3′-End maturation of histone mRNAs in higher eukaryotes requires U7 snRNA and several factors that are essential for canonical cleavage and polyadenylation. This paper reveals some interesting differences between histone 3′-end formation in Drosophila and mammalian cells. In particular, Ars2 is not essential for Drosophila histone 3′-end processing, and the Drosophila U7 snRNP is associated with several polyadenylation factors in a stable supercomplex.
Keywords: histone pre-mRNA, 3′-end processing, Drosophila U7 snRNP, Lsm11, FLASH, polyadenylation factors, CPSF73 endonuclease, SLBP, Ars2
Abstract
3′-End cleavage of animal replication-dependent histone pre-mRNAs is controlled by the U7 snRNP. Lsm11, the largest component of the U7-specific Sm ring, interacts with FLASH, and in mammalian nuclear extracts these two proteins form a platform that recruits the CPSF73 endonuclease and other polyadenylation factors to the U7 snRNP. FLASH is limiting, and the majority of the U7 snRNP in mammalian extracts exists as a core particle consisting of the U7 snRNA and the Sm ring. Here, we purified the U7 snRNP from Drosophila nuclear extracts and characterized its composition by mass spectrometry. In contrast to the mammalian U7 snRNP, a significant fraction of the Drosophila U7 snRNP contains endogenous FLASH and at least six subunits of the polyadenylation machinery: symplekin, CPSF73, CPSF100, CPSF160, WDR33, and CstF64. The same composite U7 snRNP is recruited to histone pre-mRNA for 3′-end processing. We identified a motif in Drosophila FLASH that is essential for the recruitment of the polyadenylation complex to the U7 snRNP and analyzed the role of other factors, including SLBP and Ars2, in 3′-end processing of Drosophila histone pre-mRNAs. SLBP that binds the upstream stem–loop structure likely recruits a yet-unidentified essential component(s) to the processing machinery. In contrast, Ars2, a protein previously shown to interact with FLASH in mammalian cells, is dispensable for processing in Drosophila. Our studies also demonstrate that Drosophila symplekin and three factors involved in cleavage and polyadenylation—CPSF, CstF, and CF Im—are present in Drosophila nuclear extracts in a stable supercomplex.
INTRODUCTION
Animal replication-dependent histone pre-mRNAs are processed at the 3′ end by cleavage that is not followed by polyadenylation (Muller and Schumperli 1997; Gilmartin 2005; Dominski and Marzluff 2007; Dominski et al. 2013). This processing reaction is conserved between vertebrates and invertebrates and differs from the cleavage and polyadenylation pathway that operates on canonical pre-mRNAs (Wahle and Keller 1996; Colgan and Manley 1997; Zhao et al. 1999; Mandel et al. 2008; Proudfoot 2011). 3′-End processing of histone pre-mRNAs is controlled by two sequence elements that flank the cleavage site: a highly conserved upstream stem–loop structure and a loosely conserved histone downstream element (HDE). The stem–loop structure interacts with the Stem–Loop Binding Protein (SLBP) (Tan et al. 2013), also known as the Hairpin Binding Protein (HBP) (Martin et al. 1997), whereas the HDE is a binding site for the U7 snRNP (Mowry and Steitz 1987; Cotten et al. 1988). The U7 snRNP consists of two core components: a 60- to 70-nucleotide (nt) U7 snRNA and an unusual heptameric Sm ring. In this ring, the SmD1 and SmD2 proteins are replaced by the related Lsm10 and Lsm11 proteins. The five remaining Sm proteins—B, D3, E, F, and G—are shared with the spliceosomal snRNPs (Schumperli and Pillai 2004).
The 5′ end of the U7 snRNA is partially complementary with the HDE, and the base-pair interaction between these two sequences is primarily responsible for bringing the U7 snRNP to histone pre-mRNA. Preventing this interaction in both mammalian (Cotten et al. 1989) and Drosophila extracts (Dominski et al. 2005) abolishes cleavage, indicating that the U7 snRNP is an essential component of 3′-end processing of histone pre-mRNAs in all animal cells. Mammalian SLBP functions to stabilize the binding of the U7 snRNP to histone pre-mRNA and is dispensable for processing in vitro if the HDE forms a sufficiently strong duplex with the U7 snRNA (Streit et al. 1993; Dominski et al. 1999). In contrast, Drosophila SLBP is essential for 3′-end cleavage of all histone pre-mRNAs both in vitro (Dominski et al. 2002) and in vivo (Sullivan et al. 2001).
Mammalian Lsm11 interacts through its extended N-terminal region with the N terminus of a 220-kDa protein, FLASH (Yang et al. 2009a). These two protein fragments expressed in bacteria form a platform that interacts tightly with eight mammalian proteins involved in cleavage and polyadenylation: symplekin, CstF64, and all six subunits of CPSF, including the endonuclease CPSF73 (Yang et al. 2013). We refer to this complex as the Histone pre-mRNA Cleavage Complex, or HCC (Yang et al. 2013). The interaction with the HCC critically depends on a highly conserved cluster of amino acids, LDLY, located near the N terminus of FLASH (Yang et al. 2011). Mutations within this cluster abolish the activity of FLASH in processing in vitro and render the FLASH/Lsm11 complex unable to interact with the HCC. The same polyadenylation factors associate in a FLASH-dependent manner with the mammalian U7 snRNP and are recruited to histone pre-mRNA for 3′-end processing. Thus, in mammalian cells, at least a fraction of the U7 snRNP has a highly complex structure and in addition to the U7 snRNA and the Sm ring contains FLASH and several polyadenylation factors, which are delivered to histone pre-mRNA through the base-pair interaction between the U7 snRNA and the HDE (Yang et al. 2013).
In addition to Lsm11, mammalian FLASH interacts with Ars2 (Narita et al. 2007), a versatile protein known to play a role in microRNA biogenesis (Lobbes et al. 2006; Gruber et al. 2009; Sabin et al. 2009; Xie et al. 2010). Ars2 is also required for cell proliferation (Gruber et al. 2009), and its depletion from mammalian cells results in production of small amounts of replication-dependent histone mRNAs that end in a poly(A) tail rather than with the stem–loop (Gruber et al. 2012). Thus, in mammalian cells, Ars2 may be directly involved in the U7-dependent processing.
The interaction between FLASH and Lsm11 is conserved in Drosophila (Yang et al. 2009a), and it is essential in vivo (Burch et al. 2011). However, it has not been determined whether in Drosophila these two proteins also function by recruiting polyadenylation factors to the U7 snRNP. Moreover, only three subunits of the cleavage and polyadenylation machinery—CPSF73, CPSF100, and symplekin—were shown to be essential for U7-dependent processing of histone pre-mRNAs in Drosophila cells (Wagner et al. 2007; Sullivan et al. 2009), sharply contrasting with the complexity of the mammalian U7 snRNP, which contains multiple polyadenylation factors (Yang et al. 2013).
Here, we take advantage of the fact that nuclear extracts from Drosophila Kc cells are very active in cleaving histone pre-mRNAs and enriched in processing factors in order to purify and determine the composition of the Drosophila U7 snRNP and processing complexes assembled on histone pre-mRNA. We also reinvestigated the importance of several polyadenylation factors and other proteins, including SLBP and Ars2, in 3′-end processing of histone pre-mRNAs using various in vitro and in vivo approaches.
Our results show that Drosophila U7 snRNP is associated with FLASH and a complex of polyadenylation factors that resembles the mammalian HCC. The same composite U7 snRNP is directly delivered to histone pre-mRNA for 3′-end processing. Consistent with the previous results, among the multiple polyadenylation factors that form the Drosophila HCC, only the endonuclease CPSF73, CPSF100, and symplekin are essential for generation of properly matured histone mRNA in vivo. We also show that Ars2 does not form a complex with FLASH in Drosophila cells and is not required for correct cleavage of histone pre-mRNAs, suggesting that these functions of Ars2 are limited to mammalian cells. Finally, Drosophila SLBP does not play a major role in stabilizing the interaction between the U7 snRNP and histone pre-mRNA but instead likely facilitates the recruitment of an essential and perhaps a yet-unidentified factor(s) of the processing machinery.
RESULTS
Characterization of Kc nuclear extracts used in biochemical studies on 3′-end processing
The 3′-end processing machinery that cleaves replication-dependent histone pre-mRNAs in Drosophila shares a number of similarities with the mammalian machinery (Fig. 1A), including the essential role played by the interaction of Lsm11 and FLASH (Burch et al. 2011). In mammalian cells, these two proteins form a platform that serves to recruit multiple polyadenylation factors to the U7 snRNP (Yang et al. 2013). In this study, we used large-scale nuclear extracts from Kc cells and several biochemical approaches to determine the role of the Lsm11/FLASH interaction and composition of the U7-dependent processing machinery in Drosophila.
FIGURE 1.
3′-End processing of histone pre-mRNAs in Drosophila. (A) Known factors involved in 3′-end processing of histone pre-mRNAs in Drosophila. (Black and gray lines) Histone pre-mRNA and Drosophila U7 snRNA, respectively. (Vertical lines) Base-pair interactions between the two RNAs. In the U7-specifc Sm ring, the two spliceosomal proteins SmD1 and SmD2 are replaced by Lsm10 and Lsm11 (indicated with dark gray). Lsm11 binds FLASH and in mammalian nuclear extracts these two proteins interact with CPSF73 (the endonuclease), CPSF100, symplekin, and other polyadenylation factors that together form the Histone pre-mRNA Cleavage Complex (HCC). It is unknown whether the interaction between Lsm11 and FLASH functions in the same manner in Drosophila (double arrow) and whether the HCC contains any factors in addition to CPSF73, CPSF100, and symplekin, as indicated with the question mark. (B) Levels of FLASH, SLBP, and symplekin in nuclear extract prepared from frozen (lane 1) or freshly collected Kc cells (lane 2), as determined by Western blotting. (C) 3′-End processing of Drosophila histone H3 (dH3) pre-mRNA in a Kc nuclear extract prepared from freshly harvested cells in the presence of indicated RNA competitors (lanes 3–8). The input alone and processing in the absence of any RNA competitor is shown in lanes 1 and 2, respectively. (D) 3′-End processing of the dH3 pre-mRNA in a Kc nuclear extract prepared from freshly harvested cells in the presence of indicated antibodies (lanes 3 and 4). The input alone and processing in the absence of any RNA competitor is shown in lanes 1 and 2, respectively. Inhibition of processing by the αU7 oligonucleotide is shown in lane 5. (E) Nucleotide sequence of the wild-type SL (WT SL) RNA. Regions altered in the Mut SL RNA that are essential for binding SLBP are underlined.
In our previous studies on 3′-end processing of histone pre-mRNAs in Drosophila, we typically used large-scale nuclear extracts prepared from frozen Kc cells since they had virtually the same processing activity as nuclear extracts prepared from freshly harvested cells (Dominski et al. 2002, 2005). However, closer biochemical analysis revealed that these extracts contained significantly reduced levels of two essential processing factors: SLBP and FLASH (Fig. 1B, cf. lanes 1 and 2). The reduced level of SLBP was primarily caused by its rapid leakage to the cytoplasm during cell thawing. FLASH, in addition to leaking to the cytoplasm, was degraded to shorter fragments that are fully functional in 3′-end processing in vitro (see below). As a result of leakage, cytoplasmic extracts prepared from frozen cells are active in processing, indicating that they contain complete processing machinery (Supplemental Fig. S1A, lane 3). Clearly, relatively small amounts of SLBP, FLASH, and other processing factors are sufficient to support robust processing activity of nuclear and cytoplasmic extracts when tested with a limiting amount of the dH3 pre-mRNA substrate.
Since nuclear extracts prepared from freshly harvested Kc cells were highly enriched in full-length processing factors (Supplemental Fig. S1B), our subsequent biochemical experiments were carried out using these extracts. In agreement with our previous results (Dominski et al. 2002, 2005), 3′-end processing of histone pre-mRNAs carried out in these extracts is absolutely dependent on SLBP. The presence of 10 µM wild-type stem–loop RNA competitor (WT SL, 500-fold excess over the pre-mRNA substrate), which sequesters the entire pool of SLBP in the extract inhibited cleavage of the dH3 pre-mRNA, whereas the same amount of a mutant stem–loop RNA (Mut SL) had no effect (Fig. 1C, cf. lanes 4 and 5). This mutant RNA contains alterations in the 5′-flanking adenosines and the second base pair of the stem (Fig. 1E) and does not bind SLBP (Yang et al. 2009b; Tan et al. 2013). Processing was also efficiently inhibited by 5 µM anti-U7 2′-O-methyl oligonucleotide (αU7) complementary to nucleotides 5–19 of the Drosophila U7 snRNA (Fig. 1C, lane 7) and by an antibody directed against the N-terminal portion of Drosophila FLASH (Fig. 1D, lane 3). A nonspecific 2′-O-methyl oligonucleotide targeted to the first 17 nt of the mouse U7-snRNA (αMock) (Fig. 1C, lane 8) and a nonspecific antibody (Fig. 1D, lane 4) did not reduce in vitro 3′-end processing. All these reagents had the same effect on processing in Drosophila S2 nuclear extracts prepared from cells grown either in monolayers (ML S2) or suspension cultures (Sus S2) (Supplemental Fig. S2; data not shown). We conclude that the essential role of SLBP, the U7 snRNP and FLASH in 3′-end processing of histone pre-mRNAs is a universal feature of Drosophila nuclear extracts. This conclusion is consistent with the essential role of these factors in processing in vivo (Sullivan et al. 2001; Godfrey et al. 2006; Burch et al. 2011).
Functional conservation of Drosophila FLASH
A short N-terminal fragment of human FLASH encompassing amino acids 52–139 strongly stimulates processing in diluted or poorly active mammalian nuclear extracts (Yang et al. 2009a, 2011). This fragment of FLASH tightly binds the N-terminal fragment of human Lsm11 (amino acids 1–170), and together they interact with a large nuclear complex consisting of multiple polyadenylation factors, including the endonuclease CPSF73 (Yang et al. 2013). We refer to this complex as the Histone pre-mRNA Cleavage Complex (HCC). By introducing various deletions and mutations into the N-terminal fragment of human FLASH, we demonstrated that amino acids 100–135 bind Lsm11, whereas amino acids 55–58 with the highly conserved LDLY sequence are required for the recruitment of the HCC (Yang et al. 2011).
Our previous studies in Drosophila cultured cells showed that the region located between amino acids 65 and 77 of Drosophila FLASH is essential for processing of histone pre-mRNAs in vivo, but its role has not been determined (Burch et al. 2011). This region contains an LDIY sequence (Fig. 2A), which is likely the functional counterpart of the LDLY motif in mammalian FLASH. Surprisingly, the N-terminal fragment of Drosophila FLASH (FN-ter, amino acids 1–178) did not stimulate 3′-end processing in Drosophila Kc nuclear extracts (Fig. 2B, lane 4). Moreover, a deletion mutant of this fragment lacking the first 77 amino acids (FΔ77), including the LDIY motif, did not act in a dominant-negative fashion in vitro (Fig. 2B, lane 5), although the equivalent mutant of human FLASH strongly inhibited processing in mammalian extracts (Yang et al. 2011). Note that as indicated above, FLASH is an essential component of Drosophila processing machinery, and the presence of an anti-FLASH antibody inhibits cleavage of the dH3 pre-mRNA in this and all other tested Drosophila nuclear extracts (Fig. 1D; Supplemental Fig. S2).
FIGURE 2.
The role of the LDIY motif in FLASH in processing. (A) The amino acid sequence of the first 178 residues of Drosophila FLASH. The Lsm11-binding site and the critical LDIY motif (replaced with four alanines in the LDIY-4A mutant) are underlined. (Arrow) The start point of the FΔ77N deletion mutants. (B) 3′-End processing of the dH3 pre-mRNA in a Kc nuclear extract in the presence of 100 ng of indicated recombinant proteins. The input pre-mRNA and control processing (no protein added) are shown in lanes 1 and 2, respectively. (C) The abundance of endogenous FLASH and SLBP in untreated S2 cells (No KD, lane 1) or treated with a specific double stranded (ds) RNA (FLASH KD, lane 2), as determined by Western blotting using an antibody directed against the N-terminal region of Drosophila FLASH. (*) A protein cross-reacting with the antibody that serves as a loading control. (D) 3′-End processing of the dH3 pre-mRNA in nuclear extracts prepared from untreated S2 cells (lanes 2–4) or cells depleted of FLASH (lanes 5–7). Control processing in the absence of any recombinant protein is shown in lanes 2 and 5. Processing in lanes 3–4 and 6–7 was carried out in the presence of 100 ng of indicated recombinant proteins. (E) 3′-End processing of the dH3 pre-mRNA in a nuclear extract prepared from S2 cells partially depleted of FLASH. Reactions were carried out in the presence of 100 ng of indicated recombinant FLASH proteins (lanes 2–4). The control reaction (lane 1) contains no exogenous protein. (F) Statistical analysis of 3′-end processing of the dH3 pre-mRNA in small-scale nuclear extracts from untreated S2 cells (left panel) and FLASH-depleted S2 cells (right panel). Processing was carried out in the absence of any recombinant protein (buffer) or in the presence of indicated variants of FLASH. The P-value was calculated using a paired version of the Student's t-test with a two-tailed distribution. The data represent two to four separate processing reactions and three independent nuclear extract preparations from untreated and treated S2 cells. Efficiency was measured as a ratio of the final processed product to the input (unprocessed + processed). Error bars indicate the standard deviation for measured efficiencies.
In contrast to mammalian cells, where FLASH is severely limiting, Drosophila cultured cells contain relatively high levels of this protein, either full length or its shorter proteolytic fragments, which could be precipitated by specific antibodies and visualized on SDS/polyacrylamide gels by Coomassie and silver staining (see Fig. 5B, below). We reasoned that the endogenous Drosophila FLASH is in excess to Lsm11 and quantitatively bound to the U7 snRNP, providing a potential explanation for the failure of the recombinant N-terminal FLASH to stimulate processing. To test this possibility, we reduced the endogenous level of FLASH by RNA interference (RNAi). Drosophila S2 cells were grown in monolayers for 5 d in the absence or presence of double-stranded (ds) RNA against Drosophila FLASH and used to prepare small-scale nuclear extracts. A nuclear extract from untreated cells contained readily detectable amounts of FLASH (Fig. 2C, lane 1) and was proficient for processing of the dH3 histone pre-mRNA (Fig. 2D, lane 2). Recombinant N-terminal FLASH (FN-ter, amino acids 1–178) increased the efficiency of cleavage approximately twofold in this nuclear extract (Fig. 2D, lane 3; Fig. 2F), whereas the FΔ77 FLASH mutant typically had on average a minor inhibitory effect (Fig. 2D, lane 4; Fig. 2F). It is unclear why the FN-ter and FΔ77 FLASH proteins affected processing only in the small-scale nuclear extracts from S2 cells and showed no activity when added to large-scale nuclear extracts prepared from suspension Kc cells (Fig. 2B, lanes 4, 5). S2 cells adapted to grow in suspension cultures yielded large-scale nuclear extracts that behave in the same manner as Kc nuclear extracts (data not shown). One possible explanation is that extracts from monolayer cells differ from extracts from suspension cells in the abundance of FLASH and other processing factors and their accessibility for recombinant FLASH.
FIGURE 5.
Composition of Drosophila U7 snRNP. (A) The material precipitated from a Kc nuclear extract with the anti-Ars2 (lane 2) or anti-FLASH (lane 3) antibodies was tested for the presence of indicated processing factors using specific antibodies. Lane 1 contains 10% of the input extract used for immunoprecipitation. CR indicates a protein cross-reacting with the anti-FLASH antibody. (B) Proteins precipitated with the anti-FLASH antibody from 2.0 mL of a Kc nuclear extracts were separated in an SDS/polyacrylamide gel, detected by silver staining, and identified by mass spectrometry. (C) Proteins immunoprecipitated from a Kc nuclear extract using indicated antibodies were analyzed for the presence of FLASH using the anti-FLASH antibody. Mock antibody (lane 4) was directed against a protein unrelated to 3′-end processing of histone pre-mRNA (CG9958). Lane 1 contains 2% of the input extract used in immunoprecipitation. (D) Binding of processing factors to various RNAs containing biotin for subsequent purification on streptavidin Sepharose beads. Lane 1 contains 5% of the input extract used in each binding experiment. One of the blots was simultaneously probed with the anti-FLASH and anti-SLBP antibodies; CR indicates two FLASH degradation products that migrate immediately above full-length SLBP. SLBP is virtually undetectable in this lane and highly enriched in the material purified by the Biot-SL RNA. (E) The material bound to the Biot-αU7 oligonucleotide was analyzed by Western blotting for the presence of indicated proteins. Lane 1 contains 1% of the input extract used in the binding experiment. (F,G) Major proteins bound to either the Biot-αMock (lane 1) or Biot-αU7 (lane 2) oligonucleotides were separated in 8% (F) or 12% (G) SDS/polyacrylamide gels, stained with silver, identified by mass spectrometry, and are listed to the right.
The nuclear extract prepared from S2 cells treated with dsRNA against FLASH contained a significantly reduced level of FLASH (Fig. 2C, lane 2), whereas the amounts of SLBP and the U7 snRNP (Fig. 2C, lane 2; data not shown), the two other critical processing factors, were unchanged. This extract alone poorly processed histone pre-mRNA (Fig. 2D, lane 5), and the addition of the recombinant FN-ter protein (amino acids 1–178) increased its activity more than fivefold, to a level that was higher than that of the control extract (Fig. 2D, cf. lanes 2 and 7; Fig. 2F). The stimulation of the processing efficiency by the N-terminal FLASH was highly reproducible and observed with every nuclear extract independently depleted of FLASH (Fig. 2F). The FΔ77 FLASH (amino acids 78–178) lacking the LDIY mutant was inactive (Fig. 2D, lane 6; Fig. 2F). To determine whether the lack of activity is caused by the absence of the LDIY motif, we created the LDIY-4A point mutant in which this motif was replaced by four alanines. This protein had only a residual activity in processing (Fig. 2E, lane 4; Fig. 2F), demonstrating that the LDIY motif in FLASH is critical for 3′-end processing of histone pre-mRNAs in Drosophila and supporting the notion that it is a functional counterpart of the LDLY motif in mammalian FLASH.
The interaction between a complex of the N-terminal FLASH and Lsm11 and Drosophila polyadenylation factors
We next tested whether various recombinant N-terminal FLASH fragments (GST fusion) bound to the N-terminal Lsm11 (MBP fusion) can interact with polyadenylation factors in Drosophila nuclear extracts, as previously described for the mammalian proteins (Yang et al. 2013). The bound proteins were purified on glutathione beads and analyzed by Western blotting using antibodies against the three Drosophila polyadenylation factors known to be essential for 3′-end processing of histone pre-mRNAs in Drosophila and mammalian cells: symplekin, CPSF100, and CPSF73 (Sullivan et al. 2009). The level of all recombinant protein absorbed on glutathione beads was carefully monitored by staining with Coomassie Blue (data not shown).
Initially, we used small-scale nuclear extracts prepared from untreated and FLASH-depleted S2 cells since their processing activity was increased by recombinant FLASH. A complex of the N-terminal regions of Drosophila FLASH (FN-ter, amino acids 1–178) and Lsm11 (amino acids 1–153) interacted with readily detectably amounts of symplekin and CPSF100 in extracts from both untreated (Fig. 3A, lane 3) and FLASH-depleted S2 cells (Fig. 3A, lane 7). Among the interacting proteins we also identified CPSF73, although its detection on Western blots was obscured by a bacterial protein present in the preparation of recombinant Lsm11 that strongly cross-reacted with the anti-CPSF73 antibody (see Fig. 3C, lanes 3, 4; data not shown). Importantly, the interaction of all these polyadenylation factors was nearly abolished by deleting the first 77 amino acids from the FN-ter FLASH (FΔ77) (Fig. 3A, lanes 4, 8).
FIGURE 3.

A complex of the N-terminal FLASH and Lsm11 binds Drosophila polyadenylation factors. (A–C) The interaction of indicated recombinant FLASH variants bound to the N-terminal Lsm11 with polyadenylation factors in S2 (A) or Kc (B,C) nuclear extracts. Nuclear proteins that interact with the FLASH/Lsm11 complex were collected on glutathione (GSH) beads via the GST tag attached to FLASH and analyzed by Western blotting using specific antibodies. Material bound to GSH beads in the absence of recombinant proteins was analyzed in lanes 2 and 6 (A) and lane 2 (B,C). Input (15%) of the nuclear extract is shown in lane 1 of each panel. The arrows in C indicate a contaminant from the recombinant Lsm11 protein that cross-reacts with the antibody against CPSF73.
Of multiple large-scale Kc nuclear extracts tested by this approach, only some behaved in the same fashion as the small-scale extracts prepared from S2 cells. In these Kc extracts, the FN-ter/Lsm11 complex, but not the mutant FΔ77/Lsm11 complex, interacted with symplekin, CPSF100 (Fig. 3B, cf. lanes 4 and 5) and CPSF73 (see also Fig. 3C, lane 3; data not shown). Consistent with our previous results with the mammalian system, the N-terminal FLASH alone was less efficient in binding the polyadenylation factors than its complex with Lsm11 (Fig. 3B, cf. lanes 3 and 4). To determine whether the FΔ77 FLASH mutant complexed with the N-terminal Lsm11 is unable to bind the polyadenylation factors primarily due to deletion of the LDIY motif rather than any other sequence in the first 77 amino acids of FLASH, we used the LDIY-4A mutant. The interaction of the LDIY-4A/Lsm11 complex with symplekin, CPSF100, and CPSF73 in the same Kc nuclear extract was significantly reduced (Fig. 3C, cf. lanes 3 and 4), demonstrating that the LDIY motif plays an equivalent function to the LDLY motif in mammalian FLASH and is essential for recruiting the CPSF73 endonuclease and other polyadenylation factors to the U7 snRNP.
Does Drosophila Ars2 interact with FLASH and play a role in 3′-end processing of histone pre-mRNAs?
In mammalian cells, FLASH interacts with Ars2, and this interaction is required for S-phase progression, expression of replication-dependent histone mRNAs, and efficient localization of FLASH to Histone Locus Bodies (HLBs) (Kiriyama et al. 2009). In addition, Ars2 is essential for cell proliferation (Gruber et al. 2009), and depletion of Ars2 from human cells increases the number of aberrant replication-dependent histone mRNAs that end with a poly(A) tail (Gruber et al. 2012), suggesting that Ars2 may cooperate with FLASH in U7-dependent 3′-end processing.
We tested whether Drosophila Ars2 and FLASH form a stable complex in Drosophila cells. We generated a rabbit antibody against the N-terminal portion of Ars2 to isolate complexes containing Ars2 from Kc nuclear extracts and identify interacting proteins by mass spectrometry. As determined by Western blotting, the antibody precipitated Ars2 but not FLASH (Fig. 4A, lane 3). Mass spectrometry analysis of a large-scale precipitation with the anti-Ars2 antibody additionally identified large amounts of Cap Binding Protein 80 (CBP80) in the precipitate (Fig. 4B). This 80-kDa subunit of the nuclear RNA Cap Binding Complex (CBC) and the smaller 20-kDa subunit interact with tagged versions of Ars2 in human (Gruber et al. 2009) and Drosophila cells (Sabin et al. 2009), and they likely interact with Ars2 in plants (Laubinger et al. 2008). The anti-Ars2 precipitate also contained homologs of other mammalian proteins, including the helicase Mtr4 (Fig. 4B), shown to exist in a complex with Ars2 in mammalian cells (Lubas et al. 2011). However, both Western blotting (Fig. 4A, lane 3; Fig. 5A, lane 2) and mass spectrometry (Fig. 4B) demonstrated that the antibody against Ars2 failed to coprecipitate FLASH, suggesting that these two proteins do not form a stable complex in Drosophila cells. The anti-Ars2 antibody also did not precipitate Lsm11 (Fig. 5A, lane 2). The conclusion that FLASH and Ars2 are not part of the same complex was also supported by a reciprocal experiment with anti-FLASH antibodies directed against the N-terminal (Fig. 4A, lane 2) and C-terminal regions of Drosophila FLASH (data not shown). Finally, no Ars2 was detected in the material precipitated by an anti-Lsm11 antibody, although this antibody precipitated FLASH (Fig. 4A, lane 5). The inability of the anti-FLASH and anti-Ars2 antibodies to coprecipitate Ars2 and FLASH, respectively, was highly reproducible and observed for a broad range of nuclear extracts, including those prepared from S2 cells grown in monolayers (Fig. 4A, lanes 6–9) or in high-volume suspension cultures (Supplemental Fig. S3). Collectively, these results suggest that in Drosophila, Ars2 does not interact with FLASH and is not part of the U7 snRNP.
FIGURE 4.
Analyzing the involvement of Drosophila Ars2 in 3′-end processing of histone pre-mRNAs. (A) Immunoprecipitation of Ars2 and FLASH from nuclear extracts prepared from freshly harvested Kc cells (lanes 1–5) or S2 cells grown in monolayers (MS, lanes 6–9). The antibodies used in the experiment are indicated at the top of each lane. The anti-Mock antibody is directed against 3′hExo, a human protein that has no clear homolog in Drosophila. The inputs are shown in lanes 1 and 6. (B) Proteins immunoprecipitated by the anti-Ars2 antibody from a Kc cell nuclear extract were detected by silver staining and analyzed by mass spectrometry. The major identified proteins are listed on the left. (C) Depletion of endogenous Ars2 in S2 cells by a specific dsRNA. S2 cells were treated for 5–6 d with dsRNA against various proteins and tested for depletion of Ars2 using the anti-Ars2 antibody. CR indicates a protein of ∼70 kDa that cross-reacts with the antibody and served as a loading control. Mock1 (CG7769) and Mock2 (CG8443) are two proteins unrelated to 3′-end processing of histone pre-mRNAs. (D) Northern blot analysis of endogenous histone H3 mRNA in S2 cells treated with dsRNAs shown in panel C. Species ending with the canonical stem–loop (dH3 SL) migrate as a single band. Polyadenylated species are longer and, due to the heterogeneity of the poly(A) tail, migrate as a smear. Drosophila 7SK RNA served as a loading control.
Since it was possible that Drosophila Ars2 functions in processing of histone pre-mRNAs via interacting with a different processing component, we next reduced the endogenous level of Ars2 in S2 cells by RNA interference (RNAi) and tested whether this treatment results in in vivo production of replication-dependent histone mRNAs that end with a poly(A) tail rather than the stem–loop. This phenotype was readily observed in S2 cells depleted of FLASH (Yang et al. 2009a) and other factors essential for U7-dependent 3′-end cleavage of histone pre-mRNAs in Drosophila, including symplekin, CPSF100, and CPSF73 (Wagner et al. 2007; Sullivan et al. 2009; Yang et al. 2009a) and was also observed in Ars2-depleted mammalian cells (Gruber et al. 2012). We monitored the extent of depletion by Western blotting, using the antibody against the N-terminal portion of Ars2 (Fig. 4C). The antibody detects a single band at ∼140 kDa in the whole cell lysate from untreated S2 cells, and the corresponding band is virtually missing in the lysate from S2 cells depleted of Ars2 by treatment with dsRNA (Fig. 4C, lane 6). RNAi with dsRNA specific for Drosophila FLASH or a number of proteins unrelated to 3′-end processing of histone pre-mRNA had no effect on the abundance of Ars2 in S2 cells (Fig. 4C, lanes 2–5).
Cells depleted of FLASH produced longer polyadenylated dH3 histone mRNA resulting from a defect in the U7-dependent 3′-end processing (Yang et al. 2009a) (Fig. 4D, lane 1). Importantly, depletion of Ars2 had no visible effect on this process (Fig. 4D, cf. lanes 1 and 6), although it resulted in a noticeable reduction of the overall level of the dH3 mRNA relative to the level of Drosophila 7SK RNA, a transcript generated by RNA Pol III. The same general reduction in expression of replication-dependent histone mRNAs was previously observed in Ars2-depleted human cells (Kiriyama et al. 2009; Gruber et al. 2012) and is also seen in Drosophila S2 cells depleted of mxc (Fig. 4D, lane 3), the Drosophila ortholog of NPAT (White et al. 2011). NPAT is specifically required for transcription of histone genes in mammalian cells (Ma et al. 2000).
Since Ars2 may be essential for transcription of histone genes, thereby making it difficult to assess its role in 3′-end processing of histone pre-mRNAs in vivo, we prepared nuclear extracts from Ars2-depleted S2 cells. In contrast to FLASH-depleted S2 extracts, extracts prepared from Ars2-depleted cells were consistently very active in processing the dH3 pre-mRNA (data not shown), further arguing that Ars2 is not a component of the Drosophila U7-dependent processing machinery.
Nuclear proteins associated with Drosophila FLASH
To better understand the role of FLASH in 3′-end processing of histone pre-mRNAs and to identify its binding partners in Drosophila nuclear extracts, we carried out a series of immunoprecipitations using antibodies against FLASH and other Drosophila processing factors. The antibody against the N-terminal portion of Drosophila FLASH efficiently precipitates U7 snRNA (Yang et al. 2009a) and Lsm11 (Fig. 5A, lane 3), indicating that a substantial fraction of the endogenous U7 snRNP is stably associated with FLASH. Consistently, an anti-Lsm11 antibody coprecipitates Drosophila FLASH from Kc nuclear extracts (Fig. 4A, lane 5).
We next investigated whether the endogenous Drosophila FLASH is associated with any of the polyadenylation factors known to be components of the mammalian HCC. As shown by Western blotting, the anti-FLASH antibody, but not the anti-Ars2 antibody, precipitated two polyadenylation factors that are essential for 3′-end processing of Drosophila histone pre-mRNAs: symplekin and CPSF73 (Fig. 5A, lane 3). To confirm this result and to identify other proteins associated with Drosophila FLASH, we scaled up precipitation with the anti-FLASH. In addition to FLASH, silver staining revealed several specific protein bands in the anti-FLASH precipitate that were identified by mass spectrometry as Drosophila orthologs of CPSF160 (CG10120), symplekin (CG2097), CPSF100 (CG1957), and CPSF73 (CG7698), which comigrated with the 70-kDa heat shock protein (HSC70, CG4262) (Fig. 5B). The precipitate also contained peptides from the Drosophila ortholog of WDR33 (CG1109), although this protein was not visible as a separate band on the silver-stained gel. All these proteins were either undetectable or detected with very low scores in the anti-Ars2 precipitate (Fig. 5A, lane 2; data not shown). Consistent with FLASH and symplekin forming a common complex, an antibody against Drosophila symplekin (Fig. 5C, lane 3), but not a mock antibody directed against an unrelated Drosophila protein (Fig. 5C, lane 4), coprecipitated FLASH.
We carried out additional immunoprecipitation experiments with the anti-FLASH antibody and analyzed the precipitated material in higher-percentage gels to look for smaller proteins (data not shown). These experiments failed to identify any additional polyadenylation factors associated with FLASH, including the Drosophila ortholog of CPSF30 (CG3642), which based on its amino acid sequence is expected to migrate at ∼30 kDa. The analysis did not include the area of the gel near the 50-kDa size marker that contained large amounts of the immunoglobulin heavy chain. Altogether, these results demonstrate that FLASH and Lsm11 in Drosophila nuclear extracts are tightly associated with each other and suggest that they recruit several polyadenylation factors to the U7 snRNP.
Affinity purification of Drosophila U7 snRNP from Kc nuclear extracts
We next affinity-purified Drosophila U7 snRNP from Kc nuclear extracts using a 2′-O-methyl oligonucleotide (Biot-αU7) complementary to the 5′ end of the Drosophila U7 snRNA (Dominski et al. 2005). This oligonucleotide contains biotin at the 3′ end, allowing adsorption of the associated complexes on streptavidin beads.
We initially carried out a pilot experiment with a small amount of a Kc nuclear extract (500 µL) and determined that the Biot-αU7 oligonucleotide binds both specifically and efficiently to the U7 snRNP. The material purified by this method was highly enriched in Lsm11 and also contained readily detectable amounts of FLASH (Fig. 5D, lane 3), confirming that FLASH is an integral part of the U7 snRNP. As expected, the material did not contain SLBP, which could be specifically purified by the 31-nt Biot-SL RNA, containing the sequence of the stem–loop and biotin on the 5′ end (Fig. 5D, lane 4). A nonspecific 2′-O-methyl oligonucleotide complementary to the first 17 nt of the human U7 snRNA and tagged with biotin at the 3′ end (Biot-αMock) bound neither the U7 snRNP nor SLBP (Fig. 5D, lane 2). Importantly, the material purified by the Biot-αU7 also contained detectable amounts of symplekin, CPSF100, and CPSF73 but lacked CstF50 (Fig. 5E, lane 2). These results strongly support the conclusion from the immunoprecipitation experiments that the Drosophila U7 snRNP associates with FLASH and a complex of polyadenylation factors that resembles the mammalian HCC, which also lacks the 50-kDa subunit of CstF (Yang et al. 2013).
To determine whether additional proteins associate with the U7 snRNP, we used larger amounts of both the Biot-αU7 and Kc nuclear extract (1.0 µg and 5.0 mL, respectively) and analyzed the purified material by mass spectrometry. Nuclear proteins bound to the Biot-αU7 were collected on streptavidin beads, separated on an 8% SDS/polyacrylamide gel, and visualized by silver staining (Fig. 5F, lane 1). Control purification was carried out with the Biot-αMock oligonucleotide (Fig. 5F, lane 2). Several clearly visible silver-stained protein bands were detected only in the lane containing material bound to the Biot-αU7 oligonucleotide (Fig. 5F, lane 1). These bands and other regions of the gel were analyzed by mass spectrometry (Fig. 5F, right panel). The heavily stained band near the top of lane 1 that is completely missing in lane 2 was identified as FLASH (Fig. 5F, band 2). The less-intense band migrating immediately above FLASH (band 1) was identified as Drosophila CPSF160, whereas the bands migrating faster than FLASH were identified as symplekin (band 3), CPSF100 (band 4), WDR33 (band 5), and CstF64 (band 9) (Fig. 5F). Lane 1 also contained CPSF73, although it was masked by massive amounts of another protein identified as the Drosophila ortholog of Insulin-like Growth Factor 2 mRNA Binding Protein 1 (IGF2BP1, CG1691). This protein likely directly interacts with the Biot-αU7 oligonucleotide (see below). All the polyadenylation factors purified by the Biot-αU7 oligonucleotide were undetectable by mass spectrometry in lane 2, whereas IGF2BP1 was present in this lane in trace amounts.
To analyze smaller proteins purified via the Biot-αU7, two-thirds of the same material was separated on a 12% SDS/polyacrylamide gel (Fig. 5G, lane 1) and compared with the material bound to the Biot-αMock oligonucleotide (Fig. 5G, lane 2). Two silver-stained bands were clearly visible at ∼30 and 22 kDa in lane 1, and they were absent from lane 2. These bands were identified as Drosophila Lsm11 and SmB, the two largest components of the U7-specifc Sm ring, confirming that the Biot-αU7 oligonucleotide is very efficient and specific in purifying the Drosophila U7 snRNP. Independent experiments also identified two smaller members of the ring in the material purified by the Biot-αU7 oligonucleotide: Drosophila Lsm10 and SmD3 (data not shown).
The material purified via the Biot-αU7 contained large amounts of two major forms of the Drosophila ortholog of Insulin-like Growth Factor 2 mRNA Binding Protein 1 (IGF2BP1) and several of its apparent degradation products. The high abundance of IGF2BP1 greatly exceeded the amount of Lsm11 and other core U7 snRNP components in the precipitate, suggesting that this protein is not part of the U7 snRNP. Subsequent studies and multiple immunoprecipitation experiments with the anti-FLASH antibody support the notion that IGF2BP1 directly binds the excess Biot-αU7 oligonucleotide and plays no role in 3′-end processing of histone pre-mRNAs (data not shown). Lane 1 also contained a band migrating at ∼60 kDa that was identified as a Drosophila homolog of the mammalian p62 helicase (CG10279). This protein was also present in the negative lane, and hence it likely nonspecifically binds 2′-O-methyl oligonucleotides. Altogether, the results of both immunoprecipitation and affinity purification experiments combined with mass spectrometry demonstrate that Drosophila U7 snRNP has a composite structure, and in addition to the core components of its Sm ring contains FLASH and at least six polyadenylation factors.
Characterization of processing complexes formed on histone pre-mRNA in Drosophila nuclear extracts
We next tested whether processing complexes formed on Drosophila pre-mRNA in Drosophila nuclear extracts contain the same subset of polyadenylation factors that associate with the Drosophila U7 snRNP. A 63-nt dH3 pre-RNA containing all necessary processing elements (the SL and HDE) and biotin at the 5′ end (Biot-dH3) was incubated for 20 min at room temperature with a Kc nuclear extract (750 µL), and factors that bound to this substrate were collected on streptavidin beads. To prevent cleavage, which would result in release of the U7 snRNP from the complexes, the reaction was supplemented with 0.1% NP-40. This mild detergent at low concentrations blocks processing in Drosophila nuclear extracts (Fig. 6A, lanes 3, 4), as it does in mammalian nuclear extracts (Yang et al. 2009b, 2013). However, processing complexes that assembled in the Kc nuclear extract under these conditions, while containing FLASH, SLBP, and U7 snRNP (as determined by the presence of Lsm11), lacked detectable amounts of polyadenylation factors (data not shown). Purification of Drosophila U7 snRNP by either the Biot-αU7 oligonucleotide (Fig. 6B, lane 3) or anti-FLASH antibody (data not shown) in the presence of NP-40 demonstrated that this detergent destabilized the interaction between U7 snRNP and polyadenylation factors, including symplekin (Fig. 6B, cf. lanes 2 and 3). This is the most likely cause of the toxicity of NP-40 on 3′-end processing in both Drosophila and mammalian nuclear extracts.
FIGURE 6.
Composition of processing complexes assembled on the dH3 pre-mRNA. (A) 3′-End processing of the dH3 pre-mRNA in a Kc nuclear extract in the presence of indicated concentrations of NP40 (lanes 3, 4). The input alone and processing in the absence of NP-40 is shown in lanes 1 and 2, respectively. (B) The Drosophila U7 snRNP was affinity-purified from a Kc nuclear extract using the Biot-αU7 oligonucleotide either in the absence (lane 2) or presence (lane 3) of NP-40 and subsequently probed for the presence of indicated processing factors. Lane 1 contains 1% of the input extract used for the purification. Lsm11 is virtually undetectable in this lane and highly enriched in the material purified by the oligonucleotide. (C) Binding of SLBP and Lsm11 to indicated biotinylated RNAs. Lane 1 contains 5% of the input Kc extract used in each binding experiment. Lane 2 shows the background binding to streptavidin Sepharose beads in the absence of any biotinylated RNA. (D) Binding of processing factors from a Kc nuclear extract to the Biot-dH3 pre-mRNA (lanes 3–6), either in the absence of any RNA competitor (lane 3) or in the presence of indicated RNAs (lanes 4–6). Lane 1 contains 5% of the input Kc extract used in each binding experiment. SLBP and Lsm11 are virtually undetectable in this lane and highly enriched in the material purified by the Biot-dH3 pre-mRNA. Lane 2 shows the background binding to streptavidin Sepharose beads in the absence of the Biot-dH3 pre-mRNA. (E) Binding of FLASH, symplekin, and SLBP to indicated biotinylated RNAs. Lane 1 contains 2.5% of the Kc extract used in each binding experiment. (F) Protein factors from a Kc nuclear extract bound to the Biot-dH3 pre-mRNA in the absence of any RNA competitor (lane 1) or in the presence of indicated RNAs (lanes 2, 3). The bound proteins were separated on a 12% SDS/polyacrylamide gel and stained with silver. The indicated bands and remaining portions of each lane were analyzed by mass spectrometry; the identified processing factors are listed to the right. The mass spectrometry results obtained for lane 3 were virtually identical to those obtained for lane 1 and are not shown.
Instead of using NP-40, we significantly shortened the incubation time to capture processing complexes at early stages of assembly prior to cleavage of the substrate, in a manner similar to methods previously used for canonical pre-mRNAs containing the AAUAAA sequence (Veraldi et al. 2000; Shi et al. 2009). After 4–5 min of incubation in a nuclear extract, the Biot-dH3 pre-mRNA associated with both SLBP and the U7 snRNP, as determined by the presence of Lsm11 (Fig. 6C, lane 3). Thus, at least a fraction of the input RNA remains unprocessed under these conditions. The Biot-SL and the Biot-αU7 oligonucleotides bound only to SLBP and U7 snRNP, respectively (Fig. 6C, lanes 4, 5), demonstrating that binding of the two factors to sites flanking the cleavage site in the Biot-dH3 is specific. We next carried out larger-scale experiments, either in the absence or in the presence of various RNA competitors (Fig. 6D). Western blots with available antibodies against various processing factors demonstrated that the Biot-dH3 pre-mRNA in the absence of RNA competitors assembled into a complex containing SLBP, FLASH, the U7 snRNP (as determined by the presence of Lsm11), and the three essential polyadenylation factors we assayed: CPSF73, CPSF100, and symplekin (Fig. 6D, lane 3). The presence of 2.5 µM αU7 oligonucleotide, which blocks the 5′ end of the U7 snRNA and abolishes the recruitment of the U7 snRNP to the substrate, eliminated all these proteins except SLBP from the complex (Fig. 6D, lane 6). Thus, the composite U7 snRNP carrying FLASH and polyadenylation factors is directly recruited to histone pre-mRNA through the base-pairing interaction between the U7 snRNA and the HDE.
In the presence of 1 µM WT SL RNA that sequesters SLBP, binding of SLBP to the Biot-dH3 pre-mRNA was eliminated. However, the association of the composite U7 snRNP, including FLASH and the three polyadenylation factors, with the HDE was unaffected (Fig. 6D, lane 4). Note that this concentration of the WT SL competitor is sufficient to block the cleavage reaction (Fig. 1C, lane 3), whereas the Mut SL RNA (Fig. 1E) at the same and higher concentrations has no effect on 3′-end processing (Fig. 1C, lane 5) and does not prevent binding of SLBP to the Biot-dH3 pre-mRNA (Fig. 6D, lane 5). The failure of the WT SL competitor to reduce the amount of the U7 snRNP in the complex assembled on the dH3 pre-mRNA was unexpected since previous models based on processing in mammalian and Drosophila nuclear extracts suggested that SLBP primarily functions by stabilizing the interaction of the U7 snRNP with the HDE in histone pre-mRNAs (Dominski et al. 1999, 2005).
To confirm this surprising result by a different approach, we assembled processing complexes on a derivative of the Biot-dH3 pre-mRNA that lacked 16 nt encompassing the stem–loop structure. The HDE of this new substrate designated Biot-dH3ΔSL was identical to that in the Biot-dH3 pre-mRNA. In addition to the dH3ΔSL RNA, we also used the mouse-specific Biot-H2a-614 pre-mRNA (Yang et al. 2009b). This substrate contains the stem–loop structure that normally interacts with Drosophila SLBP, but its HDE has only a limited ability to base-pair with the Drosophila U7 snRNA. Due to this reduced compatibility between the HDE and the Drosophila U7 snRNP, the H2a-614 pre-mRNA is only poorly processed in Drosophila nuclear extracts (Dominski et al. 2002, 2005).
As expected, the Biot-dH3ΔSL substrate, in contrast to the Biot-dH3 and Biot-H2a-614 pre-mRNAs, was unable to bind SLBP from the Kc nuclear extract (Fig. 6E, cf. lane 4 with lanes 2 and 3). Importantly, consistent with our conclusion based on using the stem–loop competitor, the absence of the stem–loop structure in the Biot-dH3ΔSL substrate and SLBP in the processing complex did not prevent the interaction between the composite U7 snRNP and the HDE. Both FLASH and symplekin were detected in processing complexes assembled on the Biot-dH3ΔSL, and their amount was comparable to that detected in the complexes assembled on the Biot-dH3 pre-mRNA (Fig. 6E, cf. lanes 3 and 4). The mouse-specific Biot-H2a-614 pre-mRNA interacted with the Drosophila U7 snRNP only weakly, consistent with the nature of the HDE in this substrate and its inability to form a strong duplex with the U7 snRNA (Fig. 6E, lane 2). We conclude that the interaction of the Drosophila SLBP with the stem–loop structure in histone pre-mRNA has no major effect on the recruitment of the U7 snRNP to the HDE.
Preparative scale purification (5.0 mL of Kc nuclear extract per each reaction) was next performed to analyze proteins associated with the Biot-dH3 pre-mRNA by mass spectrometry. The control reaction contained no RNA competitor, and the two other reactions contained either the αU7 oligonucleotide (1.25 µM) to block the Drosophila U7 snRNP or the WT SL RNA (1 µM) to sequester SLBP in the extract (Fig. 6F). On silver-stained gels, the three samples shared several bands, and most of these likely represented proteins nonspecifically bound to streptavidin beads and/or excess of the Biot-dH3 RNA. Compared with the control reaction (Fig. 6E, lane 1), the WT SL RNA competitor resulted in a disappearance of only one major protein band that corresponds to SLBP (Fig. 6F, lane 3). The reaction supplemented with the αU7 oligonucleotide lacked several proteins migrating in the range of ∼50–200 kDa, whereas the amount of SLBP was unchanged (Fig. 6F, lane 2), consistent with the Western data. Mass spectrometry identified these proteins (from the largest to the smallest) as CPSF160, FLASH, symplekin, CPSF100, WDR33, and CstF64 (Fig. 6F, right panel). Lanes 1 and 3, but not lane 2, also contained CPSF73 (band 7), although this polyadenylation factor was masked by an abundant protein present in all three lanes.
The material purified from the Kc nuclear extract via the Biot-dH3 pre-mRNA also contained CstF77 (CG17170) (Fig. 6F, lane 1, band 6). Results from mass spectrometry analysis suggest that the association of this protein with the Biot-dH3 substrate was only partially reduced by the presence of the αU7 oligonucleotide (Fig. 6F, right panel). CstF77 was not detected in the material purified by the anti-FLASH antibody or the Biot-αU7 oligonucleotide (see above), collectively arguing that this polyadenylation factor associates with the RNA substrate in a nonspecific manner. We also note that CstF77 was previously identified as a contaminant of the mammalian U7 snRNP affinity-purified from mouse nuclear extracts (Yang et al. 2013).
Mass spectrometry analysis of the remaining gel sections failed to identify any other major differences in protein composition between the three lanes. We conclude that the composite U7 snRNP containing FLASH and several polyadenylation factors is recruited to Drosophila histone pre-mRNA for 3′-end processing. Strikingly, SLBP does not play any detectable role in the recruitment of this composite U7 snRNP, although its binding to the stem–loop in histone pre-mRNA is absolutely required for cleavage.
CstF64 as a component of the Drosophila U7 snRNP
The mammalian HCC that interacts with FLASH and Lsm11 is composed of symplekin, all CPSF subunits, and CstF64 (Yang et al. 2013). The two remaining CstF subunits, CstF50 and CstF77, are conspicuously missing from the HCC. Based on our present studies, the equivalent complex in Drosophila also contains symplekin, most CPSF subunits, and CstF64 as the only component of the CstF subcomplex.
To confirm that CstF64 is part of the Drosophila U7 snRNP and to look more closely at its role in 3′-end processing of histone pre-mRNAs in Drosophila, we generated antibodies against bacterially expressed full-length protein (amino acids 1–437). The antibody recognizes a distinct band on Western blots of Drosophila nuclear extract that migrates at ∼50 kDa (Fig. 7A, lane 1), consistent with the mobility of CstF64 identified by mass spectrometry. We tested the material purified from Kc nuclear extracts via the Biot-αU7 oligonucleotide using the anti-CstF64 antibody and confirmed that it indeed contains CstF64 (Fig. 7A, lane 2). CstF64 was also readily detected in processing complexes assembled on the Biot-dH3 and Biot-dH3ΔSL RNAs (Supplemental Fig. S4, lanes 3, 4). Lower amounts of CstF64 were associated with the Biot-mH2a pre-mRNA, consistent with the limited complementarity of this mouse-specific substrate to the Drosophila U7 snRNA (Supplemental Fig. S4, lane 2). Thus, consistent with the mass spectrometry results, CstF64 is a stable component of the Drosophila U7 snRNP and is recruited together with other polyadenylation factors to histone pre-mRNA for 3′-end processing.
FIGURE 7.
CstF64 is a component of the Drosophila U7 snRNP. (A) Western blot analysis of proteins bound to either the Biot-αU7 (lane 2) or Biot-αMock (lane 3) oligonucleotides using an antibody directed against full-length Drosophila CstF64. Lane 1 contains 2% of the input extract used in the binding experiment. (B) Immunoprecipitation of various polyadenylation factors from a Kc nuclear extract with the anti-CstF64 antibody alone (lane 3) or in the presence of the competing recombinant protein (lane 4). (*) Recombinant CstF64 bound to the antibody and collected on protein A agarose beads. Lane 1 contains 20% of the input extract used for immunoprecipitation, and lane 2 contains proteins of the nuclear extract bound to protein A agarose in the absence of any antibody. Immunoprecipitated proteins were identified by Western blotting using specific antibodies. (C) Immunoprecipitation of various polyadenylation factors from a Kc nuclear extract with the anti-CstF64 antibody alone (lane 3) or in the presence of the competing recombinant protein (lane 4). Lane 1 contains 10% of the input extract used for immunoprecipitation. (D) Proteins immunoprecipitated with the anti-CstF64 antibody (lane 3) or an anti-Mock antibody were separated on a 12% SDS/polyacrylamide gel, silver-stained, and analyzed by mass spectrometry. Lane 1 contains material bound to protein A agarose in the absence of any antibody. (E,F) Proteins precipitated with the anti-CstF64 antibody (lane 1 in both panels) or in the presence of the competing recombinant protein (lane 2 in panel E and lane 3 in panel F) were separated on a 10% (E) or 15% (F) SDS/polyacrylamide gel, silver-stained, and analyzed by mass spectrometry. Immunoprecipitation with the anti-CstF64 antibody using the nuclear extract pre-treated with RNase A is shown in lanes 3 (E) and 2 (F). (*) The CstF64 protein competitor bound to the antibody. The recombinant CstF64 used as a competitor is additionally shown in lane 4 of panel F. (G) Levels of Drosophila CF Im68 in whole cell lysate from untreated S2 cells (lane 1) or cells treated with dsRNA against this factor (CF Im68 KD, lane 2), as determined by Western blotting using an antibody targeted to the N-terminal region of human CF Im68. CR indicates two proteins cross-reacting with the antibody that served as a loading control. (H) Immunoprecipitation of Drosophila CF Im68 from a Kc nuclear extract with the anti-CstF64 antibody (lane 4) or with a nonspecific antibody directed to a protein unrelated to 3′-end processing (lane 3). The material bound to protein A agarose in the absence of any antibody is analyzed in lane 2. Lane 1 contains 10% of the input extract used for immunoprecipitation.
We next tested the ability of the anti-CstF64 antibody to precipitate processing complexes from Kc nuclear extracts. Western blotting demonstrated that the anti-CstF64 antibody is very efficient in precipitating both CstF64 and CstF50 (Fig. 7B, lane 3), indicating that the antibody readily recognizes the entire CstF subcomplex known to consist of these three subunits in mammalian cells (Takagaki et al. 1990). Surprisingly, the anti-CstF64 precipitate also contained large amounts of CPSF73, CPSF100, and symplekin (Fig. 7B, cf. lanes 1 and 3). Precipitation of all these polyadenylation factors was blocked by recombinant CstF64 used as the antigen to generate the antibody (Fig. 7B, lane 4). Importantly, the anti-CstF64 precipitate contained readily detectable amounts of FLASH (both full-length and numerous degradation products) and Lsm11, but not Ars2 (Fig. 7C, lane 3), further confirming that in Drosophila nuclear extracts, a fraction of CstF64 exists in a complex with the U7 snRNP. Note that while at least 10% of Lsm11 was precipitated by the anti-CstF64 (Fig. 7C, lane 3), <1% of the total CstF64 was bound to U7 snRNP (Fig. 7A, lane 2), as expected from the very low amount of U7 snRNP compared with the high abundance of polyadenylation factors.
CstF64 as a component of a supercomplex of cleavage and polyadenylation factors
The ability of the anti-CstF64 antibody to precipitate a large fraction (∼50%) of symplekin and various CPSF subunits (Fig. 7B) suggested that in Drosophila distinct polyadenylation subcomplexes may associate to form a macromolecular entity resembling that previously described in mammalian cells (Takagaki and Manley 2000). To identify additional components of this hypothetical complex, we separated the precipitated proteins on either 8% (Fig. 7D,E) or 15% SDS/polyacrylamide gels (Fig. 7F) and analyzed their identity by mass spectrometry. The anti-CstF64 precipitate (Fig. 7D, lane 3) but not the control anti-Mock precipitate (Fig. 7D, lane 2) contained several proteins that were readily stained with silver. As determined by mass spectrometry, the precipitate contained all three subunits of CstF, although CstF64 and CstF50 were partially masked by the antibody heavy chain and hence not readily visible. We also identified all subunits of CPSF in the anti-CstF64 precipitate, including the smallest CPSF30 (CG3642) that migrates with the expected mobility of ∼30 kDa (Fig. 7F, lane 1). Precipitation of all these proteins was eliminated by blocking the anti-CstF64 antibody with recombinant CstF64 (Fig. 7E, lane 2; Fig. 7F, lane 3). Among CPSF subunits, WDR33 and Fip1 (CG1078) were relatively weakly stained, which may indicate that they are limiting in Drosophila nuclear extracts (Fig. 7D, lane 3; Fig. 7E, lanes 1, 3). Note that Fip1 exhibits much slower electrophoretic mobility than its mammalian counterpart (Kaufmann et al. 2004), consistent with its significantly larger size (701 amino acids vs. 550 amino acids for human Fip1).
Surprisingly, the anti-CstF64 precipitate also contained significant amounts of the Drosophila orthologs of CF Im68 (CG7185) (Fig. 7D, lane 3; Fig. 7E, lane 1) and CF Im25 (CG3689) (Fig. 7F, lane 1). In mammalian cells, these two proteins belong to a third subcomplex, CI Im (Ruegsegger et al. 1996, 1998), which was not previously detected in a tight association with the subunits of CstF or CPSF in the absence of pre-mRNA substrate. Coprecipitation of the CF Im was not affected by pre-treating the nuclear extract with RNase A (Fig. 7E, cf. lanes 1 and 3; Fig. 7F, cf. lanes 1 and 2).
To confirm this unexpected finding, we tested the precipitate with an antibody generated against the first 191 amino acids of human CF Im68. This antibody cross-reacts with the Drosophila CF Im68 (Fig. 7G), likely by recognizing the highly conserved N-terminal RRM-type RNA binding domain. Western blot analysis using this antibody confirmed that the anti-CstF64 antibody, indeed, coprecipitated CF Im68 (Fig. 7H). Thus, Drosophila nuclear extracts contain a large, preassembled 3′-end processing complex consisting of most known polyadenylation factors, with the notable exception of the Drosophila orthologs of the two subunits of CF IIm, Clp1 and Pcf11 (De Vries et al. 2000).
Components of the HCC essential for 3′-end processing of histone pre-mRNAs
In mammalian cells, CstF64 interacts with symplekin, and this interaction is mutually exclusive with the interaction between CstF64 and CstF77 (Takagaki and Manley 2000; Bai et al. 2007) and essential for 3′-end processing of histone pre-mRNAs (Ruepp et al. 2011). Moreover, our in vitro studies with mammalian nuclear extracts suggest that CstF64 and symplekin may function as an adaptor that directly interacts with FLASH and Lsm11 and brings other polyadenylation factors, including the endonuclease CPSF73 and its homolog CPSF100, to the U7 snRNP (Yang et al. 2013). Surprisingly, previous in vivo studies in S2 cells based on using a GFP reporter driven by a histone gene-specific promoter (Wagner et al. 2007) and direct analysis of histone mRNA 3′ ends (Sullivan et al. 2009) led to the conclusion that in Drosophila only CPSF73, CPSF100, and symplekin are essential for histone pre-mRNA processing, whereas CstF64 and several other polyadenylation factors are dispensable.
We reasoned that CstF64 and some other proteins, in addition to playing essential roles in 3′-end processing, might also be absolutely required for transcription in vivo. This possibility is supported by the known association of polyadenylation factors with the promoter region of protein-encoding genes (McCracken et al. 1997; Proudfoot et al. 2002; Calvo and Manley 2003; Zorio and Bentley 2004; Bentley 2005). Depletion of these factors instead of activating GFP expression as a result of misprocessing could primarily result in general transcriptional silencing of genes transcribed by RNA Pol II (including the reporter gene), as recently demonstrated for Pcf11 (Mapendano et al. 2010; Andersen et al. 2013). We therefore constructed a reporter gene that relies on a promoter derived from the Drosophila U7 snRNA gene. snRNA genes associate with a distinct set of transcriptional factors, and primary transcripts generated from these genes (pre-snRNAs) are cleaved at the 3′ end by a distinct machinery termed Integrator that shares no common components with the two types of 3′-end processing machineries operating on pre-mRNAs (Baillat et al. 2005; Egloff et al. 2008; Ezzeddine et al. 2011).
The transcribed region of the new reporter starts with a 54-nt upstream portion of the U7 snRNA (Fig. 8A). The construct is lacking the last 17 nt of the U7 snRNA, including most of its terminal stem–loop structure, and the 3′ box, the two sequence elements required for 3′-end processing of pre-snRNAs via the Integrator. The snRNA region is followed by a short open reading frame for Drosophila histone H3, the two necessary signals for the U7-dependent processing, the coding region for GFP, and all necessary polyadenylation sequence elements, as previously described (Wagner et al. 2007). The transcript encodes a protein of 350 amino acids, with the last 239 residues corresponding to GFP, unless it is cleaved downstream from the stem–loop by the U7-dependent machinery (Fig. 8A).
FIGURE 8.

A GFP reporter driven by an snRNA gene promoter. (A) A schematic structure of the GFP reporter gene used in the experiment. (Vertical arrows) The two potential cleavage sites, one cleaved by the U7-dependent processing machinery and the other by the cleavage and polyadenylation machinery. Two alternative proteins can be expressed from the reporter gene, both starting at ATG, with the longer protein containing GFP. (B) Fluorescent microscopy images of S2 cells expressing GFP after depletion of indicated processing factors. (C,D) Expression of selected processing factors in lysates of S2 cells depleted (KD) of indicated proteins, as determined by Western blotting using specific antibodies. CR indicates cross-reacting protein interacting with antibodies against CPSF73 (C) and FLASH (D) that served as the loading control. (E) Expression of CstF64 and GFP in cells depleted (KD) of indicated processing factors. Note that efficient depletion of CstF64 (lane 2, top panel) resulted in no detectable expression of GFP (lane 2, bottom panel).
When expressed either stably or transiently in S2 cells, the snRNA-driven reporter showed no expression of GFP, likely resulting from efficient cleavage at the U7-dependent site prior to reaching the GFP coding region (Fig. 8B). Indeed, following 3 d of treatment with double-stranded RNAs against FLASH, S2 cells exhibited strong fluorescence (Fig. 8B) and expressed GFP that was readily detectable on Western blots (Fig. 8E, lane 5). Depletion of the presumptive catalytic component of Drosophila Integrator, Int9, did not result in any detectable activation of GFP expression (data not shown). Thus, the transcript generated from the reporter gene under the control of the snRNA-type promoter is exclusively processed by the U7-dependent processing machinery.
We used this reporter gene to screen CstF64 and other polyadenylation factors that associate with the Drosophila U7 snRNP for their role in 3′-end processing of histone pre-mRNAs. In agreement with the previous reports, depletion of CPSF73, CPSF100, and symplekin resulted in a strong gain of fluorescence by S2 cells (Fig. 8B), and this was paralleled by readily detectable GFP expression on Western blots (Fig. 8E, lanes 3, 4; data not shown), further confirming that these three polyadenylation factors are essential for 3′-end processing of histone pre-mRNAs. Depletion of CstF64 (Fig. 8B,E, lane 2) and the two remaining components of the HCC found in the complex with the Drosophila U7 snRNP, WDR33 and CPSF160 (data not shown), had no effect on expression of GFP from the reporter gene. We also did not observe any expression of GFP upon depleting Drosophila Ars2 (data not shown).
DISCUSSION
We recently showed that in mammalian nuclear extracts supplemented with the bacterially expressed N-terminal portion of FLASH, a subset of polyadenylation factors that we refer to as the Histone Cleavage Complex (HCC) associates with the U7 snRNP. The association depends on the interaction between FLASH and Lsm11, the largest protein of the U7-specific Sm ring (Yang et al. 2013). The mammalian HCC contains symplekin, all six CPSF subunits, and CstF64, but lacks the two remaining CstF subunits. This composite U7 snRNP, containing the CPSF73 endonuclease among other components of the HCC, is subsequently recruited to the Histone Downstream Element (HDE) in histone pre-mRNA for the cleavage reaction (Yang et al. 2013). FLASH is present at a very low level in mammalian cells and is highly susceptible to proteolysis during preparation of nuclear extracts, likely explaining the failure of initial studies to detect its association with U7 snRNP and polyadenylation factors (Smith et al. 1991; Pillai et al. 2001).
FLASH and U7 snRNP are relatively abundant in a broad range of Drosophila nuclear extracts and likely not limiting for 3′-end processing of histone pre-mRNAs in vitro. In this study, we took advantage of this fact to partially purify the U7 snRNP from the Drosophila Kc nuclear extracts and directly analyzed its proteome by mass spectrometry. Our main goal was to determine whether Drosophila U7 snRNP associates with the same set of polyadenylation factors that form the HCC in mammalian nuclear extracts and whether this association depends on the interaction between Drosophila FLASH and Lsm11.
Drosophila U7 snRNP has a composite structure
We affinity-purified the Drosophila U7 snRNP from Kc nuclear extracts via a 2′-O-methyl oligonucleotide complementary to the 5′ end of the U7 snRNA. The purified material, in addition to Lsm11 and other proteins of the U7-specifc ring, contained readily detectable amounts of endogenous FLASH, confirming that this protein indeed constitutes an integral component of the U7 snRNP. The purified U7 snRNP also contained several polyadenylation factors. Among them we identified symplekin, four out of six CPSF subunits (CPSF160, CPSF100, CPSF73, and WDR33), and CstF64 as the only component of the CstF subcomplex. We confirmed that CstF64 is indeed a component of the U7 snRNP in Drosophila by demonstrating that an antibody directed against full-length CstF64 efficiently coprecipitates both Lsm11 and FLASH.
Consistent with FLASH being an integral component of U7 snRNP, the anti-FLASH antibody precipitates Lsm11, and the antibody directed against Lsm11 precipitates FLASH. Importantly, the anti-FLASH precipitate contained the same polyadenylation factors that were identified in the affinity-purified U7 snRNP fraction. Thus, based on using two independent purification protocols, we conclude that the Drosophila U7 snRNP has a composite structure, consisting of the U7 snRNP core (U7 snRNA and the Sm ring), FLASH, and the Drosophila counterpart of the HCC. Compared with the mammalian HCC, the Drosophila HCC seems to lack orthologs of two CPSF components: Fip1 and CPSF30. Our immunoprecipitation experiments with an anti-CstF64 antibody indicate that Fip1 may be present in low levels in Drosophila nuclear extracts, and it is possible that this subunit is also underrepresented in the HCC and escaped our detection. CPSF30, on the other hand, is readily detectable as a component of the Drosophila CPSF complex involved in cleavage and polyadenylation, and its absence in the HCC is surprising. Interestingly, RNAi-mediated knockdown of Drosophila CPSF30 in S2 cells in contrast to knockdown of CPSF160 does not result in codepletion of CPSF73, CPSF100, and symplekin, suggesting that CPSF30 may not be an essential component of CPSF in Drosophila cells, explaining the absence of this subunit in the HCC (Sullivan et al. 2009).
We found no evidence for an association of the Drosophila U7 snRNP with the Drosophila ortholog of CF Im68. CF Im68 was recently reported to stably associate with the mammalian U7 snRNP (Ruepp et al. 2010), although previous results did not find CF Im68 as part of the mammalian HCC and the endogenous U7 snRNP (Yang et al. 2013) or the highly related Heat Labile Factor (HLF) purified from mammalian cells (Kolev and Steitz 2005). Our present study argues that CF Im68 has no role in 3′-end processing of histone pre-mRNAs in Drosophila nuclear extracts.
The composition of the HCC is much different from the composition of a related Drosophila complex that functions in cleavage and polyadenylation. This complex consists of symplekin, all three CstF subunits, all six CPSF subunits, and the two CF Im subunits of 25 and 68 kDa. The supercomplex was purified by precipitation with the anti-CstF64 antibody and is not disrupted by treatment with RNase A, indicating that its assembly involves multiple protein–protein interactions rather than binding of individual subcomplexes (CstF, CPSF, and CF Im) to a single pre-mRNA molecule. A similar complex was previously described in mammalian nuclear extracts (Takagaki and Manley 2000).
In 3′-end processing of mammalian histone pre-mRNAs, a critical interaction between the HCC and FLASH bound to Lsm11 is mediated by the LDLY motif located in the N-terminal region of FLASH (Yang et al. 2013). Recombinant N-terminal FLASH lacking this motif is unable to interact with the HCC or to stimulate 3′-end processing in mammalian nuclear extracts. It is yet unknown which component of the mammalian HCC directly contacts the LDLY motif, although symplekin is the most likely candidate to perform this function. Drosophila FLASH contains a highly related motif, LDIY, within its N-terminal region (amino acids 71–74), and our previous studies demonstrated that deleting the first 77 amino acids impairs the wild-type function of FLASH in processing in Drosophila cultured cells (Burch et al. 2011). We now show that recombinant N-terminal FLASH with the LDIY motif being replaced by four alanines is unable to restore processing activity of nuclear extracts prepared from S2 cells depleted of the endogenous FLASH. We also showed that Lsm11 and this LDIY mutant form a complex that is unable to interact with CPSF73, CPSF100, and symplekin. Thus, the mammalian LDLY and Drosophila LDIY motifs are functionally conserved and play a critical role in bringing the HCC to the U7 snRNP.
Most HCC components and Ars2 are dispensable for U7-dependent processing
Previous studies that used a sensitive GFP-based reporter gene driven by a histone gene promoter (Wagner et al. 2007) and direct analysis of histone mRNA 3′ ends (Sullivan et al. 2009) indicated that only three polyadenylation factors are essential for 3′-end processing of histone pre-mRNAs in Drosophila cells: (1) CPSF73, which functions as the endonuclease in cleavage and polyadenylation and 3′-end processing of histone pre-mRNAs; (2) CPSF100, which is a homolog of CPSF73; and (3) symplekin, which plays an unknown role in processing but likely functions as a scaffold or adaptor linking CPSF73 and CPSF100 with the rest of each processing machinery.
We reinvestigated the importance of all components of the HCC in formation of histone mRNAs in Drosophila by using a GFP reporter driven by a structurally distinct promoter from the U7 snRNA gene. However, consistent with the previously used reporter gene, the new reporter also suggests that polyadenylation factors other than CPSF73, CPSF100, and symplekin have no function in 3′-end processing of histone pre-mRNAs. Depletion of only these three subunits resulted in a robust expression of GFP. The remaining polyadenylation factors may be simply passive components of the Drosophila U7 snRNP. Alternatively, they may coordinate 3′-end processing of histone pre-mRNAs with important developmental events and these functions cannot be observed in cultured cells.
The lack of any essential role in 3′-end processing of Drosophila histone pre-mRNAs for CstF64 is surprising. CstF64 directly interacts with symplekin (Takagaki et al. 1990) and plays an important role in 3′-end processing of histone pre-mRNAs in human cells (Ruepp et al. 2011). In addition, our recent studies in mammalian nuclear extracts suggested that CstF64 in conjunction with symplekin may directly link the HCC with FLASH bound to Lsm11 (Yang et al. 2013). This is further evidence that U7-dependent processing machineries in mammalian and Drosophila cells are not identical (Table 1).
TABLE 1.
Major differences in 3′-end processing of replication-dependent histone pre-mRNAs between mammals and Drosophila
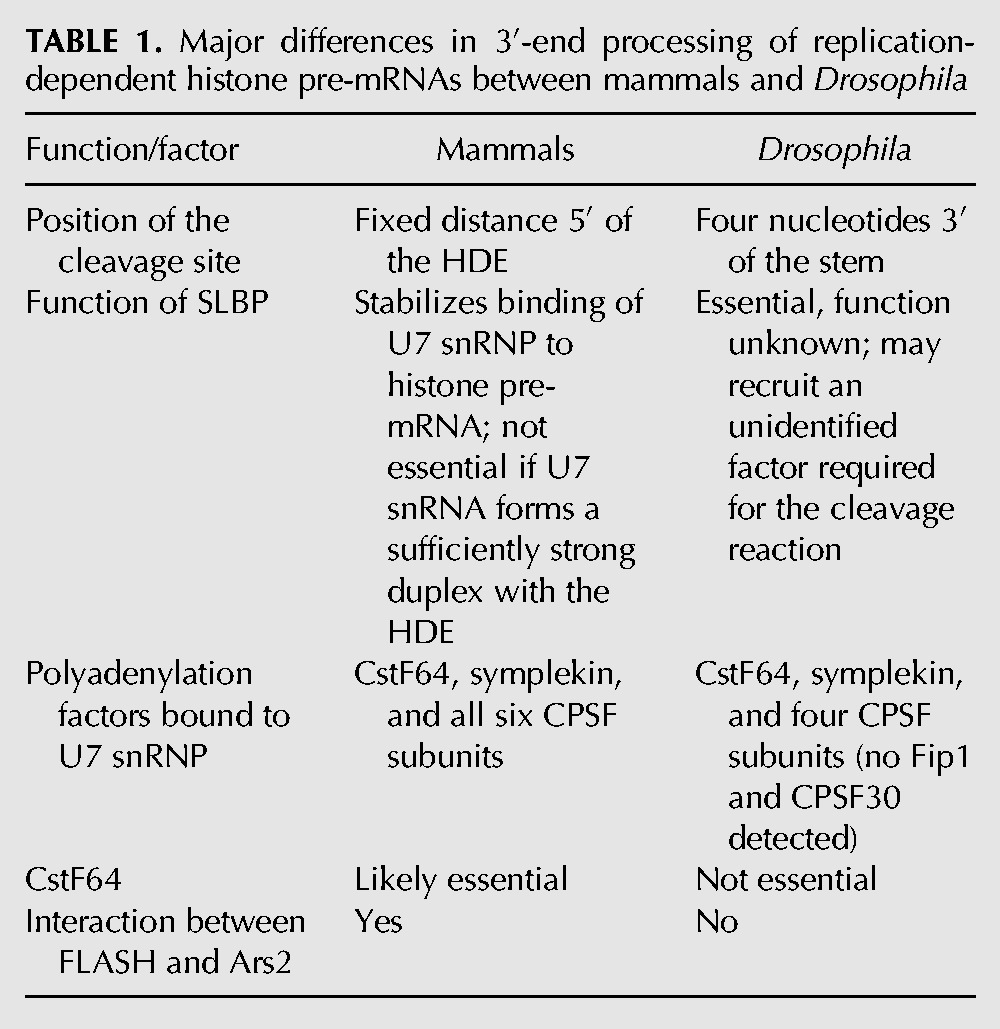
The GFP reporter driven by the snRNA gene promoter as well as direct analysis of endogenous histone mRNAs in Ars2-depleted S2 cells did not reveal any role for Ars2 in 3′-end processing of Drosophila histone pre-mRNAs in vivo. In the absence of Ars2, the U7-dependent processing is fully operative, and no replication-dependent histone mRNAs with a poly(A) tail accumulate. This is in sharp contrast to depletion of Ars2 in human cells, which resulted in redirecting a fraction of nascent histone pre-mRNAs to the cleavage and polyadenylation pathway (Gruber et al. 2012). The lack of any major role in 3′-end processing of histone pre-mRNAs for Ars2 in Drosophila is supported by our biochemical studies, which failed to detect this protein in association with U7 snRNP or any other processing factors known to operate on histone pre-mRNA. Overall, our results suggest that the interaction between Ars2 and FLASH and the role of Ars2 in promoting correct 3′-end cleavage of histone pre-mRNAs are specific for higher metazoans (Table 1).
U7 snRNP as an RNA-guided nuclease and the role of SLBP in processing
The composite Drosophila U7 snRNP containing FLASH and multiple polyadenylation factors is recruited to histone pre-mRNA for 3′-end processing, resembling the situation recently described for mammalian extracts (Yang et al. 2013). An antisense oligonucleotide complementary to the 5′ end of the Drosophila U7 snRNA that prevents the interaction between the U7 snRNP and the HDE completely eliminates FLASH and the polyadenylation factors from the complex formed on histone pre-mRNA. Thus, the U7 snRNA can be considered as a vehicle that identifies a target substrate by base-pairing and delivers the endonuclease CPSF73, CPSF100, symplekin, and several accessory polyadenylation factors to the site of cleavage. This mode of action is highly reminiscent of snoRNAs, which use an RNA-guided mechanism to locate target sequences for enzymatic modifications (Kiss 2002).
We note that in purified fractions of the U7 snRNP and processing complexes, FLASH stains with silver significantly stronger than the polyadenylation factors. It is therefore possible that a fraction of the U7 snRNP contains FLASH, but not the HCC (Fig. 9). Alternatively, more than one molecule of FLASH is required to interact with Lsm11, as suggested (Yang et al. 2013), or binding of the HCC to the FLASH/Lsm11 complex is relatively weak and partially disrupted during purification of the U7 snRNP.
FIGURE 9.
Potential forms of the U7 snRNP in Drosophila. The core particle consists of the U7 snRNA and the U7-specific Sm ring. This form is predominant in mammalian cells. In Drosophila cultured cells, most if not all U7 snRNP contains FLASH that interacts with Lsm11. As a result of this interaction, at least a fraction of the U7 snRNP assembles with a subset of polyadenylation factors (light gray) that form the Histone pre-mRNA Cleavage Complex (HCC). CPSF73, CPSF100, and symplekin are essential for 3′-end processing of histone pre-mRNAs in vivo, whereas the remaining polyadenylation factors (CPSF160, WDR33, and CstF64) are dispensable and may play accessory/regulatory functions. The arrangement of polyadenylation factors within the HCC is arbitrary.
The most notable difference between the mechanism of 3′-end processing of histone pre-mRNAs in Drosophila and mammals is that Drosophila processing is absolutely dependent on SLBP (Dominski et al. 2002), and cleavage invariably occurs 4 nt after the stem (Table 1; Dominski et al. 2005). These observations suggest that in Drosophila, SLBP and U7 snRNP are equally important in determining the site of cleavage. In mammalian processing, SLBP is an auxiliary rather than an obligatory factor and functions to stabilize the interaction between histone pre-mRNA and the U7 snRNP, which is capable of directing cleavage of the pre-mRNA on its own (Streit et al. 1993; Dominski et al. 1999). Consistently, in mammalian nuclear extracts, the site of cleavage is determined by the U7 snRNP (Scharl and Steitz 1994, 1996).
An excess of the stem–loop RNA added to Drosophila nuclear extracts results in a complete inhibition of in vitro processing due to preventing the interaction between SLBP and the stem–loop in histone pre-mRNAs (Dominski et al. 2002). Strikingly, this treatment had no effect on the recruitment of the composite Drosophila U7 snRNP to histone pre-mRNA. The core U7 snRNP, FLASH, and all the polyadenylation factors that form the HCC, including the CPSF73 endonuclease, are present in the processing complex assembled on histone pre-mRNA in the absence of free SLBP or on a pre-mRNA that lacks an SLBP binding site. Thus, in contrast to our previous model (Dominski et al. 2005), the most critical role in recruiting the composite U7 snRNP to Drosophila histone pre-mRNA is played by the sequence of the HDE. SLBP (and hence the stem–loop) is yet indispensable for processing in Drosophila nuclear extracts and may function by delivering an unknown essential factor(s) to the U7 snRNP positioned near the cleavage site.
Mass spectrometry did not detect any major differences in the composition of processing complexes assembled either in the presence or the absence of Drosophila SLBP. One possibility is that the SLBP-interacting factor exists in Drosophila nuclear extracts at a very low concentration or its interaction with SLBP is weak or transient and cannot be detected biochemically. What could be the nature and function of this factor? It may, for example, represent a hypothetical activator of the CPSF73 endonuclease, which was speculated to depend on a structural rearrangement prior to acquiring the catalytic activity (Mandel et al. 2006; Dominski 2010). Alternatively, it may function by bypassing the requirement for CstF64 or Ars2 in 3′-end processing in Drosophila. Elucidating the precise role of Drosophila SLBP in activating cleavage of histone pre-mRNA is the main focus of our ongoing studies.
MATERIALS AND METHODS
Drosophila nuclear extracts and 3′-end processing
The 106-nt Drosophila H3 pre-mRNA used as a substrate for 3′-end processing was generated by T7 RNA polymerase, treated with calf intestinal phosphatase (NEB), and labeled at the 5′ end with 32P using T4 polynucleotide kinase (NEB). 3′-End processing reactions were carried out in nuclear extracts prepared from either S2 or Kc Drosophila cells, as previously described (Dominski et al. 2002, 2005). Unless otherwise indicated, the extracts were prepared from freshly harvested cells. The cells were grown at room temperature in the serum-free D-22 medium (US Biological) using spinner flasks.
Synthetic RNAs
The following RNAs (written in the 5′–3′ orientation) were used in this study:
-
-Biot-dH3 pre-mRNA (63-mer):
(Bi)(18S)(18S)CUCUAUAAUCGGUCCUUUUCAGGACCACAAACCAGAUUCAAUGAGAUAAAAUUUUCUGUUGCC
-
-Biot-dH3ΔSL (47-mer):
(Bi)(18S)(18S)CUCUAUAAUCACAAACCAGAUUCAAUGAGAUAAAAUUUUCUGUUGCC
-
-Biot-mH2a pre-mRNA (62-mer):
(Bi)(18S)(18S)CUCCCAAAAAGGCUCUUUUCAGAGCCACCCACUGAAUCAGAUAAAGAGUUGUGUCACGGUAG
-
-Biot-SL (31-mer):
(Bi)(18S)(18S)GUGCCAAAAAGGCUCUUUUCAGAGCCACCCA
-
-Biot-αU7 (20-mer):
mCmAmAmAmGmAmGmAmAmUmAmAmAmAmAmUmUmUmUmC(18S)(18S)(Bi)
-
-Biot-αMock (17-mer):
mAmAmAmGmAmGmCmUmGmUmAmAmCmAmCmUmU(18S)(18S)(Bi)
-
-αU7 (17-mer):
mCmAmAmAmGmAmGmAmAmUmAmAmAmAmAmUmU
-
-WT SL (31-mer):
GUGCCAAAAAGGCUCUUUUCAGAGCCACCCA
-
-Mut SL (31-mer):
GUGCCAAUUAGCCUCUUUUCAGAGGCACCCA
where (Bi) biotin; (18S) 18-atom spacer; and (m) 2′-O-methyl group. The Biot-dH3ΔSL RNA was synthesized by IDT. The remaining RNAs were ordered from Dharmacon.
RNAi in Drosophila S2 cells
For preparation of nuclear extracts from RNAi-depleted cells, a total of 106 Drosophila S2 cells were plated in serum-free SF900 medium (GIBCO) and treated daily with 10 µg of double-stranded (ds) RNAs against selected Drosophila genes. After 5 d of treatment, cells were collected and used to prepare small-scale nuclear extracts. Each preparation yielded ∼100 µL of a nuclear extract. For analyzing the expression of GFP reporters, Drosophila S2 cells were plated as described above and treated with dsRNA for 48–72 h. Following the treatment, cells were transfected with appropriate GFP reporter plasmids, as described (Wagner et al. 2007; Burch et al. 2011) and 24 h later viewed on an Olympus IX81-ZDC inverted fluorescence microscope.
Mutagenesis and protein expression
Mutations in the N-terminal fragment of Drosophila FLASH (amino acids 1–178) were generated using PCR and appropriately altered oligonucleotide primers. Drosophila FLASH fused N-terminally to glutathione S-transferase (GST) was expressed in bacteria from the pET-42a vector. Drosophila Lsm11 (amino acids 1–153) fused N-terminally to maltose binding protein (MBP) was expressed in bacteria from the pDEST566 vector. Both recombinant proteins were purified on nickel beads (QIAGEN) using the His-tag present in each protein.
Binding of Drosophila nuclear proteins to Drosophila FLASH/Lsm11 complex
The Drosophila FLASH/Lsm11 complex was formed by mixing 100 pmol of each recombinant protein in 100 µL of buffer D (100 mM KCl, 20 mM HEPES at pH 7.9, 20% glycerol, 0.2 mM EDTA at pH 8, 0.5 mM DTT). All the subsequent steps were carried out as described previously (Yang et al. 2013), with the exception that 100 µL of Drosophila nuclear extracts from either Kc or S2 cells was used instead of mammalian nuclear extracts.
Formation of processing complexes and purification of the Drosophila U7 snRNP
Processing complexes were assembled in a final volume of 4 mL that contained 3 mL of a nuclear extract from Kc cells, 2.5 µg of the 63-nt Biot-dH3 pre-mRNA, 50 µg of yeast tRNA (Invitrogen), and 10 mM EDTA. The samples were incubated for 5 min at 22°C followed by 1 h of rotation at 4°C. The RNA substrate and associated proteins were bound to streptavidin beads, washed several times with buffer D/10 mM EDTA for a total of 1.5 h, and separated on a SDS/polyacrylamide gel. A similar approach was used for the single-step purification of the endogenous U7 snRNP with the exception that the Biot-αU7 2′-O-methyl oligonucleotide containing biotin at the 3′ end was used instead of the Biot-dH3 pre-mRNA.
Immunoprecipitations
A single sample typically contained 1 mL of a nuclear extract (∼10 mg of total protein) and 3–5 µg of an affinity-purified specific antibody, and it was rotated for 4–14 h at 4°C. Following incubation, the extract was spun down at 10,000 rpm in a table-top microcentrifuge and rotated for 2 h with ∼30 µL of protein A plus agarose beads (Pierce) to purify protein complexes bound to the antibody. The beads were washed with buffer D (see above) for a total of 2 h, moved to a new tube, and resuspended in an SDS sample buffer. The material immobilized on the beads was separated in a single SDS/polyacrylamide gel for analysis by silver staining/mass spectrometry or in three or four gels for analysis by Western blotting using various antibodies.
Mass spectrometry
Protein associated with the Drosophila U7 snRNP, Biot-dH3 pre-mRNA, or precipitated by specific antibodies was separated on SDS/polyacrylamide gels, excised from the gel, treated with trypsin, and identified by LC-MS/MS using the Nano-Acquity LC system (Waters) and Orbitrap Velos mass spectrometer (Thermo Electron Corp.), as described (Yang et al. 2013).
Antibodies
Custom-made antibodies against Drosophila Lsm11 and Drosophila CstF50 were kindly provided by J. Gall and John Lis, respectively. Antibodies against the N-terminal portion of Ars2 (amino acids 1–167 of total 769 residues), full-length Drosophila CstF64 (amino acids 1–437), and the N-terminal portion of human CF Im68 (amino acids 1–191), all N-terminally tagged with 6xHis and GST, were generated in rabbits by Pacific Immunology and affinity-purified using their respective protein antigens. Antibodies against Drosophila FLASH, CPSF73, CPSF100, and symplekin were described previously (Sullivan et al. 2009; Yang et al. 2009a).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to J. Gall (Carnegie Mellon) and J. Lis (Cornell University) for their generous gift of antibodies. We thank D. Price (Iowa University) for Kc cells, B. Burch (UNC at Chapel Hill) for advice on designing the reporter driven by the U7 snRNA gene-specific promoter, and S. Meaux for help with statistical analysis. We also thank J. Debski, J. Oledzki, and A. Fabijanska for mass spectrometry analysis. This work was funded by NIH grant GM58921. A.S. and M.D. were supported by the grants POIG.02.02.00-00-025/09, CEPT, and TEAM/2011-7/1.
REFERENCES
- Andersen PK, Jensen TH, Lykke-Andersen S. 2013. Making ends meet: Coordination between RNA 3′-end processing and transcription initiation. Wiley Interdiscip Rev RNA 4: 233–246. [DOI] [PubMed] [Google Scholar]
- Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L. 2007. Crystal structure of murine CstF-77: Dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell 25: 863–875. [DOI] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123: 265–276. [DOI] [PubMed] [Google Scholar]
- Bentley DL. 2005. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256. [DOI] [PubMed] [Google Scholar]
- Burch BD, Godfrey AC, Gasdaska PY, Salzler HR, Duronio RJ, Marzluff WF, Dominski Z. 2011. Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA 17: 1132–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo O, Manley JL. 2003. Strange bedfellows: Polyadenylation factors at the promoter. Genes Dev 17: 1321–1327. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766. [DOI] [PubMed] [Google Scholar]
- Cotten M, Gick O, Vasserot A, Schaffner G, Birnstiel ML. 1988. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro RNA processing reaction. EMBO J 7: 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M, Schaffner G, Birnstiel ML. 1989. Ribozyme, antisense RNA, and antisense DNA inhibition of U7 small nuclear ribonucleoprotein-mediated histone pre-mRNA processing in vitro. Mol Cell Biol 9: 4479–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries H, Rüegsegger U, Hübner W, Friedlein A, Langen H, Keller W. 2000. Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J 19: 5895–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z. 2010. The hunt for the 3′ endonuclease. Wiley Interdiscip Rev RNA 1: 325–340. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. 2007. Formation of the 3′ end of histone mRNA: Getting closer to the end. Gene 396: 373–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Zheng LX, Sanchez R, Marzluff WF. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol Cell Biol 19: 3561–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang X, Raska CS, Santiago CS, Borchers CH, Duronio RJ, Marzluff WF. 2002. 3′ end processing of Drosophila histone pre-mRNAs: Requirement for phosphorylated dSLBP and co-evolution of the histone pre-mRNA processing system. Mol Cell Biol 22: 6648–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Purdy M, Marzluff WF. 2005. Differences and similarities between Drosophila and mammalian 3′ end processing of histone pre-mRNAs. RNA 11: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Carpousis AJ, Clouet-d'Orval B. 2013. Emergence of the β-CASP ribonucleases: Highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim Biophys Acta 1829: 532–551. [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Murphy S. 2008. Expression of human snRNA genes from beginning to end. Biochem Soc Trans 36: 590–594. [DOI] [PubMed] [Google Scholar]
- Ezzeddine N, Chen J, Waltenspiel B, Burch B, Albrecht T, Zhuo M, Warren WD, Marzluff WF, Wagner EJ. 2011. A subset of Drosophila Integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3′-end formation. Mol Cell Biol 31: 328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin GM. 2005. Eukaryotic mRNA 3′ processing: A common means to different ends. Genes Dev 19: 2517–2521. [DOI] [PubMed] [Google Scholar]
- Godfrey AC, Kupsco JM, Burch BD, Zimmerman RM, Dominski Z, Marzluff WF, Duronio RJ. 2006. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA 12: 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber JJ, Zatechka DS, Sabin LR, Yong J, Lum JJ, Kong M, Zong WX, Zhang Z, Lau CK, Rawlings J, et al. 2009. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell 138: 328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber JJ, Olejniczak SH, Yong J, La Rocca G, Dreyfuss G, Thompson CB. 2012. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol Cell 45: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J 23: 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriyama M, Kobayashi Y, Saito M, Ishikawa F, Yonehara S. 2009. Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol Cell Biol 29: 4729–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA. 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev 19: 2583–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Ratsch G, Weigel D. 2008. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci 105: 8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. 2006. SERRATE: A new player on the plant microRNA scene. EMBO Rep 7: 1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43: 624–637. [DOI] [PubMed] [Google Scholar]
- Ma TL, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. 2000. Cell cycle-regulated phosphorylation of p220NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev 14: 2298–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. 2008. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65: 1099–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. 2010. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 40: 410–422. [DOI] [PubMed] [Google Scholar]
- Martin F, Schaller A, Eglite S, Schümperli D, Müller B. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J 16: 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan GH, Greenblatt J, Patterson SD, Wickens M, Bentley DL. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361. [DOI] [PubMed] [Google Scholar]
- Mowry KL, Steitz JA. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science 238: 1682–1687. [DOI] [PubMed] [Google Scholar]
- Muller B, Schumperli D. 1997. The U7 snRNP and the hairpin binding protein: Key players in histone mRNA metabolism. Semin Cell Dev Biol 8: 567–576. [DOI] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell 26: 349–365. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Will CL, Lührmann R, Schümperli D, Müller B. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J 20: 5470–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. 2011. Ending the message: Poly(A) signals then and now. Genes Dev 25: 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. [DOI] [PubMed] [Google Scholar]
- Ruegsegger U, Beyer K, Keller W. 1996. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem 271: 6107–6113. [DOI] [PubMed] [Google Scholar]
- Ruegsegger U, Blank D, Keller W. 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell 1: 243–253. [DOI] [PubMed] [Google Scholar]
- Ruepp MD, Vivarelli S, Pillai RS, Kleinschmidt N, Azzouz TN, Barabino SM, Schumperli D. 2010. The 68 kDa subunit of mammalian cleavage factor I interacts with the U7 small nuclear ribonucleoprotein and participates in 3′-end processing of animal histone mRNAs. Nucleic Acids Res 38: 7637–7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp MD, Schweingruber C, Kleinschmidt N, Schumperli D. 2011. Interactions of CstF-64, CstF-77, and symplekin: Implications on localisation and function. Mol Biol Cell 22: 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. 2009. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 138: 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharl EC, Steitz JA. 1994. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J 13: 2432–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharl EC, Steitz JA. 1996. Length suppression in histone messenger RNA 3′-end maturation: Processing defects of insertion mutant premessenger RNAs can be compensated by insertions into the U7 small nuclear RNA. Proc Natl Acad Sci 93: 14659–14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumperli D, Pillai RS. 2004. The special Sm core structure of the U7 snRNP: Far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci 61: 2560–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR III, Frank J, Manley JL. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HO, Tabiti K, Schaffner G, Soldati D, Albrecht U, Birnstiel ML. 1991. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligoribonucleotides. Proc Natl Acad Sci 88: 9784–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A, Koning TW, Soldati D, Melin L, Schümperli D. 1993. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res 21: 1569–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev 15: 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Steiniger M, Marzluff WF. 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell 34: 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 20: 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. 1990. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev 4: 2112–2120. [DOI] [PubMed] [Google Scholar]
- Tan D, Marzluff WF, Dominski Z, Tong L. 2013. Structure of histone mRNA stem–loop, human stem–loop binding protein, and 3′hExo ternary complex. Science 339: 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraldi KL, Edwalds-Gilbert G, MacDonald CC, Wallace AM, Milcarek C. 2000. Isolation and characterization of polyadenylation complexes assembled in vitro. RNA 6: 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF. 2007. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell 28: 692–699. [DOI] [PubMed] [Google Scholar]
- Wahle E, Keller W. 1996. The biochemistry of polyadenylation. Trends Biochem Sci 21: 247–250. [PubMed] [Google Scholar]
- White AE, Burch BD, Yang XC, Gasdaska PY, Dominski Z, Marzluff WF, Duronio RJ. 2011. Drosophila histone locus bodies form by hierarchical recruitment of components. J Cell Biol 193: 677–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Khanna K, Ruan S. 2010. Expression of microRNAs and its regulation in plants. Semin Cell Dev Biol 21: 790–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. 2009a. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell 36: 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Torres MP, Marzluff WF, Dominski Z. 2009b. Three proteins of the U7-specific Sm ring function as the molecular ruler to determine the site of 3′-end processing in mammalian histone pre-mRNA. Mol Cell Biol 29: 4045–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Xu B, Sabath I, Kunduru L, Burch BD, Marzluff WF, Dominski Z. 2011. FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5′ exonucleolytic degradation of the downstream cleavage product. Mol Cell Biol 31: 1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Sabath I, Debski J, Kaus-Drobek M, Dadlez M, Marzluff WF, Dominski Z. 2013. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol Cell Biol 33: 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Bentley DL. 2004. The link between mRNA processing and transcription: Communication works both ways. Exp Cell Res 296: 91–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.









