In this paper, the authors present a novel method to generate RNA (sRNA) expression libraries, in which sRNAs isolated from cells are converted to cDNA and directly cloned into a lentiviral expression vector containing two convergent RNA-pol III promoters. This offers the opportunity to identify novel small RNA-coding genes with particular physiological functions. A functional screening using this library led to the identification of hsa-miR-30b and hsa-miR-30c as negative regulators of cell death induced by loss of attachment (anoikis). This sRNA expression and screening strategy is apparently independent of the miRNA/siRNA biogenesis pathway and constitutes an important methodological addition to the sRNA functional analysis toolbox.
Keywords: small RNAs, functional screens, libraries, miR-30b/c, anoikis
Abstract
MicroRNAs (miRNAs) have been widely studied in order to elucidate their biological functions. MicroRNA microarrays or miRNA overexpression libraries generated by synthesis and cloning of individual miRNAs have been used to study their different roles. In this work, we have developed a novel methodology to express mature miRNAs and other small RNAs from a double convergent RNA polymerase III promoter. We show that the generated miRNAs function similarly to those processed from primary transcripts or pri-miRNAs. This system allowed us to produce a lentiviral library expressing the whole population of small RNAs present in a metastatic cell line. A functional screening using this library led to the identification of hsa-miR-30b and hsa-miR-30c as negative regulators of cell death induced by loss of attachment (anoikis). Importantly, we demonstrated that the acquisition of anoikis resistance via these miRNAs is achieved through down-regulation of caspase 3 expression. Moreover, overexpression of these miRNAs resulted in a decrease of other types of caspase 3-dependent cell death and enhanced the survival of MCF10A acinar cells in morphogenesis assays, suggesting a putative role as oncomirs. In summary, this novel methodology provides a powerful and effective way for identifying novel small RNAs involved in a particular biological process.
INTRODUCTION
It is estimated that ∼98% of the transcriptional output of the human genome represents RNA that does not encode proteins. Therefore, these noncoding RNAs (ncRNAs) are either largely useless transcripts or they fulfill a wide range of unexpected functions in eukaryotic biology (Taft et al. 2010). More specifically, microRNAs (miRNAs), the most studied ncRNAs, regulate the expression of hundreds of genes through sequence-specific binding to mRNA (Bartel 2009; Kim et al. 2009). MicroRNAs are transcribed by RNA polymerase II or III, which produce a primary transcript containing the mature miRNA sequence and a varying amount of flanking regions. For most mammalian miRNAs, the pri-miRNA folds into a hairpin structure characteristic of miRNA genes. Subsequently, a complex formed by the RNase III enzyme Drosha and its cofactor Dgcr8 recognizes the hairpin and cleaves the primary miRNA into a precursor miRNA, which is exported from the nucleus by Exportin-5. After reaching the cytoplasm, the precursor miRNA is further processed by Dicer nuclease and loaded into the RNA-induced silencing complex (RISC). Next, a specific single strand of the miRNA duplex in the RISC is selected as a guide to direct sequence-specific targeting of mRNAs, usually through their 3′ untranslated regions (UTRs), leading to translational repression and/or mRNAs destabilization (Kim et al. 2009). MicroRNA target genes can regulate several biological processes such as cell development, differentiation, cell cycle and apoptosis (Inui et al. 2010). Detachment of normal epithelial cells from the extracellular matrix triggers a type of programmed cell death called anoikis. Anoikis is physiologically relevant during development, tissue homeostasis, and disease progression (Chiarugi and Giannoni 2008). Several miRNAs have been reported to play a crucial role in tumorigenesis by regulating cell proliferation and apoptosis as oncogenes (also called oncomirs) or as tumor suppressors (Bueno et al. 2008). Recently, some of these miRNAs have been specifically involved in anoikis, such as miR-214, miR-200c, or miR-221 (Gramantieri et al. 2009; Howe et al. 2011; Penna et al. 2011). MiR-200c promotes anoikis, but miR-214 and miR-221 inhibit it in melanoma and in hepatocellular carcinoma-derived cell lines, respectively.
Approaches to identify miRNAs involved in a particular biological process have been largely based on miRNA microarray analysis or on the use of miRNA expression libraries. Currently, these libraries are generated by cloning of individual miRNAs reported in different databases. In the present work, we have developed a methodology to generate a lentiviral library expressing the whole population of 19- to 29- nt RNAs (including miRNAs and other RNAs, such as small nucleolar RNAs, small nuclear RNAs, or small ribosomic RNAs) present in the metastatic breast cancer and anoikis-resistant cell line MDA-MB-231 (Fukazawa et al. 2004). We demonstrate that the processed miRNAs expressed by dual convergent promoters used in this library are functionally similar to full-length expressed miRNAs and operate independently of the components involved in miRNA biogenesis. A functional screening using this library uncovered hsa-miR-30b (miR-30b) and hsa-miR-30c (miR-30c) as inhibitors of anoikis. These miRNAs promoted survival of the immortalized retinal pigment epithelial cell line hTERT RPE1 (RPE-1) growing under anoikis-inducing conditions by suppressing Casp3 translation. Furthermore, miR-30b and miR-30c also restrained other types of caspase 3-dependent cell death mechanisms as demonstrated by a delay in proper mammary acinar morphogenesis, an important event in the progression of epithelial tumors.
RESULTS
Design and validation of the processed miRNAs expression system
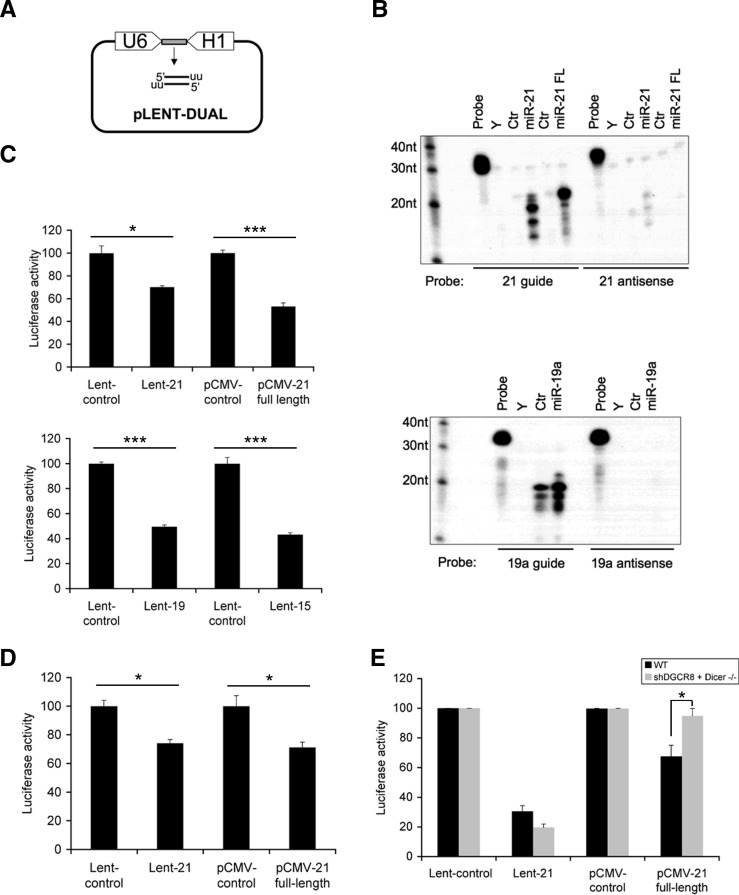
The design of the small RNA (sRNA) libraries was based on the transcription of both strands of a short DNA fragment flanked by two human RNA polymerase III promoters, generating a duplex RNA molecule (Zheng et al. 2004). The lentiviral vector pLENT-DUAL contains two RNA polymerase III terminators just downstream from the transcriptional start sites of H1 and U6 promoters (Fig. 1A; Supplemental Fig. S1). After transcription, the resulting small RNA has a similar structure as the “natural” double-stranded processed miRNA. DNA sequences corresponding to miR-21 (Lent-21), miR-19a (Lent-19), and miR-15a (Lent-15) mature miRNAs were cloned into the pLENT-DUAL vector to validate our technology. RNase protection assays in HEK-293T cells demonstrated that the guide strand of the double-stranded miRNA was stably overexpressed. None, or very low signal, was detected for the complementary strand (Fig. 1B; Supplemental Fig. S2A). In the case of miR-21, similar expression profiles were observed between the expression of miR-21 transcribed from pLENT-DUAL or from pCMV-21 full-length expressing the complete pri-miR-21 (Fig. 1B, upper panel), although shorter isomirs were detected when the mature miRNA was directly expressed (Berezikov 2011). The ability of the pLENT-DUAL system to suppress gene expression was examined by analyzing the activity of the luciferase gene containing specific miRNA-binding sites for miR-21, miR-15a, or miR-19a in its 3′ UTR in HEK-293T cells. A depletion of 35%–50% of luciferase activity could be observed for both the processed and full-length versions of miR-21 (Fig. 1C, upper panel). Although the most abundant mature miR-21 expressed from pLENT-DUAL was 2 nt shorter than that expressed from a full-length version, luciferase activity assays demonstrated that the level of expression of mature miR-21 with the expected length and/or the shorter isomirs depleted a target efficiently. The reduced luciferase activity was also observed in similar experiments using pLENT-DUAL-19 or -15 plasmids (Fig. 1C, lower panel). In addition, real time PCR experiments showed that the loss of luciferase activity using pLENT-DUAL-21, -15, or -19 was the result of a translational inhibition similar to that of the pCMV-21 full-length construct (Supplemental Fig. S2B). Furthermore, the processed miRNA-21 expressed from the pLENT-DUAL system down-regulated the expression of the luciferase open reading frame (ORF) fused to the complete 3′ UTR of the Pdcd4 gene, a well-known target of this miRNA (Fig. 1D; Asangani et al. 2008). Next, we checked whether the pLENT-DUAL expression system was independent of the miRNA-processing pathway. In order to ensure a potent reduction of the production of mature miRNAs from pri-miRNAs, we needed to abolish both the cytoplasmatic and nuclear processing steps. To this purpose, Dicer-deficient HCT116 cells (Cummins et al. 2006) were infected with a lentivirus carrying a small hairpin RNA against DGCR8 mRNA (Kumar et al. 2007), which encodes a RNA-binding protein required for pri-miRNA biogenesis. In these cells, miR-21 expressed from the pCMV-21 full-length plasmid failed to deplete luciferase expression, whereas the processed miR-21 expressed from pLENT-DUAL maintained the level of repression (Fig. 1E). This indicates that our miRNA expression system seems to be independent of the miRNA processing machinery. Taken together, our data demonstrate the general and versatile applicability of pLENT-DUAL to drive functional miRNA (and other types of small RNA functioning as miRNA or siRNA) expression experiments.
FIGURE 1.
Mature miRNAs expressed under the control of dual convergent promoters are functionally similar to full-length expressed miRNAs. (A) Schematic drawing showing the plasmid used to express mature miRNAs and others small RNAs. (B) RNase protection assay of miR-21, miR-19a transiently overexpressed from the pLENT-DUAL plasmid in HEK-293T cells (upper and lower panel). miR-21 full-length (miR-21 FL) was expressed from a pCMV-miR-21 plasmid. Control (Ctr) cells expressed the empty vectors pLENT-DUAL or pCMV. Y corresponds to yeast RNA. (C) Transient transfections were carried out in HEK-293T cells, using pLENT-DUAL-21, -19, -15 (expressing the mature form of miR-21, miR-19a or miR-15a, respectively) or pCMV-miR-21 full-length, pLuc-BS carrying a binding site for miR-21, miR-19a or miR-15a and pCMV-Renilla (normalization control) at 100:10:1 proportions, respectively. Firefly and Renilla activities were measured 40 h after transfection. (D) Similar luciferase assay as described in C, using pLENT-DUAL-miR-21 and pCMV-miR-21 full-length and a plasmid harboring a 652- nt wild-type sequence corresponding to the 3′ UTR of PDCD4 at the 3′ position of the luciferase reporter gene. (E) Similar luciferase assay as described in C in HCT116 DICER −/− cells, using pLENT-DUAL-miR-21 and pCMV-miR-21 full-length and a plasmid carrying a binding site for miR-21. For these experiments, HCT116 DICER −/− cells infected with a lentivirus expressing a small hairpin RNA against DGCR8 were used. All transfections were performed in triplicate, and results are shown as the averages ± standard errors of the means from at least three independent experiments. The data were subjected to two-tailed Student's t-test [(*) P < 0.05, (***) P < 0.001], with the exception of section E, where a two-way ANOVA followed by Bonferroni's post-test was used. (*) P < 0.05.
Generation of a lentiviral small RNA-expressing library and its use in a functional screening
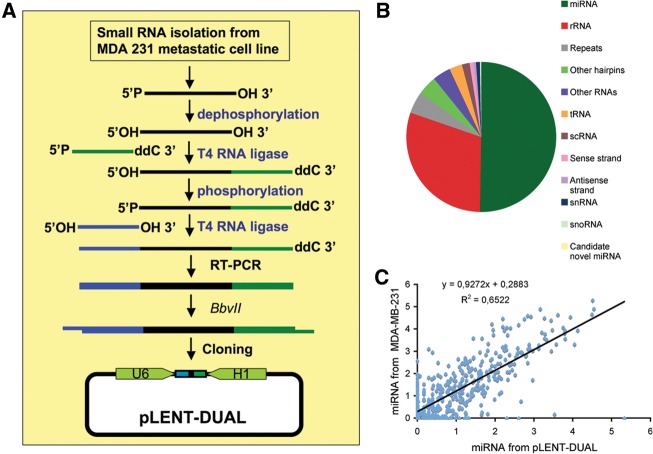
MDA-MB-231 (clone 4175) human breast cancer cells were selected to generate a library of small RNAs because of their high invasive ability and metastatic potential. Small 19- to 29-nt RNAs from this anoikis-resistant cell line were isolated by polyacrylamide gel electrophoresis and subjected to sequential modifications including 3′- and 5′-adapter ligation, RT-PCR amplification, and directional cloning into the pLENT-DUAL plasmid (Fig. 2A; Supplemental Fig. S1). Escherichia coli cells were electroporated and more than 8 × 105 independent colonies were pooled to compose the library. In order to check the complexity and quality of the library, we carried out a deep-sequencing analysis of the small RNAs cloned into the pLENT-DUAL vector as well as the small RNA population from MDA-MD-231 cells (Fig. 2B,C; Supplemental Table S1). We found that not only miRNAs were present in our library but also other small noncoding RNAs, such as snoRNAs, tRNA-derived small RNAs, small RNAs from coding and noncoding regions, rRNA, small RNAs from repeat regions, and other types of sRNAs (Fig. 2B). In addition, the comparison between miRNAs present in the small RNA population of MDA-MB-231 cells and miRNAs from small RNAs cloned into pLENT-DUAL showed a high level of correlation (R2 = 0.652) even though it was difficult to compare libraries generated by different protocols (Fig. 2C; Supplemental Table 1; Hafner et al. 2011; Toedling et al. 2012; Zhuang et al. 2012). Notably, ∼71% of miRNAs with more than five reads per million (rpm) in the MDA-MB-231 small RNA library were present at five or more rpm in the pLENT-DUAL library. Additionally, the pLENT-DUAL library contained a moderately high percentage (62%) of isomirs present in MDA-MB-231, indicating that the library was free of major sequence/size biases. These results demonstrated that our library contained most of the miRNAs highly or moderately expressed in MDA-MB-231 and showed a high degree of complexity, including other types of small RNAs, some of them being able to function as miRNAs.
FIGURE 2.
Generation of a small RNA library from MDA-MB-231 (clone 4175) cells. (A) Scheme illustrating the small RNA cloning strategy to generate expression libraries based on the pLENT-DUAL lentiviral vector. (B) Pie chart representing the percentage of the different classes of small RNAs in the pLENT-DUAL library. (Repeats) Regions annotated as repeats (of any type, excluding simple repeats and primarily transposable elements), (scRNA) small cytoplasmic RNA, (other hairpins) loci that look like hairpins but do not pass all the criteria for annotation as miRNAs, (sense strand) small RNA matching sense strand to coding and noncoding regions, (antisense strand) small RNA matching antisense strand to coding and noncoding regions. (C) A correlation between known miRNAs expressed in MDA-MB-231 cells and known miRNAs cloned in the library (log [1 + number of reads normalized to total number of mapped reads]). R2 derived from Pearson's correlation of the data is indicated.
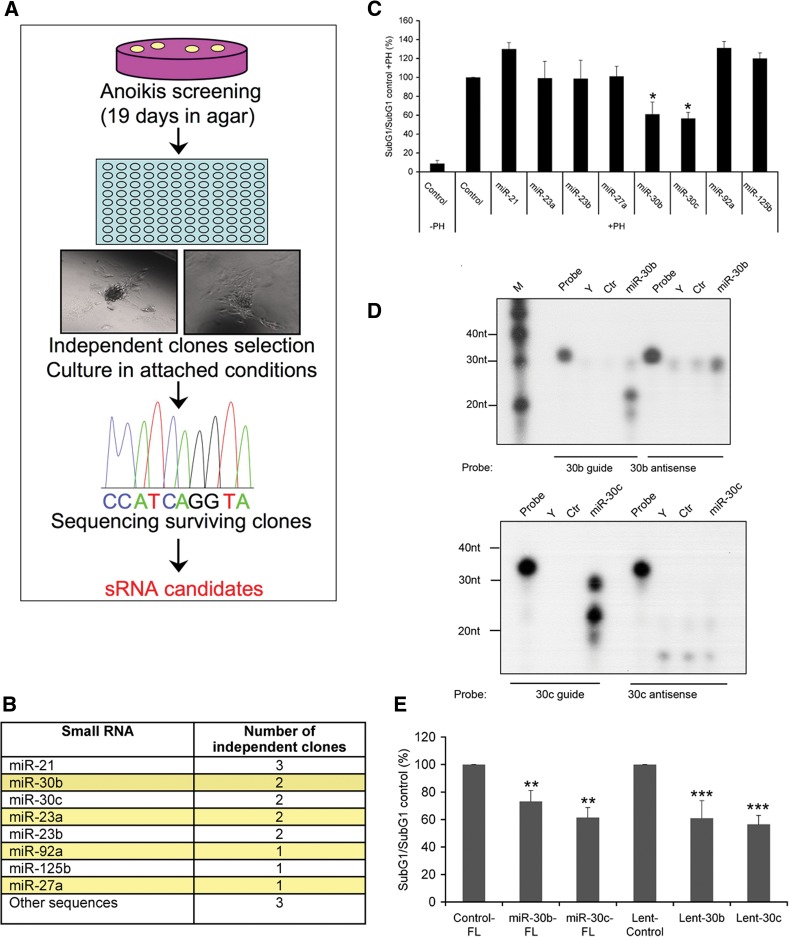
To perform a functional screen for anoikis suppression, we used nonmalignant, anoikis-sensitive RPE-1 cells (Supplemental Fig. S3; Alfano et al. 2006). These cells were infected with the lentiviral library (MOI = 1) or empty pLENT-DUAL lentiviruses as a control for 72 h, transferred to agar-covered Petri dishes, and kept in culture for 19 d at low confluence to avoid any contact (Fig. 3A; see Materials and Methods section for details). Seventeen clones from cells infected with the lentiviral library containing at least 50 cells were isolated and transferred to adhesive culture dishes to allow the expansion of anoikis-resistant colonies. Only one clone was isolated from cells infected with empty pLENT-DUAL lentivirus, which was unable to proliferate in an adhesive culture dish. In order to know which sRNAs were responsible for the observed anoikis-resistant phenotype, lentiviral DNA from independent clones was amplified and sequenced (Fig. 3B). Notably, although in these long-term anoikis assays, cells overexpressing the selected small RNAs formed colonies in soft agar, miR-30b- and miR-30c (miR-30b/c)-overexpressing clones seemed to proliferate much more rapidly (data not shown).
FIGURE 3.
A functional screening using a novel small RNAs expression library reveals hsa-miR-30b and hsa-miR-30c as repressors of anoikis. (A) Immortalized RPE-1 cells were infected with a lentiviral expression library containing a small RNAs collection from the MDA-MB-231 cell line. RPE-1-infected cells were cultured for 19 d in agar dishes at low confluence avoiding any contact and surviving clones transferred to adhesive culture dishes to allow the growth of anoikis-resistant cells. DNA from these clones was isolated and sequenced. (B) Table showing the small RNA sequences overexpressed in anoikis-resistant clones. (C) Flow cytometry analysis of the levels of apoptosis in RPE-1 cells (SubG1/SubG1 control + polyHEMA) infected with lentivirus containing candidate mature miRNAs or empty vector (control). Three days after infection, cells were deprived of serum for 24 h and then cultured in polyhema (PH) plates plus methylcellulose (2%) in serum-free medium for another 24 h. (D) RNase protection assay of miR-30b and miR-30c transiently overexpressed in HEK-293 cells from pLENT-DUAL plasmid. Control (Ctr) cells expressed the empty vector pLENT-DUAL; Y corresponds to yeast RNA; M, Decade RNA markers (Ambion). (E) Levels of apoptosis in RPE-1 cells (SubG1/SubG1 control) infected with lentivirus carrying full-length miR-30b, miR-30c-1 (FL), miR-30b/c mature miRNAs (Lent-30b/c), and the corresponding empty vectors as controls in a similar assay as described in C. Results are shown as the averages ± standard errors of the means from at least three independent experiments and were analyzed by one-way ANOVA, followed by Bonferroni post-test for significance versus control cells. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
Additionally, RPE-1 cells were infected with empty pLENT-DUAL or pLENT-DUAL carrying individual mature miRNA candidates and the number of apoptotic cells measured in a short-term anoikis experiment (Supplemental Fig. S3). Among all the candidates, only miR-30b/c-expressing clones achieved a solid and significant anoikis-resistant phenotype, showing a 40%–45% reduction in the number of cells with a subG1 DNA content, compared to control cells (Fig. 3C). Thus, miR-30b/c induced anoikis resistance in both short- and long-term experiments, and they were selected for a detailed study. Interestingly, miR-30c and, more notably, miR-30b, were present in the pLENT-DUAL library at a low percentage (0.187% and 0.0173%, respectively) (Supplemental Table S1), meaning that low-abundant small RNAs can be isolated and identified in a well-designed functional screening using our novel libraries. RNase protection assays demonstrated that the guide strands of the double-stranded miR-30b and miR-30c were stably overexpressed from pLENT-DUAL plasmids. None or very low nonspecific signal was detected for the complementary strands (Fig. 3D). Similar results were observed when RPE-1 cells were infected with lentiviruses overexpressing full-length miR-30b/c (Fig. 3E; Supplemental Fig. S4A). Therefore, the identification of miR-30b/c as regulators of anoikis validated our novel small RNA expression library and its application for functional screenings.
Caspase 3 is the main target of miR-30b/c for anoikis regulation
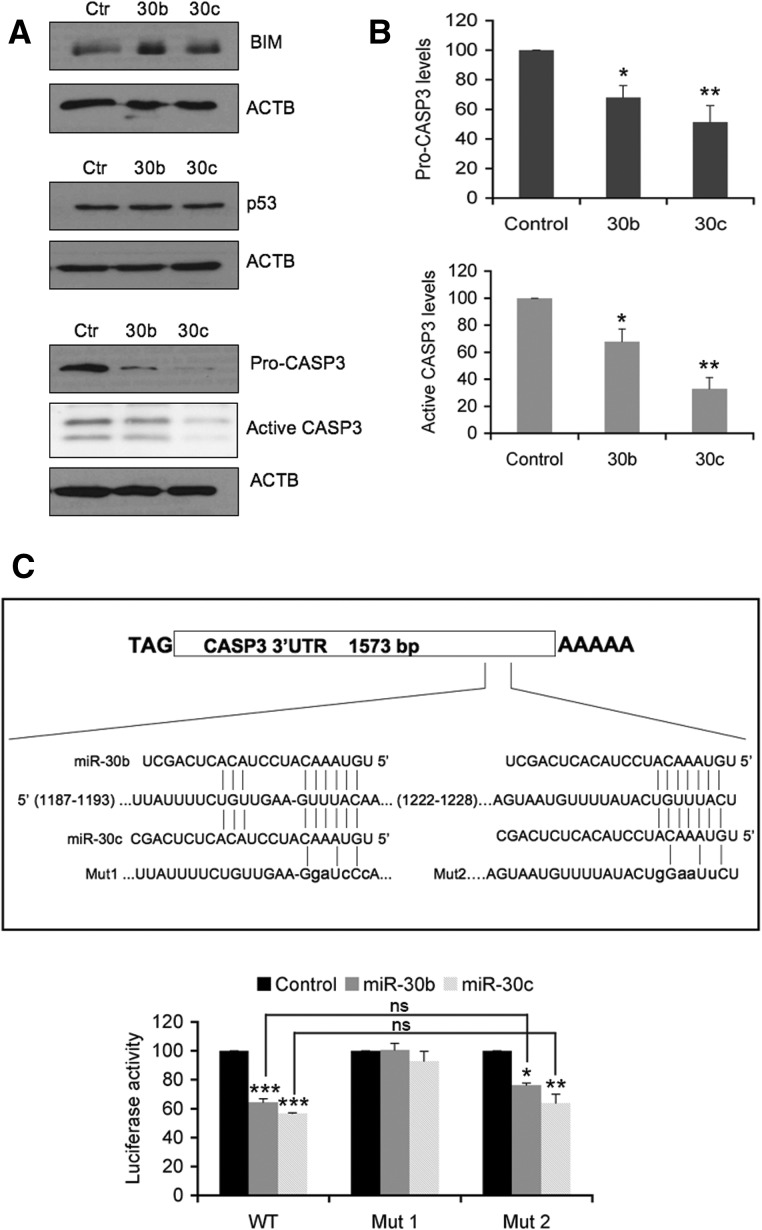
Anoikis is a very complex process that involves signalling at various levels (Chiarugi and Giannoni 2008). In order to identify putative targets of miR-30b/c, different computational algorithms including those applied in software such as miRanda (Betel et al. 2008), TargetScan (Friedman et al. 2009) and DIANA-microT (Maragkakis et al. 2009) were used. We found several genes related to the induction of cell death containing at least one putative miR-30b/c binding site in their 3′ UTR predicted by all three resources. Among these genes, we selected BCL2-like 11 (BIM), TP53 (p53), and caspase 3 (CASP3) because they had already been shown to be involved in the anoikis process (Chiarugi and Giannoni 2008). As shown in Figure 4A, Western blot experiments confirmed that among these three candidates, only caspase 3 expression was depleted by miR-30b/c. In cells growing under adherent conditions and overexpressing miR-30b/c, the expression levels of pro-CASP3 were reduced to 25%–40% of the levels in control cells infected with a lentivirus carrying an empty vector. Notably, active CASP3 protein levels in nonadherent conditions were measured in these cells, and a significant reduction (25%–50%) was observed in miR-30b/c-overexpressing cells (Fig. 4A,B). Furthermore, quantitative PCR analysis determined that CASP3 mRNA levels remained constant in both control and miR-30b/c-expressing cells, suggesting that depletion of the protein was caused by translational inhibition (Supplemental Fig. S5).
FIGURE 4.
Hsa-miR-30b and hsa-miR-30c down-regulate CASP3 expression through its 3′ UTR-binding. (A) Western blot analysis of p53, CASP3, and BCL2-like 11 (BIM), three putative target proteins of miR-30b/c related to anoikis. RPE-1 cells were infected with lentivirus expressing full-length miR-30b, miR-30c-1 (miR-30c), or empty vector as a control. Active CASP3 was analyzed in cells cultured under anoikis conditions as described in Figure 3C. Actin (ACTB) was used as the loading control. (B) Densitometric analysis of pro-CASP3 and active CASP3 levels. ACTB protein was used as the normalization control. Results are shown as the averages ± standard errors of the means from at least three independent experiments and were analyzed by one-way ANOVA, followed by Bonferroni post-test (*P < 0.05; **P < 0.01). (C) Schematic representation of the caspase 3 3′ UTR containing two putative binding sites for miR-30b/c. The 3′ UTR of CASP3 mRNA was cloned downstream from the open reading frame of luciferase (pLuc-BS). Both broadly (1187–1193) and poorly (1222–1228) conserved putative miRNA regulatory elements of the CASP3 3′ UTR were mutated (Mut1 and Mut2, respectively) and cloned along with the wild-type 3′ UTR (WT). HEK-293T cells were transiently cotransfected with plasmids overexpressing full-length miRNAs (miR-30b, miR-30c-1, or empty vector as control), pLuc-3′ UTR CASP3 (WT, Mut1, or Mut2) and pCMV-Renilla at 100:10:1 proportions, respectively. Firefly and Renilla activities were measured 40 h after transfection. The luciferase activity normalized to Renilla is shown (lower panel). Transfections were performed in triplicate, and results are shown as the averages ± standard errors of the means from at least three independent experiments and were analyzed by one-way and two-way ANOVA, followed by Bonferroni post-test. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (ns) not significant.
Using the TargetScan software (Friedman et al. 2009), two putative binding sites for miR-30b/c located in the 3′ UTR of caspase 3 were found. One of them was highly conserved among different mammalian species, while the other showed low conservation. To verify the functionality of these sites, the 3′ UTR of CASP3 was cloned into a reporter vector, and luciferase assays, using the wild-type sequence (WT) and sequences mutated in the highly (Mut1) or poorly (Mut2) conserved sites, were carried out (Fig. 4C). Cells were transfected with the control or miR-30b/c plasmid and the different versions of the 3′ UTR of CASP3. Figure 4C shows that miR-30b and miR-30c decreased luciferase activity only when the highly conserved site was intact, showing that the regulation of caspase 3 expression by miR-30b/c was achieved directly through a conserved binding site in its 3′ UTR.
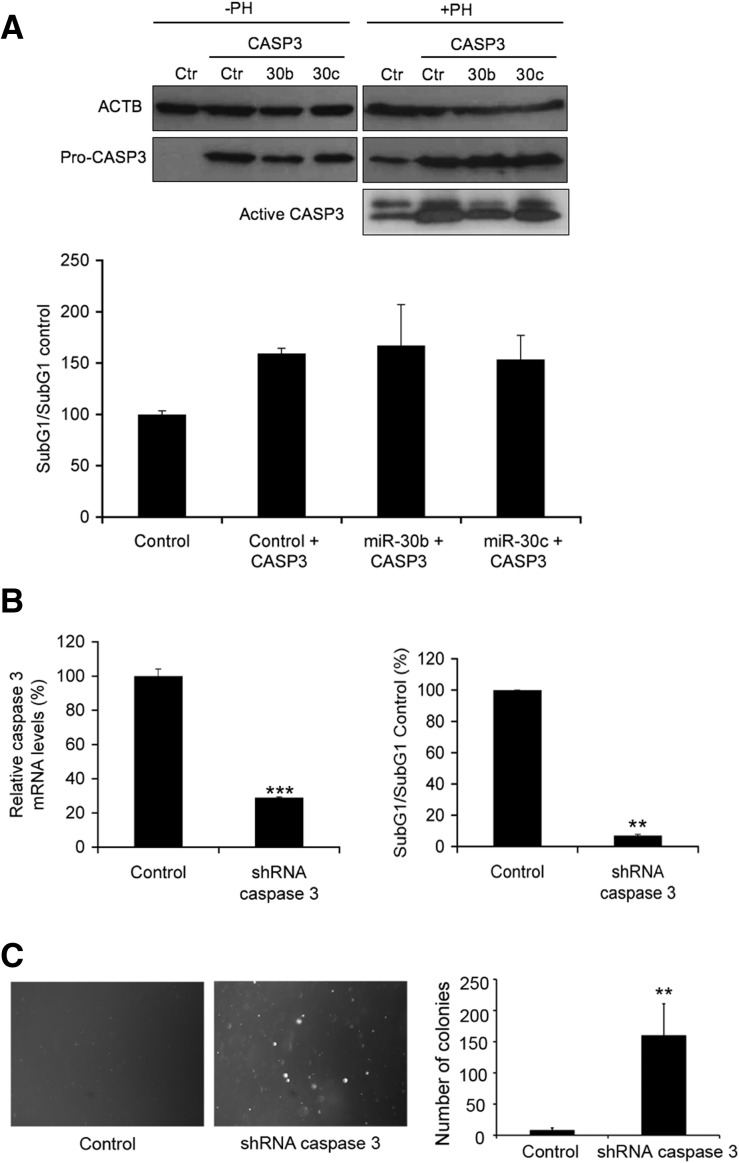
In addition, the expression of a CASP3 mRNA lacking the 3′ UTR rescued CASP3 levels in RPE-1 cells overexpressing miR-30b/c (Fig. 5A, upper panel). The reconstitution of CASP3 protein levels led to a reversion of the anoikis-resistant phenotype in miR-30b/c-overexpressing RPE-1 cells (Fig. 5A, lower panel). Moreover, the depletion of caspase 3 expression by short hairpin RNA in RPE-1 cells was sufficient to mimic or even improve the anoikis resistance described in cells overexpressing miR-30b/c (Fig. 5B). Importantly, RPE-1 cells stably expressing shRNA targeting caspase 3 formed anchorage-independent colonies (Fig. 5C), pointing out caspase 3 as the key target for the control of cell detachment-induced apoptosis by these miRNAs.
FIGURE 5.
Caspase 3 is the main target of miR-30b/c in the anoikis-resistance context. (A) RPE-1 cells were co-infected with both lentiviruses expressing full-length miR-30b/c and the ORF of caspase 3. Western blot analysis of pro-CASP3, active CASP3, and ACTB is shown for RPE-1 cells growing either in complete medium under adherent conditions (−PH) or in anoikis-inducing conditions (+PH). Flow cytometry analysis of the levels of apoptosis (SubG1/SubG1 control) in CASP3-infected cells growing under anoikis-inducing conditions (lower panel). (B) RPE-1 cells were infected with either lentivirus expressing a shRNA to silence the expression of caspase 3 or a scramble shRNA (control). Experiments were carried out with RPE-1 infected cells previously selected in puromycin for 12 d. The diagram to the left shows CASP3 mRNA levels measured by real time PCR after puromycin selection. HPRT mRNA was used as the normalization control. On the right, levels of apoptotic cells in anoikis-inducing conditions are shown. Results are shown as the averages ± standard errors of the means from three independent experiments. The data were subjected to two-tailed Student's t-test. (**) P < 0.01, (***) P < 0.001. (C) Number of colonies after plating 2 × 104 cells from B per well in six-well plates with a bottom layer of 0.5% agar and a top layer of 0.35% agarose. Colonies were photographed and counted after 4 wk. Results are shown as the averages ± standard errors of the means from three independent experiments. The data were subjected to two-tailed Student's t-test. (**) P < 0.01.
Other members of miR-30 family do not confer anoikis resistance in RPE-1 cells
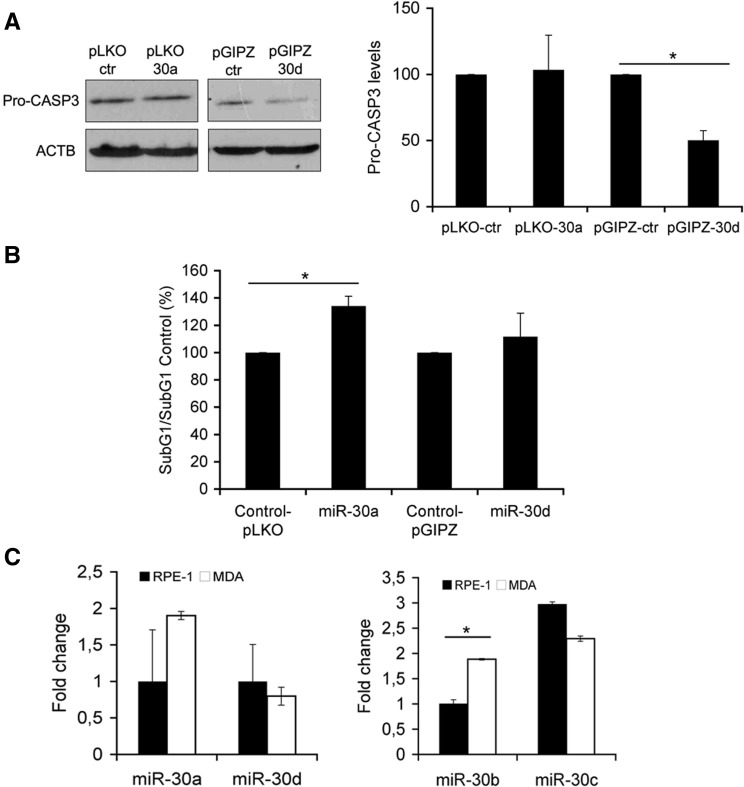
We have shown that all members of miR-30 family are present in our library (Supplemental Table S1) and presumably share most of the targets because they contain the same seed sequence at the 5′ end of their mature sequence. Based on this fact, we investigated if other miRNAs belonging to miR-30 family were able to deplete caspase 3 expression and reproduce the phenotype observed following miR-30b/c overexpression. As RNase protection assays showed, miR-30a and miR-30d were expressed properly in cells transfected with pLENT-DUAL-30a and pLENT-DUAL-30d plasmids (Supplemental Fig. S6). Using previously described lentiviral systems (Gaziel-Sovran et al. 2011; Baraniskin et al. 2012), miR-30a and miR-30d were overexpressed in RPE-1 cells (Supplemental Fig. S7), and pro-CASP3 depletion was checked by Western blot (Fig. 6A). Enforced expression of miR-30d depleted pro-CASP3 levels, but this effect was not able to attenuate apoptosis induced by lack of cell adhesion (Fig. 6B). Interestingly, the levels of pro-CASP3 were not affected by miR-30a overexpression but significantly increased anoikis in RPE-1 cells (Fig. 6A,B). Furthermore, luciferase assays using the described reporter vector (Fig. 4C) showed that overexpression of miR-30a did not deplete luciferase expression (data not shown). Notably, miR-30b, but not miR-30a/c/d, was expressed at significantly higher levels in MDA-MB-231 cells in comparison to RPE-1 cells (Fig. 6C), indicating that at least one of the miR-30 family members identified in the screening is up-regulated in this anoikis-resistant cell line.
FIGURE 6.
Hsa-miR-30a and -30d do not confer anoikis resistance in RPE-1 cells. (A) Western blot analysis of pro-CASP3 in RPE-1 cells infected with lentivirus expressing full-length miR-30a, miR-30d, or their corresponding empty vectors as control. Actin (ACTB) was used as the loading control. On the right, densitometric analysis of pro-CASP3 is shown. ACTB protein was used as a normalization control. (B) Flow cytometry analysis of the levels of apoptosis (SubG1/SubG1 control) in infected cells growing under anoikis-inducing conditions. (C) Endogenous levels of mature miR-30a/d (left panel) and miR-30b/c (right panel) expressed in RPE-1 and MDA-MB-231 (clone 4175) cells. miR-16 and miR-324 were used as normalization controls for miR-30b/c and miR-30a/d, respectively. Similar results were observed when human U6 snRNA primer and U6-snRNA (RNU6B) were used as controls. Results are shown as the averages ± standard errors of the means from three independent experiments. The data were subjected to two-tailed Student's t-test. (*) P < 0.05.
miR-30b/c negatively modulate other CASP3-dependent biological processes
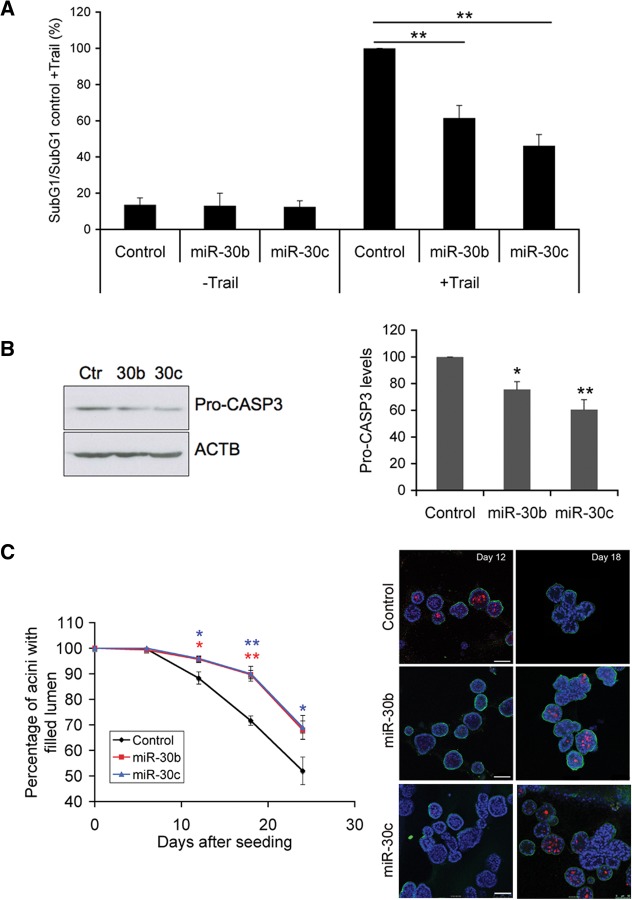
CASP3 is an executioner Caspase involved in the last stage of the apoptotic-signalling pathway (Strasser et al. 2011). Therefore, depletion of CASP3 mediated by miR-30b/c overexpression could affect diverse apoptotic mechanisms activated by other stimuli beyond the lack of cell adhesion. To this purpose, we studied CASP3-dependent apoptosis mediated by TNF-related apoptosis-inducing ligand (TRAIL) in the immortalized human mammary epithelial cell line MCF-10A. As we have previously described for anoikis, overexpression of miR-30b/c in MCF-10A cells (Supplemental Fig. S4B) led to a reduction of 35%–50% in TRAIL-mediated apoptosis compared to control cells (Fig. 7A). Importantly, a correlation between the reduction of apoptotic cells and the depletion of CASP3 protein level in cells overexpressing miR-30b/c could be observed (Fig. 7B), suggesting a direct link between the overexpression of these miRNAs and cell death resistance following down-regulation of CASP3. In order to investigate the physiological role of miR-30b/c in tumor formation, we carried out a morphogenesis assay using MCF-10A cells. These cells, when plated on a bed of extracellular matrix, undergo morphogenesis characterized by a proliferative phase and an apoptotic phase resulting in formation of proliferation-arrested 3D structures made up of polarized epithelial cells surrounding a central hollow lumen (Debnath and Brugge 2005). Interestingly, lumen formation appears to be driven by the removal of cells that have been detached from the extracellular matrix, rendering them susceptible to anoikis and other caspase-independent cell death mechanisms. To determine whether miR-30b/c interferes with morphogenesis of 3D acini, MCF10A overexpressing miR-30b/c and MCF10A control cells were seeded in Matrigel. As reported (Debnath and Brugge 2005), we observed virtually no apoptosis in the developing acini until day 5. Thereafter, as the structure grew, a delay on hollow lumen formation in acini overexpressing miR-30b/c was observed (Fig. 7C, left panel). This delay was associated with a decreased apoptotic rate in these acini in comparison with control acini. In fact, after day 12, control acini contained an increased amount of apoptotic cells, as was determined by immunofluorescence of cleaved CASP3. In contrast to this pattern, very low immunofluorescence signal of cleaved CASP3 was detected in mir-30b/c-overexpressing acini. After day 18, a significant percentage of the control acini were hollow, whereas in miR-30b/c-overexpressing acini, the luminal space was densely populated with apoptotic cells (Fig. 7C, right panel). These results revealed that miR-30b/c overexpression during mammary acini development in 3D results in a deregulation of the process, leading to a delay of acini clearance associated with a decrease of CASP3 activation.
FIGURE 7.
Hsa-miR-30b and hsa-miR-30c reduce cell death induced by TRAIL and delay acini clearance in MCF10A cells. (A) Cells were infected with lentivirus containing full-length miR-30b, miR-30c, and empty vector as a control. Four days after infection, cells were cultured in the presence of Trail (500 ng/mL) for 7 h, and the levels of apoptosis were determined (SubG1/SubG1 control + Trail) by flow cytometry. Values represent the average ± standard errors of the mean from three independent experiments. The data were analyzed by one-way ANOVA, followed by Bonferroni post-test. (**) P < 0.01. (B) Western blot analysis of pro-CASP3 levels in infected cells. ACTB was used as a loading control. On the right, a densitometric analysis of pro-CASP-3 protein levels from three independent experiments is shown. ACTB was used as a normalization control. Values represent the average ± standard errors of the mean from at least three independent experiments. The data were analyzed by one-way ANOVA, followed by Bonferroni post-test. (*) P < 0.05, (**) P < 0.01. (C) Effects of overexpression of miR-30b/c on MCF10 acini morphogenesis assays. Cells were infected as in A and assayed for acinar clearance. For 24 d, acini were fixed and analyzed by confocal microscopy every 6 d. Activation of CASP3 was determined with an antibody specific to cleaved CASP3 (red). Acinar morphology was visualized using an α6-integrin antibody (green), and 4′,6-diamidino-2-phenylindole was used to stain nuclei (blue). Results are shown as the averages ± standard errors of the means from three independent experiments, and 100 acini were analyzed in each condition. The data were analyzed by one-way ANOVA, followed by Bonferroni post-test. (*) P < 0.05, (**) P < 0.01. Percentage of acini containing four or less intact nuclei in lumen was measured at the indicated time points during morphogenesis. On the right, representative images of acini on day 12 and 18 corresponding to control or miR-30b/c-overexpressing cells. Scale bars, 75 μM.
DISCUSSION
In this work, we have developed a novel system for the direct expression of mature miRNAs. Using different approaches, we have demonstrated that these mature miRNAs have a comparable length and function similar to processed full-length miRNAs expressed from RNA polymerase II or endogenous promoters, although some particular isomir variants (Fig. 1B, upper panel; Berezikov 2011) and differences in the efficiency of gene down-regulation were found (Fig. 1E). Our system has enabled the generation of a novel type of lentiviral expression library containing a significant representation of the population of mature miRNAs from a particular cell line as well as other types of small RNAs that can function as miRNAs. At present, other miRNA expression libraries have been developed to carry out gain-of-function genetic screens in different formats such as double-stranded miRNA or expressing full-length pri-miRNA in a plasmid or a viral-based vector (Izumiya et al. 2011). Lentiviral libraries have overcome the problems derived from double-stranded miRNAs and plasmid libraries, such as efficiency of transfection and transient expression. However, all these strategies have shown limitations due to the high cost and/or technical difficulties. Moreover, the content of these libraries is limited to miRNAs annotated in the different databases. In our libraries, the small RNAs have been cloned and tested to identify those RNA molecules leading to a particular phenotype, avoiding tedious steps such as identification of putative miRNAs and the subsequent isolation and cloning of DNA precursors sequences.
pLENT-DUAL lentiviral particles are produced in a lower rate (about 10-fold less) than other conventional lentiviruses (see Materials and Methods for details). In order to get a higher yield and to guard against possible artefacts, the selected candidates in the functional screen were generated and tested as pre-miRNAs in a canonical vector. Despite this, the libraries reported here provide several significant improvements. They include known and novel small RNAs, which can be stably expressed as double-stranded RNA. As was expected, highly or moderately expressed miRNAs in MDA-MB-231 cells were found in a higher proportion in the pLENT-DUAL library. Remarkably, deep-sequencing analysis to test the quality of the library revealed that not only miRNAs but also other types of small RNAs, such as small nuclear RNAs, small nucleolar RNAs, and small RNA derived from tRNAs, were represented in the library (Fig. 2B; Supplemental Table 1). Some of these small RNAs interact with the RISC complex and have been described as “miRNA-like” RNAs capable of regulating gene expression (Berezikov 2011; Rother and Meister 2011; Dueck et al. 2012; Li et al. 2012b). Therefore, this novel approach progresses toward the generation of small RNA (especially those functioning like a siRNA or miRNA) expression libraries, and its use constitutes a valuable resource tool for gene analysis and discovery in mammalian systems. Of great interest is the application of this technology for functional screenings in cell types from less well- or noncharacterized organisms with poor small RNA sequencing data. The fact that a significant amount of miRNAs are expressed at saturating levels and/or can function redundantly in a wild-type background often complicates the study of individual miRNAs function (Wang et al. 2008). Using this novel technology, we have demonstrated that the expression of miRNAs from the pLENT-DUAL system is independent of the biogenesis of primary and precursor forms that precedes the generation of the mature miRNA, enabling the performance of long-term functional screens in different genetic backgrounds where mature endogenous miRNAs (or other small RNAs) are not being produced. Moreover, the protocol to generate the library could be adaptable to short synthetic oligonucleotides, allowing the generation of a library at much lower cost than when producing the long oligonucleotides required for classical microRNA or shRNA hairpins. In addition, once miRNAs are expressed, they result in two different mature molecules annotated as miRNA/miRNA* or miRNA-5p/miRNA-3p. An extra advantage of our libraries is to permit the specific and stable overexpression of one of the miRNA species and not its natural associated molecule.
In order to validate our technology, a functional screening was carried out in the anoikis-sensitive cell line RPE-1 infected with a lentiviral library comprising a good representation of the population of miRNAs and other small RNAs expressed in the cell line MDA-MB-231. This screening enabled us to identify hsa-miR-30b and hsa-miR-30c as novel negative regulators of cell death induced by loss of attachment even when these miRNAs were present at low percentage in our library. This supports the high potential of our libraries to uncover miRNAs and other “miRNA-like” small RNAs. The phenotype described for the mature miRNAs overexpressed using our system was confirmed by expression of the full-length miRNA versions, validating our previous data on the novel small RNAs expression system and demonstrating that the phenotype observed was due to the function of the guide strands of miR-30b/c and not to the complementary or passenger strands. These results are consistent with previously published data that classify miR-30b and miR-30c as oncomirs (Volinia et al. 2010; Garofalo et al. 2011). In addition, a paper demonstrating a role of miR-30b/c as oncomirs in glioma and lung cancer cells has been recently published (Quintavalle et al. 2013). In these tumor-derived cells, miR-30b/c conferred TRAIL-induced apoptosis resistance targeting caspase 3 through the conserved binding site in its 3′ UTR, similarly as we have demonstrated for immortalized epithelial cells. These recent and independent results confirm and reinforce this novel small RNA expression system as a powerful technique for uncovering new small RNAs (miRNAs, among others) involved in a particular biological process. In addition, our results point out that not all members belonging to a particular miRNA family are functionally similar, and they can even be involved in antagonistic processes. In fact, no additional members of the miR-30 family sharing the seed sequence were identified in this screening, although they were present in the library. Despite the fact that miR-30d depleted pro-CASP3, as has been recently proposed for cancer cells (Li et al. 2012a), this does not seem to be sufficient to block anoikis in these immortalized cells. The supplementary pairing outside of the seed region of miRNAs affects the target specificity, decreasing the dissociation rate of the bound silencing complex (Brennecke et al. 2005; Grimson et al. 2007). Interestingly, miR-30d and miR-30b/c differ significantly outside of the seed region (nt 13,14, and 18–20). This fact could generate, for each miRNA, specific targets involved in other apoptotic or anti-apoptotic processes. Because under our experimental conditions, anoikis seemed to be mainly reduced by only down-regulation of CASP3 in RPE-1, miR-30d overexpression might affect other unknown targets in this cell line that offset the depletion of CASP3 levels. Notably, only one mismatch at the 12th nucleotide was found between miR-30a and miR-30d. This nucleotide may be important because, in cells overexpressing miR-30a, neither a depletion of pro-CASP3 nor an inhibition of cell death induced by lack of attachment was observed. In addition, luciferase assays using the 3′ UTR of CASP3 cloned into a reporter vector showed that overexpression of miR-30a did not deplete luciferase expression (data not shown). In contrast, a significant increase of the cell death (Fig. 6B) as well as cell growth inhibition was clearly visible in RPE-1 cells overexpressing miR-30a, suggesting that ectopic expression of this miRNA was functional and related to the observed tumor suppression. Consequently, miR-30a has been described as a suppressor of tumor growth and inhibitor of cell migration and invasion (Baraniskin et al. 2012; Cheng et al. 2012; Kumarswamy et al. 2012; Zhang et al. 2013). Altogether, our results indicate that miR-30b/c and not other miR-30 family members are specifically involved in anoikis resistance. Interestingly, mature miR-30b and miR-30c shared the highest degree of identity beyond the seed sequence within miR-30 family members, and miR-30b/c were expressed more abundantly in the metastatic MDA-MB-231 cell line than in immortalized MCF-10A cells (Wang et al. 2009). MiR-30b, and not other miR-30 family members, was also overexpressed in MDA-MB-231 cells compared to RPE-1. Although the depletion of miR-30b/c did not affect the anoikis resistance in MDA-MB-231 cells (data not shown), these miRNAs seem to collaborate positively in tumor progression. Importantly, miR-30a and miR-30d are more abundant than miR-30b and miR-30c in the generated library from MDA-MB-231 cells (miR-30a = 0.416%; miR-30d = 0.19%; miR-30b = 0.0173%; miR-30c = 0.187%), but only miR-30b and miR-30c were selected in the functional screening. This reinforces the idea that the library represents a bona fide technology for functional screenings, making possible the isolation of specific members belonging to a particular miRNA family and the identification of small RNAs that are underrepresented.
One of the mechanisms behind the progression of tumors is acquired resistance to apoptosis (Hanahan and Weinberg 2011). In particular, anoikis has been shown to play an important role during tumorigenesis (Chiarugi and Giannoni 2008). The regulation of apoptotic cell signalling by miRNAs is still not fully understood despite considerable research during recent years (Lima et al. 2011). Several miRNAs have been shown to repress anoikis by targeting different genes (Gramantieri et al. 2009; Penna et al. 2011). In this work, we have identified caspase 3 as a main target down-regulated by miR-30b/c conferring an anoikis-resistance phenotype in immortalized epithelial cells. A single miRNA can block the expression of many target mRNAs. However, the anoikis-resistant phenotype observed with miR-30b/c overexpression could be mimicked using a specific CASP3 shRNA and overcome by CASP3 expression, suggesting that cell death inhibition by miR-30b/c was mainly mediated through down-regulation of pro-CASP3 levels. In fact, previous reports have demonstrated that down-regulation of pro-CASP3 expression leads to reduced apoptosis induced by different stimuli (Kugler et al. 2005; Ovcharenko et al. 2007).
Other cell death processes depending on caspase 3 were controlled by miR-30b/c, including TRAIL-induced cell death, which was partially abolished in immortalized epithelial MCF10A cells overexpressing these miRNAs. In agreement with this result, a subset of miRNAs that targets pro-CASP3 in other cellular and genetic backgrounds, such as miR-155, miR-Let-7a, miR-378, and more notably, miR-30b/c/d, enhanced cell viability and suppressed apoptosis in different physiological contexts, suggesting that their deregulation may lead, in the case of some of them, to tumorigenesis (Tsang and Kwok 2008; Fang et al. 2011; Wang et al. 2011; Li et al. 2012a; Quintavalle et al. 2013). Strikingly, Kaposi's sarcoma herpesvirus miR-K12-1, -3, and -43p miRNAs targeted endogenous caspase 3. This resulted in decreased apoptosis activity in different cell types, which could contribute to the malignant phenotype triggered by KSHV infection (Suffert et al. 2011). These results point to the possible role of miR-30b/c as oncomirs through blocking caspase 3 expression. In this regard, the overexpression of miR-30b/c led to a delay in acini clearance linked to decreased CASP3 activation when a three-dimensional Matrigel-induced acinar morphogenesis assay system was used. This model resembles important histopathological features observed in epithelial cancers in vivo (Debnath and Brugge 2005). Interestingly, miR-30c has been reported as highly expressed in different types of solid cancers such as prostate, colon, and pancreas (Volinia et al. 2010), and the overexpression of miR-30b has been related to high metastatic potential of melanoma cells (Gaziel-Sovran et al. 2011) and to medulloblastoma (Lu et al. 2009). Recently, miR-30b/c have been described as important factors in the resistance to gefinitib by targeting BIM in lung cancers (Garofalo et al. 2011). However, we could not observe a depletion of BIM in the immortalized cell line RPE-1. Probably, this could be explained by differences in the genetic backgrounds of these cell types. On the other hand, lack of expression of caspase 3 has been described in breast cancer (Devarajan et al. 2002) and in human prostatic tumors. Remarkably, a significant loss of CASP3 protein, but not mRNA levels, has been reported in human prostatic tumors (Winter et al. 2001), suggesting a post-transcriptional control of caspase 3 expression as it has been described in this work, mediated by miR-30b/c. Altogether, these findings indicate that the overexpression of miR-30b/c can represent a potential step during the loss of apoptotic control during tumorigenesis.
Furthermore, high levels of miR-30b/c could also play an important role in resistance to death ligand-induced and drug-induced apoptosis in cancer cells, as described here for human mammary epithelial cells and in other cell types in which caspase 3 expression is inhibited by these and other miRNAs (Suffert et al. 2011; Wang et al. 2011; Quintavalle et al. 2013), confirming the network miR-30b/c-caspase 3 as a broad contributing factor for tumorigenesis. However, it cannot be ruled out that other effects arising from the overexpression of miR-30b/c could affect tumor progression.
In summary, our data highlight the power of our system to generate novel small RNA expression libraries that can be used to perform large-scale screens for gain-of-function phenotypes in any given biological phenomenon. As a proof of principle, and as an example of the identification of novel functions for known miRNAs, we isolated two miRNAs that confer cellular resistance to anoikis. Furthermore, the unbiased nature of our technology provides the possibility to discover previously undescribed miRNAs and other types of small RNAs functioning as miRNA or siRNA with regulatory functions, which constitutes an important breakthrough when compared to current approaches. We believe that this methodology will lead to a better understanding of the complex mechanisms underlying the control of gene expression by small noncoding RNAs.
MATERIALS AND METHODS
Cell culture
Immortalized human retinal pigment epithelial cell line hTERT RPE-1 (Clontech) was maintained in DMEM/F12 supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and antibiotics (50 units/mL penicillin, 50 μ/mL streptomycin). HEK293T transformed cell line and colorectal carcinoma HCT116 cell line (obtained from the American Type Culture Collection) were grown in DMEM and McCoy's 5A, respectively, with the same supplements as RPE-1 cells. The human breast epithelial cell line MCF-10A was grown in DMEM/F12 supplemented with 5% donor horse serum, 2 mM L-glutamine, 20 ng/mL epidermal growth factor (EGF), 10 μ/mL insulin, 100 ng/mL cholera toxin/mL, and 0.5 µg/mL hydrocortisone. MDA-MB-231 (clone 4175) cells were grown in high-glucose DMEM medium with 10% fetal bovine serum.
Constructs
The pLENT-DUAL plasmid has been derived from pFIV-H1/U6-Puro (System Biosciences) with the following modifications. The puromycin gene was replaced by EGFP as a marker. In addition, the modified lentiviral vector pLENT-DUAL contains two RNA polymerase III terminators downstream from the transcriptional start sites of H1 and U6 promoters and two BbvII restriction sites as a cloning site. Processed sequences (mature miRNA based-DNA oligonucleotides annealed to their complementary sequences) of different small RNAs were cloned into the BbvII cloning sites flanked by both RNA polymerase III promoters H1 and U6 (Supplemental Fig. S1). The pLuc-BS plasmid was derived from pCDNA3.1 and contains the ORF of the luciferase gene luc2 (Photinus pyralis) obtained from the pGL4 plasmid (Promega), followed by a multicloning site (XbaI, KpnI, BamHI, BstXI, EcoRI, EcoRV, NotI, XhoI, and PmeI restriction sites) where binding sites sequences for the different miRNAs or complete 3′ UTR of caspase 3 were cloned. The 3′ UTR of caspase 3 was amplified by PCR using the primers 5′-CCGAATTCAGAAATGGTTGGTTGGTGGTT-3′ and 5′-CCCTCGAGTAATTTTAAGTTTGAATGTATATTTTG-3′, containing EcoRI and XhoI restriction sites, respectively.
Transfections, lentivirus production, and infections
Transfection of HCT116 cells was performed using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's protocol. Transfection of HEK293T cells was performed using the CalPhost Kit (Clontech). The HIV packaging (pCMVDR8.91) and VSV-G (pMDG) plasmids and the lentiviral vectors pHRSIN-DUAL-GFP (also known as pHRSIN-CSGWdINotI_pUb_Em [Mazzon et al. 2009]) were kindly provided by Mary K. Collins (Windeyer Institute, London). The full-length miR-30b/c and ORF sequence of caspase 3 were cloned into the BamHI/NotI restriction sites of the pHRSIN-DUAL-GFP plasmid. The lentiviral vectors pGIPZ-30d (Gaziel-Sovran et al. 2011) and pLKO-30a (Baraniskin et al. 2012), containing miRNA-30d and miR-30a precursors, respectively, and their corresponding controls were kindly provided by Eva Hernando (New York University Medical Center) and Stephan A. Hahn (Molecular GI-Oncology, Center of Clinical Research, Ruhr-University, Bochum, Germany), respectively.
Fragments of 675 bp and 665 bp containing pre-miR-30b and pre-miR-30c-1, respectively, were amplified from RPE-1 genomic DNA using the primers 5′-CGGGATCCCCTGGGCAATATAGTGAGACC-3′ and 5′-CGGCGGCCGCATTGAGTAACAAATTTAAAGG-3′ for pre-miR-30b and 5′-CGGGATCCGATGGGGTCAATACCAGCTTG-3′ and 5′-CGGCGGCCGCGGTTAATGGGAAACAGGGCTTTTCC-3′ for pre-miR-30c-1. Caspase 3 ORF sequence was amplified using the primers 5′-GCGGATCCATGGAGAACACTGAAAACTCAG-3′ and 5′-CACGCGGCCGCTTAGTGATAAAA ATAGAGTTC-3′. Insertion and orientation of DNA fragments were confirmed by restriction enzyme digestions and sequencing. For the production of lentivirus, 3 × 106 293T cells were seeded onto a 10-cm Petri dish and transfected with Lipofectamine 2000 (Invitrogen) using 13.5 µg of the transfer vector pHRSIN carrying caspase 3 ORF, miR-30b, or miR-30c-1, pGIPZ-30d, pLKO-30a, or their corresponding controls, 9 µg of pCMVDR8.91, and 4.5 µg pf pMDG. Lentiviruses were harvested 48 h post-transfection, passed through a 0.45-µm filter, and concentrated by ultracentrifugation at 100,000g for 90 min. Virus particles were resuspended in serum-free DMEM-F12 (Invitrogen) and stored at −80°C. Lentiviral particle titers were determined by flow cytometry in RPE-1 and MCF10A infected cells yielding 10 mL of 0.1–1 × 106 infection units per mL of nonconcentrated supernatant from one 10-cm culture dish (pLKO, pSIN-DUAL, pGIPZ). For infection with the lentiviral stock, RPE-1 or MCF10A cells were seeded at 8 × 104 cells in a six-well plate 24 h prior to infection, then lentiviral particles (MOI = 4 for miRNAs expression and MOI = 2 for CASP3 expression) were added, and the culture medium was replaced 24 h after infection. In the case of MCF10A cells, 8 µg/mL polybrene were added. Production of FIV pseudoviral particles (pLENT-DUAL) was performed using the pFIV Packaging kit (System Biosciences) following the manufacturer's protocol with minor modifications. The yield of recombinant dual promoter lentiviral particles was 10 mL of 5–9 × 104 infection units per mL of nonconcentrated supernatant from one 10-cm culture dish, resulting in titers about 10-fold lower, on average, than other conventional lentiviruses. Transduction of cells with FIV lentiviruses was carried out as described above for pHRSIN-DUAL at MOI = 2. The lentiviral DNA was amplified by PCR using the primers H1: 5′-AATGTCTTTGGATTTGGGAATCTTATA-3′ and U6: 5′-AAAGTAATAATTTCTTGGGTAGTTTGC-3′. Lentiviruses for shRNA interference of CASP3 mRNA were purchased from Sigma-Aldrich. RPE-1 cells were infected at MOI = 4 and grown for 12 d in puromycin at 4 µg/mL. Experiments were carried out uniquely with puromycin-resistant RPE-1 cells.
Library construction
Total RNA from MDA-MB-231 human breast cancer cells was extracted by using TRIZOL Reagent (Invitrogen) according to the manufacturer's instruction. RNAs of 19 to 29 nt in length were size-selected on a 15% denaturing polyacrylamide gel, eluted, and ethanol-precipitated in siliconized tubes, with glycogen as carrier. Gel-purified RNA was first dephosphorylated to prevent circularization and incubated with 5 µM 3′-adapter oligonucleotide (5′-puuuuuuGTCTTCACTGAGCGGGCATA/3ddC/-3′; p, phosphate; ddC, Dideoxycytidine; lowercase, RNA; uppercase, DNA) with a blocked 3′ terminus to prevent the participation in the ligation reaction (50 mM Tris-HCl, pH 7.8, 10 mM Mg Cl2, 10 mM DTT, 1 mM ATP, 15% DMSO, 1 unit/µL T4 RNA ligase). The ligation products of 46 to 56 nt were purified on a 15% denaturing polyacrylamide gel. The eluted product was phosphorylated and added to a secondary T4 RNA ligase reaction, similar to the first one except that 5 µM 5′-adapter oligonucleotide (5′-CGGCTCAACTCTCTGGAAGACTCaaag-3′; lowercase, RNA; uppercase, DNA) replaced the 3′-adapter oligonucleotide. The new ligation products were gel- purified and reverse transcribed into cDNAs in a standard 30-µL reverse transcription reaction (SuperScript III; Invitrogen) with the biotinylated RT primer (5′-/5Bio/GTATGCCCGCTCAGTGAA). The cDNA was amplified, using 10 µL from the reverse transcription reaction as a template, the RT primer, and a biotynilated oligonucleotide within the 5′-adapter (5′-/5Bio/CGGCTCAACTCTCTGGAA-3′) following this PCR protocol: 2 min at 94°C, 20 cycles of 45 sec at 94°C, 30 sec at 50°C, and 30 sec at 72°C, and a final step of 7 min at 72°C. PCR products were gel-purified, ethanol-precipitated, and digested with BbvII restriction enzyme. Digested products were incubated with streptavidin-coupled Dynabeads (Invitrogen), and the supernatant, containing the internal fragments, was ethanol-precipitated. The small cDNA fragments were directionally cloned into the BbvII-digested pLENT-DUAL lentiviral vector. E. coli cells were electroporated and more than 8 × 105 independent colonies were pooled to comprise the library.
RNA and DNA samples for deep-sequencing libraries
Fifty nanograms of size-selected miRNA (19–29 nt, using acrylamide gel separation) was used for miRNA library construction with the Ion Total RNA-Seq Kit v2 following the Ion Total RNA-Seq Kit v2 for Small RNA Libraries protocol. One hundred nanograms of amplicons (10 cycles of amplification using the primers H1: 5′-AATGTCTTTGGATTTGGGAATCTTATA-3′ and U6: 5′-AAAGTAATAATTTCTTGGGTAGTTTGC-3′) from the pLENT-DUAL library with an average length of 250 nt was used for PCR products library construction with the Ion Xpress Plus Fragment Library kit following the protocol for >400-nt PCR products detailed on the Ion Xpress Plus gDNA and Amplicon Library. Library concentrations were estimated with Q-PCR with the Ion Library Quantification kit (Life Technologies) prior to clonal amplification.
Emulsion PCR and sequencing
The emulsion PCR was carried out applying the Ion Xpress Template kit V2.0 (Life Technologies) as described in the appropriate user guide provided by the manufacturer. Quality and quantity of the enriched spheres were checked on a Qubit 2.0 instrument using the Qubit assay (Life Technologies) as described in the appendix of the Ion Xpress Template Kit User Guide. Sequencing of the libraries was carried out on the Ion Torrent Personal Genome Machine (PGM) system (Lifesequencing S.L.) using the Ion Sequencing 200 kit following the corresponding protocol. In brief, the chip was washed, and the spheres were loaded twice onto a 318 chip (Life Technologies); each loading was followed by four rounds of centrifugation at maximum speed for 1 min (Mini Star; VWR International GmbH), and the complete sample was loaded. For every sample a separate 318 chip was used.
Bioinformatics
Analysis of the sequencing data was performed by miR-Intess small RNA pipeline (InteRNA Genomics B.V.) (Berezikov et al. 2010). Reads were preprocessed to trim the adapter sequences and mapped against the human genome assembly GRCh37. Annotations of the mapped loci were retrieved from Ensemble database (v. 69) and from miRBase (v. 19), and aligned reads were classified according to these annotations. Prediction of novel miRNA candidates was performed by miR-Intess as described previously (Berezikov et al. 2010).
RNase protection assay
The expression of processed miRNAs from pLENT-DUAL was analyzed by RNase protection assay (RPA). HEK293T cells were seeded in six-well plates at 50% confluence the day before transfection. Forty hours after transfection, RNA was collected and enriched in small RNA using the mirVana miRNA Isolation kit (Ambion). The mirVana miRNA probe construction kit (Ambion) was used to synthesize 32P-labeled miR-21, miR-19a, miR-15a, miR-30a, miR-30b, miR-30c, and miR-30d sense (guide strand) and antisense (complementary strand) probes. Primers to make the RPA probes are listed in the Supplemental Material. Probe hybridization and RNase protection were carried out using the mirVana miRNA detection kit (Ambion) according to the manufacturer's instructions. After hybridization and RNase treatment, the protected RNAs were resolved on a 15% denaturing urea polyacrylamide gel and visualized using phosphoimaging and autoradiography. RNA markers were γ-32P-end-labeled Decade RNA markers (Ambion) prepared according to the manufacturer's instructions.
Luciferase assays
Cells were harvested 40 h post-transfection and luciferase activity was measured using a luciferase assay kit (Promega) according to the manufacturer's protocol. Renilla activity was used for normalization control.
Soft-agar anoikis assay
Cells were plated at a density of 105 cells per 100-mm plate with a bottom layer of 3% agarose and a top layer of 0.5% agarose. The functional screening was carried out plating 1.2 × 106 and 2 × 105 cells infected with lentiviral library and pLENT-DUAL (empty) as a control, respectively. Cells were grown for 19 d at 37°C in 5% CO2, and clones were picked up under a dissecting microscope and transferred to adhesive plates to allow their expansion. Seventeen anoikis-resistant clones were finally obtained from cells infected with the library and one clone from control cells that was unable to grow on adhesive plates.
Short-term cell death assays
Anoikis was induced in vitro by transferring serum-deprived RPE-1 cells, to plates treated with polyHEMA (to prevent cellular-matrix adhesion) in the presence of 2% methylcellulose (to inhibit cell-cell interactions) and serum-free medium for an additional 24 h.
For TRAIL-mediated cell death experiments, MCF10A cells were treated with this apoptosis inducer at 500 ng/mL for 7 h. Soluble human His-tagged recombinant TRAIL (residues 95–281) was produced as previously described (Yerbes et al. 2011).
Immunoblotting
Cell lysates were prepared by lysing cells in RIPA buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1% NP40, 0.25% Na-deoxycholate) containing a complete cocktail of protease inhibitors (Roche Diagnostics) plus 1 mM PMSF and resolved on SDS-PAGE. Immunoblotting was performed on nitrocellulose membrane according to the manufacturer's instructions. Commercially available antibodies to full-length and cleaved CASP3 (Cell Signaling), β-actin (Sigma-Aldrich), p53 (Santa Cruz Technology), and BIM-EL (Calbiochem) were used as recommended by the manufacturers. Signals were detected by using the Enhanced Chemilumiescence Reagent ECL kit (GE Healthcare). Protein levels were quantified using LabScan and ImageQuant TL software (GE Healthcare).
Quantitative RT-PCR
Real time PCR experiments were carried out using 1 µL of cDNAs generated from 1 µg of total RNA using the Qiagen OneStep RT-PCR kit (Qiagen). The Quantimir RT Small RNA Kit (System Biosciences) was used for small RNA quantification following the instructions of the manufacturer. Expression of CASP3, HPRT, mature miR-30b/c, miR-16, and U6 snRNA (System Biosciences) were determined by using the LightCycler-FastStart DNA SYBR Green I Kit (Roche) and the ABI PRISM 7500 instrument (Applied Biosystems). The PCR cycling profile consisted of incubation at 50°C for 2 min, followed by a denaturing step at 95°C for 10 min, and 40 cycles at 95°C for 15 sec, and 60°C for 1 min for CASP3 and HPRT and at 63°C for 1 min for miRNAs. The following primers were used: CASP3: forward, 5′-CAGAGGGATCGTTGTAGAA-3′ and reverse, 5′-GTATGCATACAAGAAGTCGGC-3′; HPRT: forward, 5′-GGACTAATTATGGACAGGACTG-3′ and reverse, 5′-TCCAGCAGGTCAGCAAAGAA-3′. For miR-30b/c and miR-16, the forward primers corresponded to the mature sequence of these miRNAs, and the 3′ universal reverse primer was provided with the Quantimir RT Small RNA Kit. Alternatively, for quantitative real-time PCR of miRNA-30a and miRNA-30d, 15 ng of total RNA was reverse-transcribed in a 10-μL total reaction using specific stem–loop RT primers (Applied Biosystems), and 2.5 μL of cDNA was subjected to a TaqMan miRNA assay (Applied Biosystems) in a 20-μL total reaction using primers and a probe specific for miR-30a or miR-30d, according to the manufacturer's protocol. Expression values were calculated using a comparative Ct method. miR-324 and U6-snRNA (RNU6B) were used as endogenous controls.
Flow cytometry
For flow cytometry analysis, floating and adherent cells were washed in phosphate-buffered saline (PBS), fixed in 70% ethanol, and stained with propidium iodide (50 µg/mL). EGFP-expressing cells were analyzed without previous fixation. Cells were analyzed by using a FACScalibur (Becton Dickinson Immunocytometry Systems).
Morphogenesis assay and immunofluorescence
Assays were performed as previously described (Yerbes et al. 2011). Briefly, MCF-10A cells were resuspended in assay medium (DMEM/F12 supplemented with 2% donor horse serum, 10 μg of insulin/mL, 100 ng of cholera toxin/mL, 0.5 μg of hydrocortisone/mL, and 5 ng of EGF/mL) 4 d after infection with different lentiviruses. Eight-well RS glass slides (BD Falcon) were coated with 40 μL of Matrigel per well. Five thousand cells per well were plated in assay medium containing a final concentration of 2% Matrigel and replaced every 4 d. For the immunofluorescence experiments, acini were fixed in 4% formalin for 25 min at room temperature. Fixed structures were washed with PBS-glycine (130 mM NaCl, 7 mM Na2HPO4, 100 mM glycine, pH 7.4) three times for 15 min each time. The structures were then blocked in IF buffer (130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 7.7 mM NaN3, 0.1% bovine serum albumin, 0.2% Triton X-100, 0.05% Tween-20) plus 10% goat serum for 1 to 2 h, followed by 2% blocking buffer (i.e., IF buffer containing 10% goat serum and 20 μg of goat anti-mouse F(ab)2/mL) for 40 min. Primary antibodies against cleaved CASP3 (Cell Signaling) and α6-Integrin (Chemicon) were diluted in 2% blocking buffer, followed by incubation overnight at 4°C. Structures were washed three times in IF buffer for 15 min each. Anti-mouse or anti-rabbit secondary antibodies coupled with Alexa Fluor dyes (Molecular Probes) were diluted in IF buffer containing 10% goat serum, followed by incubation for 60 min. After a wash with IF buffer as described above, structures were incubated with 0.5 ng of DAPI (4′,6-diamidino-2-phenylindole)/mL (Sigma) before being mounted with the anti-fade agent Prolong Gold (Molecular Probes). Confocal analysis was performed in a Leica TCS-SP5 laser microscope. Active CASP3 was determined in GFP-positive acini.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Llamazares for proofreading of the manuscript, I. Luque (IBVF-CSIC) for helping with the generation of the E. coli library, I. Palmero (IIB-CSIC), Y.Y. Mo (Southern Illinois University), Eva Hernando (New York University Medical Center) and Stephan A. Hahn (Ruhr-University) for providing pSicoR-DGCR8, pCMV-miR21 full-length plasmid, pGIPZ-30d, and pLKO-30a. We also thank B. Vogelstein (Johns Hopkins University) and J. Massagué (Memorial Sloan-Kettering Cancer Center) for providing HCT116 DICERex5 and MDA-MB-231 (clone 4175) cells, respectively. J.A.P.-T. was supported by grants from the Ministerio de Educación y Ciencia of Spain and the Dirección General de Universidades e Investigación of Junta de Andalucía. M.A.M.-M. and C.M.-V. were recipients of a post-doctoral contract from the Junta de Andalucía and Spanish National Research Council respectively. C.R.-M. was recipient of a FPI fellowship from the Ministerio de Economia y Competitividad.
REFERENCES
- Alfano D, Iaccarino I, Stoppelli MP. 2006. Urokinase signaling through its receptor protects against anoikis by increasing BCL-xL expression levels. J Biol Chem 281: 17758–17767. [DOI] [PubMed] [Google Scholar]
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. 2008. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27: 2128–2136. [DOI] [PubMed] [Google Scholar]
- Baraniskin A, Birkenkamp-Demtroder K, Maghnouj A, Zollner H, Munding J, Klein-Scory S, Reinacher-Schick A, Schwarte-Waldhoff I, Schmiegel W, Hahn SA. 2012. MiR-30a-5p suppresses tumor growth in colon carcinoma by targeting DTL. Carcinogenesis 33: 732–739. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E. 2011. Evolution of microRNA diversity and regulation in animals. Nat Rev 12: 846–860. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Liu N, Flynt AS, Hodges E, Rooks M, Hannon GJ, Lai EC. 2010. Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat Genet 42: 6–9; author reply 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. 2008. The microRNA.org resource: Targets and expression. Nucleic Acids Res 36: D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. 2005. Principles of microRNA-target recognition. PLoS Biol 3: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MJ, Perez de Castro I, Malumbres M. 2008. Control of cell proliferation pathways by microRNAs. Cell Cycle 7: 3143–3148. [DOI] [PubMed] [Google Scholar]
- Cheng CW, Wang HW, Chang CW, Chu HW, Chen CY, Yu JC, Chao JI, Liu HF, Ding SL, Shen CY. 2012. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res Treat 134: 1081–1093. [DOI] [PubMed] [Google Scholar]
- Chiarugi P, Giannoni E. 2008. Anoikis: A necessary death program for anchorage-dependent cells. Biochem Pharmacol 76: 1352–1364. [DOI] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. 2006. The colorectal microRNAome. Proc Natl Acad Sci 103: 3687–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Brugge JS. 2005. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer 5: 675–688. [DOI] [PubMed] [Google Scholar]
- Devarajan E, Sahin AA, Chen JS, Krishnamurthy RR, Aggarwal N, Brun AM, Sapino A, Zhang F, Sharma D, Yang XH, et al. 2002. Down-regulation of caspase 3 in breast cancer: A possible mechanism for chemoresistance. Oncogene 21: 8843–8851. [DOI] [PubMed] [Google Scholar]
- Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G. 2012. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res 40: 9850–9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Song XW, Tian J, Chen HY, Li DF, Wang JF, Ren AJ, Yuan WJ, Lin L. 2011. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis 17: 410–423. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa H, Noguchi K, Masumi A, Murakami Y, Uehara Y. 2004. BimEL is an important determinant for induction of anoikis sensitivity by mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitors. Mol Cancer Ther 3: 1281–1288. [PubMed] [Google Scholar]
- Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon YJ, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G, et al. 2011. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med 18: 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, Vega-Saenz de Miera E, Rakus JF, Dankert JF, Shang S, Kerbel RS, et al. 2011. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 20: 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L, Negrini M. 2009. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res 15: 5073–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. 2007. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, Pena JT, Nusbaum JD, Morozov P, Ludwig J, et al. 2011. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA 17: 1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Howe EN, Cochrane DR, Richer JK. 2011. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res 13: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S. 2010. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Izumiya M, Tsuchiya N, Okamoto K, Nakagama H. 2011. Systematic exploration of cancer-associated microRNA through functional screening assays. Cancer Sci 102: 1615–1621. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139. [DOI] [PubMed] [Google Scholar]
- Kugler W, Buchholz F, Kohler F, Eibl H, Lakomek M, Erdlenbruch B. 2005. Downregulation of Apaf-1 and caspase-3 by RNA interference in human glioma cells: Consequences for erucylphosphocholine-induced apoptosis. Apoptosis 10: 1163–1174. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. 2007. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39: 673–677. [DOI] [PubMed] [Google Scholar]
- Kumarswamy R, Mudduluru G, Ceppi P, Muppala S, Kozlowski M, Niklinski J, Papotti M, Allgayer H. 2012. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer 130: 2044–2053. [DOI] [PubMed] [Google Scholar]
- Li N, Kaur S, Greshock J, Lassus H, Zhong X, Wang Y, Leminen A, Shao Z, Hu X, Liang S, et al. 2012a. A combined array-based comparative genomic hybridization and functional library screening approach identifies mir-30d as an oncomir in cancer. Cancer Res 72: 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. 2012b. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 40: 6787–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima RT, Busacca S, Almeida GM, Gaudino G, Fennell DA, Vasconcelos MH. 2011. MicroRNA regulation of core apoptosis pathways in cancer. Eur J Cancer 47: 163–174. [DOI] [PubMed] [Google Scholar]
- Lu Y, Ryan SL, Elliott DJ, Bignell GR, Futreal PA, Ellison DW, Bailey S, Clifford SC. 2009. Amplification and overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22-q24.23 in medulloblastoma. PLoS One 4: e6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. 2009. DIANA-microT web server: Elucidating microRNA functions through target prediction. Nucleic Acids Res 37: W273–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. 2009. Dengue virus NS5 inhibits interferon-α signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis 200: 1261–1270. [DOI] [PubMed] [Google Scholar]
- Ovcharenko D, Kelnar K, Johnson C, Leng N, Brown D. 2007. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res 67: 10782–10788. [DOI] [PubMed] [Google Scholar]
- Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De Pitta C, et al. 2011. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J 30: 1990–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintavalle C, Donnarumma E, Iaboni M, Roscigno G, Garofalo M, Romano G, Fiore D, De Marinis P, Croce CM, Condorelli G. 2013. Effect of miR-21 and miR-30b/c on TRAIL-induced apoptosis in glioma cells. Oncogene 32: 4001–4008. [DOI] [PubMed] [Google Scholar]
- Rother S, Meister G. 2011. Small RNAs derived from longer non-coding RNAs. Biochimie 93: 1905–1915. [DOI] [PubMed] [Google Scholar]
- Strasser A, Cory S, Adams JM. 2011. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J 30: 3667–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suffert G, Malterer G, Hausser J, Viiliainen J, Fender A, Contrant M, Ivacevic T, Benes V, Gros F, Voinnet O, et al. 2011. Kaposi's sarcoma herpesvirus microRNAs target caspase 3 and regulate apoptosis. PLoS Pathog 7: e1002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. 2010. Non-coding RNAs: Regulators of disease. J Pathol 220: 126–139. [DOI] [PubMed] [Google Scholar]
- Toedling J, Servant N, Ciaudo C, Farinelli L, Voinnet O, Heard E, Barillot E. 2012. Deep-sequencing protocols influence the results obtained in small-RNA sequencing. PLoS One 7: e32724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WP, Kwok TT. 2008. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis 13: 1215–1222. [DOI] [PubMed] [Google Scholar]
- Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu Jarour R, et al. 2010. Reprogramming of miRNA networks in cancer and leukemia. Genome Res 20: 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. 2008. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40: 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rathinam R, Walch A, Alahari SK. 2009. ST14 (suppression of tumorigenicity 14) gene is a target for miR-27b, and the inhibitory effect of ST14 on cell growth is independent of miR-27b regulation. J Biol Chem 284: 23094–23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis D, Jia LT, Wu SX, Huang J, Chen J, Luo ZJ. 2011. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J Pathol 225: 232–242. [DOI] [PubMed] [Google Scholar]
- Winter RN, Kramer A, Borkowski A, Kyprianou N. 2001. Loss of caspase-1 and caspase-3 protein expression in human prostate cancer. Cancer Res 61: 1227–1232. [PubMed] [Google Scholar]
- Yerbes R, Palacios C, Reginato MJ, Lopez-Rivas A. 2011. Cellular FLIPL plays a survival role and regulates morphogenesis in breast epithelial cells. Biochim Biophys Acta 1813: 168–178. [DOI] [PubMed] [Google Scholar]
- Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, Hu G, Yang Q. 2013. MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene 10.1038/onc.2013.286. [DOI] [PubMed] [Google Scholar]
- Zheng L, Liu J, Batalov S, Zhou D, Orth A, Ding S, Schultz PG. 2004. An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc Natl Acad Sci 101: 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang F, Fuchs RT, Robb GB. 2012. Small RNA expression profiling by high-throughput sequencing: Implications of enzymatic manipulation. J Nucleic Acids 2012: 360358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.