performed using cells that were not expressing any FLAG-tagged protein. The lower panel shows Western blot analysis of Myc-tagged LIN28A expression and the efficiency of FLAG tag-mediated immunoprecipitation of proteins used for RIP analyses. “Input” shows the whole cell lysates used for the IPs. The ectopic expression of Myc-tagged LIN28A was monitored with specific anti-Myc antibodies. DIS3L2 was detected with anti-FLAG antibodies. (B) Sequencing analysis of the 3′ termini of pre-let-7 RNAs coprecipitated with the D391N DIS3L2 mutant in HEK293T-Rex cells ectopically overexpressing LIN28A and pri-let-7a-1. The analysis suggests that the DIS3L2 mutant binds to uridylated pre-let-7a-1 and pre-let-7i because 31 out of 35 sequenced clones contained nontemplate oligo(U)-tails. The gray box schematically represents the body of pre-let-7 miRNAs. A graphical representation of the (U)-extension statistics is shown at the right. (C) Oligouridylation stimulates DIS3L2-mediated degradation of pre-let-7 miRNA. An in vitro degradation assay with FLAG-DIS3L2 purified from HEK293T-Rex cells and pre-let-7 RNA (represented by the schematic stem–loop) containing either no 3′ extension or 3′ oligo(U) and oligo(A) extensions of increasing lengths (indicated at the top) as substrates. D391N indicates a reaction with purified catalytically inactive DIS3L2. (D) DIS3L2 exhibits a preference for RNA substrates with oligo(U) extensions. A 5′ end-labeled pre-let-7-U8 RNA was incubated with purified DIS3L2 in the presence of a 10-fold excess of different unlabeled competitor RNAs (indicated at the top) for the time periods shown.
The lack of pre-let-7+U coprecipitation with WT DIS3L2 was likely due to high turnover of the bound substrate in vivo. Importantly, overexpression of WT DIS3L2 caused a reduction in the uridylated pre-let-7 miRNA level compared with that in untransfected cells or cells overexpressing the D391N mutant (Fig. 3A, pre-let-7a+U, RNA input). Interestingly, D391N was also able to pull down pre-let-7U+ fragments without the ectopic expression of LIN28A or pri-let-7a-1 (Supplemental Fig. S4A). This result suggests that either low endogenous levels of LIN28A (Heo et al. 2012) or other TUT cofactor(s) are sufficient to promote efficient pre-let-7 oligouridylation in epithelial cells. We, therefore, examined whether DIS3L2 targets pre-let-7 in HeLa cells that exhibit higher endogenous levels of let-7 miRNA expression. Northern blot analysis of RNAs isolated from cells treated with two different sets of siRNAs did not indicate pre-let-7 stabilization. On the contrary, Northern blot and Q-PCR analyses revealed reduced levels of mature let-7 and, less significantly, miR-30 miRNAs (Supplemental Fig. S4B,C). This miRNA down-regulation was not due to altered levels of Dicer (Supplemental Fig. S4D). Currently, the connection between DIS3L2 down-regulation and let-7 reduction in somatic cells is unknown. Future studies will reveal whether miRNA dysregulation may contribute to the development of Perlman syndrome or increased tumor incidence in individuals bearing the mutant DIS3L2 allele (Astuti et al. 2012).
In summary, we have demonstrated that the cytoplasmic exoribonuclease DIS3L2 specifically recognizes uridylated pre-let-7 miRNAs in vivo and that pre-let-7 uridylation is important for DIS3L2-mediated degradation. This establishes DIS3L2 as a strong candidate for the sought-after nuclease targeting uridylated pre-let-7 in embryonic stem cells (Fig. 4). Because the TUT-DIS3L2 mechanism strongly resembles the nuclear polyadenylation-mediated RNA degradation pathway (Kadaba et al. 2004; LaCava et al. 2005; Vanacova et al. 2005), future studies should examine to what extent it operates as a general cytoplasmic (mi)RNA surveillance and decay pathway.
FIGURE 4.
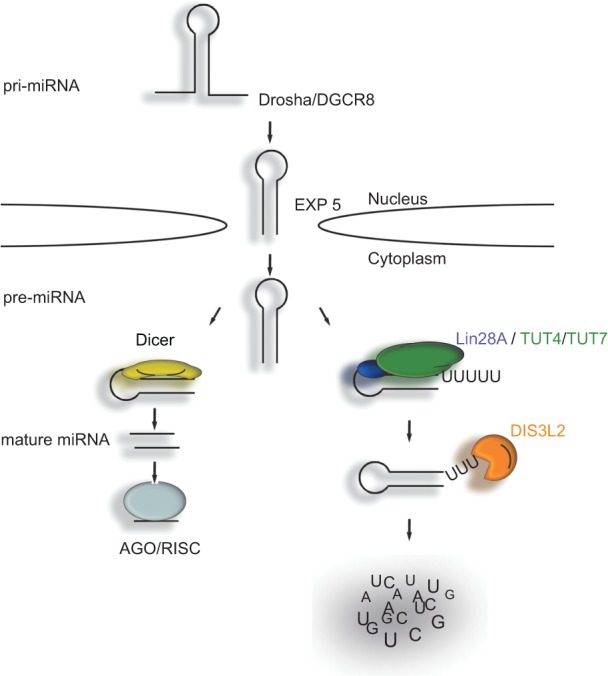
A model for the regulation of let-7 miRNA biogenesis via the LIN28A-TUT4/7-DIS3L2 pathway. The primary transcripts (pri-let-7) are processed by the microprocessor complex in the nucleus to pre-let-7, which is exported to the cytoplasm. In the cytoplasm, pre-let-7 is either processed by Dicer to mature let-7 miRNA, or it is oligouridylated in the presence of LIN28A. The oligo(U) tails are then recognized by the DIS3L2 exonuclease, which initiates the degradation of RNA in the 3′ to 5′ direction.
Over the course of preparing this manuscript, two other groups reported their findings on the role of DIS3L2 in the degradation of uridylated RNAs. Malecki et al. identified DIS3L2 as an oligo(U)-specific enzyme targeting uridylated mRNAs in Schizosaccharomyces pombe (Malecki et al. 2013). Chang et al. revealed the role of mouse Dis3l2 in the LIN28A-TUT4/7 pathway (Chang et al. 2013). These authors showed the in vitro activity of Dis3l2 toward uridylated pre-let-7 miRNAs and demonstrated that down-regulation of Dis3l2 in mESCs can lead to elevated levels of uridylated pre-let-7 without affecting the level of mature let-7 miRNA. Because we observed down-regulation of mature let-7 upon DIS3L2 knockdown, it is possible that DIS3L2 has two distinct cell type-specific mechanisms operating during let-7 biogenesis.
MATERIALS AND METHODS
LC-MS/MS analysis
Thin slices excised from a stained SDS-PAGE gel were de-stained, washed, and incubated with trypsin. Liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis was performed using the EASY-nLC system (Thermo Fisher Scientific) coupled with an HCTultra PTM Discovery System ion trap mass spectrometer (Bruker Daltonik). The MASCOT 2.3.02 (MatrixScience) search engine was used for processing the MS and MS/MS data. Database searches were performed against the NCBI database (nonredundant, taxonomy: Mus musculus). (For details, see Supplemental Material.)
siRNA-mediated knock down of DIS3L2
On the day prior to transfection, 1.7 × 105 cells were seeded. siRNAs were transfected using INTERFERin transfection reagent (Polyplus transfections) at a 20-nM final concentration, following the manufacturer's instructions. The siRNA treatment was repeated after 24 h, and the cells were collected for further analysis on the following day. (See Supplemental Table S4 for the list of siRNAs used.)
In vitro degradation assay
In vitro degradation assays were performed in 10-μL reaction volumes containing 10 mM Tris (pH 8.0), 50 mM KCl, 5 mM MgCl2, and 10 mM DTT (modified from Lorentzen et al. 2008; Staals et al. 2010; Tomecki et al. 2010). Typically, 150 nM of purified recombinant protein and 20 pmol of 5′ end-labeled RNA substrate were incubated at 37°C for the time periods indicated. The reactions were terminated with one volume of formamide loading buffer (80% formamide, 0.1% bromphenol blue, 0.1% xylene cyanol, 5 mM EDTA). The reactions were resolved on denaturing 20% polyacrylamide gels containing 8 M urea. The radioactively labeled substrate was detected using a phosphorimaging screen and an FLA-9000 phosphorimager (FujiFilm).
RNA binding assay
The binding reactions were performed in 10-μL volumes. Recombinant DIS3L2 was incubated with 5′ end-labeled U30 RNA for 20 min at room temperature in binding buffer (50 mM KCl, 10 mM Tris pH 8.0, 10 mM DTT, 10% glycerol, 0.1 μg of BSA). Proteins and RNAs were resolved on native 8% polyacrylamide gels at 100 V for ∼8 h and visualized by phosphorimaging.
Northern blot analysis
Total RNA was resolved on a 15% denaturing polyacrylamide gel and transferred to a Hybond-N+ membrane (GE Healthcare) by electroblotting (BioRad). Hybridization with radioactively labeled oligonucleotides was performed in ULTRAhyb-oligo hybridization buffer (Ambion) at 42°C. Prior to the addition of the labeled probe, the membrane was prehybridized at 42°C for 2 h. The radioactive signal was monitored using an FLA-9000 phosphorimager (FujiFilm).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ger Pruijn and Petr Svoboda for the hRRP41, hRRP40, and Dicer antibodies; Torben Jensen for the HEK293T-Rex cell lines expressing FLAG-DIS3 and FLAG-DIS3L; and Narry Kim for the pri-miRNA DNA constructs. We also thank Leona Svajdova for excellent technical support and Ivana Horvathova for providing the LIN28A construct. This work was supported by the Wellcome Trust (084316/Z/07/Z to S.V.), the Czech Science Foundation (305/11/1095 to S.V., D.H. was supported by P305/12/G034, and P206-12-G151 to D.P. and Z.Z.), and the CEITEC-Central European Institute of Technology (CZ.1.05/1.1.00/02.0068) from the European Regional Development Fund.
Author contributions: D.U. designed, performed, and analyzed most of the experiments; D.H. identified DIS3L2 using RNA-based protein precipitation; J.P. provided the mESC material; K. Chalupnikova performed immunofluorescence studies; K.H. prepared DIS3L2 constructs and specific antibodies; K. Cetkovska and S.U. assisted with the initial HEK293T manipulations; and D.P. and Z.Z. performed the MS analysis. D.U. and S.U. were also involved in the preparation of the manuscript. S.V. designed the project and experiments, provided funding, supervised students, and prepared the manuscript.
Footnotes
Freely available online through the RNA Open Access option.
REFERENCES
- Astuti D, Morris MR, Cooper WN, Staals RH, Wake NC, Fews GA, Gill H, Gentle D, Shuib S, Ricketts CJ, et al. 2012. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet 44: 277–284. [DOI] [PubMed] [Google Scholar]
- Bussing I, Slack FJ, Grosshans H. 2008. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med 14: 400–409. [DOI] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. 2013. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 497: 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Patena W, Leavitt AD, McManus MT. 2012. Widespread RNA 3′-end oligouridylation in mammals. RNA 18: 394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. 2009. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10: 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, et al. 2011. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol 9: e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B. 2007. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14: 15–22. [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16: 1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. 2008. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 32: 276–284. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708. [DOI] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. 2012. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 151: 521–532. [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Wilce MC, Yoga YM, Pendini NR, Gunzburg MJ, Cowieson NP, Wilson GM, Williams BR, Gorospe M, Wilce JA. 2012. Different modes of interaction by TIAR and HuR with target RNA and DNA. Nucleic Acids Res 39: 1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724. [DOI] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Seraphin B. 2008. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456: 993–996. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. 2008. Structure of the active subunit of the yeast exosome core, Rrp44: Diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell 29: 717–728. [DOI] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43: 624–637. [DOI] [PubMed] [Google Scholar]
- Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. 2013. Exonuclease hDIS3L2 specifies an exosome-independent 3′-5′ degradation pathway of human cytoplasmic mRNA. EMBO J 32: 1855–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. 2013. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J 32: 1842–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minasaki R, Eckmann CR. 2012. Subcellular specialization of multifaceted 3′end modifying nucleotidyltransferases. Curr Opin Cell Biol 24: 314–322. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. 2008. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem 283: 21310–21314. [DOI] [PubMed] [Google Scholar]
- Rissland OS, Mikulasova A, Norbury CJ. 2007. Efficient polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol 27: 3612–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MJ, Norbury CJ. 2010. Polyadenylation and beyond: Emerging roles for noncanonical poly(A) polymerases. Wiley Interdiscip Rev RNA 1: 142–151. [DOI] [PubMed] [Google Scholar]
- Schmidt MJ, West S, Norbury CJ. 2011. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA 17: 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Anderson JT, Tollervey D. 2007. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell 27: 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Leung E, Brown J, Tollervey D. 2009. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res 37: 1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltaninassab SR, McAfee JG, Shahied-Milam L, LeStourgeon WM. 1998. Oligonucleotide binding specificities of the hnRNP C protein tetramer. Nucleic Acids Res 26: 3410–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJ, Raijmakers R, Pruijn GJ. 2010. Dis3-like 1: A novel exoribonuclease associated with the human exosome. EMBO J 29: 2358–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano JE. 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36: 145–154. [DOI] [PubMed] [Google Scholar]
- Su W, Slepenkov SV, Slevin MK, Lyons SM, Ziemniak M, Kowalska J, Darzynkiewicz E, Jemielity J, Marzluff WF, Rhoads RE. 2013. mRNAs containing the histone 3′ stem-loop are degraded primarily by decapping mediated by oligouridylation of the 3′ end. RNA 19: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. 2012. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 18: 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. 2010. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J 29: 2342–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. 2006. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12: 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom KH, Heo I, Lee J, Hohng S, Kim VN, Joo C. 2011. Single-molecule approach to immunoprecipitated protein complexes: Insights into miRNA uridylation. EMBO Rep 12: 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.