Abstract
Background
Our previous study indicated that gene expression profiling of intestinal metaplasia (IM) or spasmolytic polypeptide-expressing metaplasia (SPEM) can identify useful prognostic markers of early stage gastric cancer, and seven metaplasia biomarkers (MUC13, CDH17, OLFM4, KRT20, LGALS4, MUC5AC, and REG4) were selectively expressed in 17-50% of gastric cancer tissues. We investigated whether the combined expression of these metaplasia biomarkers could predict the prognosis of advanced stage gastric cancer.
Methods
The expression of seven metaplasia biomarkers was evaluated immunohistochemically using tissue microarrays comprised of 450 gastric cancer patients. The clinicopathologic correlations and the prognostic impact were analyzed according to the expression of multiple biomarkers.
Results
MUC13, CDH17, LGALS4 and REG4 were significant prognostic biomarkers in univariate analysis. No expression of four markers was found in 56 cases (14.2%); 1 marker was seen in 67 cases (17.0%), 2 in 106 cases (27.0%), 3 in 101 cases (25.7%), and 4 in 63 cases (16.0%). Patients in which ≤ 2 proteins were expressed (Group B) showed younger age, undifferentiated or diffuse type cancer, a larger tumor size, a larger number of metastatic lymph nodes and more advanced stage than those in which ≥ 3 proteins were expressed (Group A). In undifferentiated or stage II/III gastric cancer, the prognosis of Group B was significantly poorer than that of Group A by multivariate analysis.
Conclusion
The combined loss of expression of multiple metaplasia biomarkers is considered as an independent prognostic indicator in undifferentiated or stage II/III gastric cancer.
Introduction
Multiple gene expression profiling using cDNA microarrays and/or tissue arrays has been widely used for the prediction of prognosis in breast cancer, lung cancer, lymphoma and colon cancer.[1-4] While the application of cDNA microarrays allows for the precise analysis of the expression pattern of many suspected genes in each tissue, tissue microarray technology can accelerate the complex tissue analysis of genes with clinicopathologic findings, such as diagnostic or prognostic data.[5-7]
Recently several studies have reported useful biomarkers which might be related to gastric cancer as well as premalignant gastric lesions.[8, 9] Compared with the cancer tissue, which often shows heterogenous cell morphologies, the metaplastic lineages are often more uniform, even compared with the lineages from the normal stomach.[10] We have previously reported gene microarray studies analyzing laser capture-microdissected samples of both intestinal metaplasia (IM) and spasmolytic polypeptide-expressing metaplasia (SPEM) in humans as well as SPEM in mice.[11, 12] Using this approach, we also reported that CDH17, which was selectively expressed in both IM or SPEM, is a single, independent metaplasia biomarker for the prediction of prognosis in stage I or node-negative gastric cancer.[11]
The utility of single biomarker for cancer detection or prognosis has often been fraught with difficulties due to various interfering conditions.[8, 13-15] Nevertheless, several studies have employed multiple genes or proteins as useful biomarkers for tumor progression as well as cancer prediction.[16, 17] However, few previous investigations have evaluated how markers of metaplasia could reflect prognosis in gastric cancer with advanced stages. In our previous studies about biomarkers of metaplasia, we found that seven proteins selectively expressed in either IM or SPEM including MUC13, CDH17, OLFM4, KRT20, LGALS4, MUC5AC and REG4 were also expressed in 17-50% of human gastric cancer tissues compared with normal gastric mucosa.[11]
The purpose of this study was to analyze the expression profiles of multiple proteins selectively expressed in IM and/or SPEM as well as gastric cancer, and to investigate the prognostic impact of the combination of protein biomarkers in advanced stage gastric cancer.
Methods
To identify the expression profiles of seven metaplasia biomarkers, MUC13, CDH17, OLFM4, KRT20, LGALS4, MUC5AC and REG4, which were found from the previous study, immunostaining was performed on tissue microarray (TMAs) comprised of 450 gastric adenocarcinomas resected at Seoul National University Hospital in 2004 (SNUH-2004-GC; SuperBioChips. During the operation, D2 lymph node dissection was performed on the patients who underwent curative resection, irrespective of the type of gastrectomy.[18] None of the patients received neoadjuvant chemotherapy or radiation therapy before the surgery. Adjuvant chemotherapy, most commonly a 5-fluorouracil-based combination was usually indicated in patients with stage II or higher stages.
In addition to the 3 proteins (CDH17, MUC13 and OLFM4) which were evaluated in our previous study, we performed immunohistochemical staining for an additional four proteins (MUC5AC, KRT20, LGALS4 and REG4) with commercially available antibodies.[11]
TMAs were assembled according to the following procedure: Core tissue biopsies (diameter 2 mm) were obtained from individual paraffin-embedded gastric tumors (donor blocks) and arranged in new recipient paraffin blocks (tissue array blocks) using a trephine apparatus (Superbiochips Laboratories, Seoul, Korea). The tissue array blocks contained up to 60 cores on 8 arrays, for a total of 450 cases for immunohistochemistry (IHC) staining. Tumors occupying more than 10% of the core area were considered adequate. Each paraffin block contained internal controls, which consisted of non-neoplastic gastric mucosa from the body and antrum as well as intestinal metaplasia. IHC was performed using a Leica Bond-max automated immunostainer (Leica Microsystems, Newcastle, UK), as described by the manufacturer's protocol.
After tissues were sampled from in each core, staining patterns were scored as 0 (negative), 1 (weakly to moderately positive) and 2 (strongly positive), which was separated into negative (0) and positive (1 or 2) groups. For statistical analysis, cellular staining was considered positive only when more than 10% of the total cancer cells within 1 core were stained. The immunohistochemical staining for each core was assessed and scored without any clinical information.
The clinicopathologic and prognostic significance of the expression of these proteins was analyzed using SNUH-2004-GC TMA for the following variables: age, sex, the WHO differentiation, the Lauren classification, tumor size, the number of metastatic lymph nodes and retrieved lymph nodes, lymphatic/ venous/ perineural invasion, TNM stage, radicality and disease-specific survival.[19] Regarding the WHO differentiation, papillary, well differentiated and moderately differentiated types were categorized as the differentiated group, and poorly differentiated, signet ring cell and mucinous types were categorized as the undifferentiated group.
The correlations between multiple protein staining results and clinicopathologic characteristics were analyzed. The Student's t test and chi-square test were used for comparative statistical analyses. Disease- specific survival rates were evaluated by the Kaplan-Meier method and the prognoses were compared using the log-rank test. All clinicopathologic variables as well as the expression of the combination of multiple protein biomarkers with a log-rank P value less than 0.05 were entered into the multivariate analysis. The Cox proportional hazards model was used for the multivariate analysis to identify independent prognostic factors. All tests were 2-sided and were performed at the 5% level of significance using SPSS version 17.0 (SPSS Inc, Chicago, IL, USA). The median follow-up period was 49.1 months (range, 0.4-64.4 months).
This study was approved by the Institutional Review Board of Seoul National University Hospital (H-1102-062-351).
Results
Table 2 shows the demographic data for the patients samples on the SNUH-2004-GC tissue microarray, including age, sex, tumor location, tumor size, the number of metastatic lymph nodes, the number of retrieved lymph nodes, the Lauren classification, the WHO classification, lymphatic/venous/perineural invasion and TNM stage. The staining patterns of MUC13, CDH17, OLFM4, KRT20, LGALS4, MUC5AC and REG4 in the SNUH-2004-GC TMA are shown in Fig. 1.
Table 2.
Patients’ demographic data of SNUH-2004-GC for tissue microarrays.
| SNUH-2004-GC | n=450 | |
|---|---|---|
| Age (yr) | 57.5±12.7 | |
| Male to Female ratio | 2.7 :1 | |
| Location | Upper | 83 (18.4%) |
| Mid | 126 (28.0%) | |
| Lower | 220 (48.9%) | |
| Entire | 20 (4.4%) | |
| Remnant | 1 (0.2%) | |
| WHO | Papillary | 2 (0.4%) |
| Well differentiated | 30 (6.7%) | |
| Moderately differentiated | 155 (34.4%) | |
| Poorly differentiated | 159 (35.3%) | |
| Mucinous | 14 (3.1%) | |
| Signet Ring cell | 82 (18.2%) | |
| Undetermined | 8 (1.8%) | |
| Lauren | Intestinal | 185 (41.1%) |
| Diffuse | 185 (41.1%) | |
| Mixed | 77 (17.1%) | |
| Undetermined | 3 (0.7%) | |
| Lymphatic invasion | 262 (58.2%) | |
| Venous invasion | 77 (17.1%) | |
| Perineural invasion | 217 (48.2%) | |
| Size (cm) | 5.6±3.1 | |
| metastatic LN | 5.6±8.7 | |
| Retrieved LN | 31.4±12.9 | |
| Radicality | R0 | 403 (89.6%) |
| R1/R2 | 47 (10.4%) | |
| T stage | T1 | 128 (28.4%) |
| T2 | 71 (15.8%) | |
| T3 | 155 (34.4%) | |
| T4 | 96 (21.3%) | |
| N stage | N0 | 192 (42.7%) |
| N1 | 63 (14.0%) | |
| N2 | 60 (13.3%) | |
| N3 | 135 (30.0%) | |
| M stage | M0 | 397 (88.2%) |
| M1 | 53 (11.8%) | |
| TNM stage | Stage I | 157 (34.9%) |
| Stage II | 102 (22.7%) | |
| Stage III | 138 (30.7%) | |
| Stage IV | 53 (11.8%) | |
| F/U month (mo) | 49 (0-64) |
SNUH-2004-GC indicates the collection of 450 gastric adenocarcinomas resected at Seoul National University Hospital in 2004 (SuperBioChips) used for tissue microarrays.
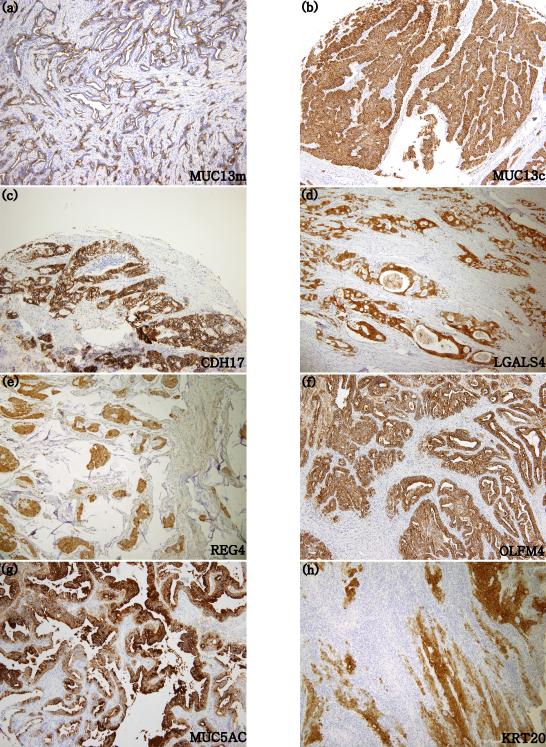
Figure 1.
Protein expression using immunohistochemical staining (100x). membranous type MUC13; MUC13m (1a), cytoplasmic type MUC13; MUC13c (1b), CDH17 (1c), LGALS4 (1d), REG4 (1e), OLFM4 (1f), MUC5AC (1g) and KRT20 (1h).
Among the biomarkers tested, MUC13 showed two distinct expression patterns: one is the membranous type which is usually expressed in intestinal type tumors (MUC13m), and the other is the diffuse cytoplasmic type (MUC13c) as previously described.[11]
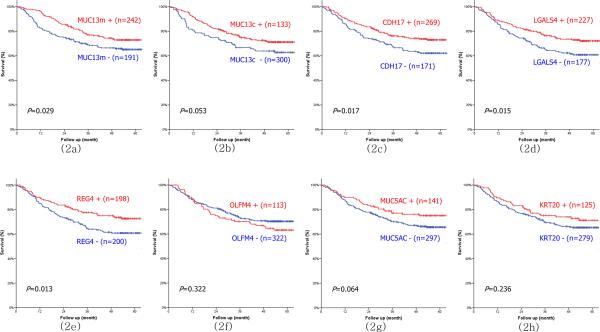
Among the seven proteins, four markers including MUC13m, CDH17, REG4 and LGALS4, were significantly related to the prognosis of gastric cancer by univariate survival analysis, whereas MUC13c, OLFM4, KRT20 and MUC5AC were not significantly related to survival rates (Fig. 2) The correlations between the expression profiles and clinicopathologic parameters of former four protein markers were analyzed (Table 3). The loss of CDH17 expression was more frequently associated with undifferentiated type and diffuse type cancer . The loss of REG4 expression showed significant association with younger age, diffuse type cancer, perineural invasion, advanced T stage and M1 stage. The loss of LGALS4 expression was significantly associated with lymphatic invasion, N stage and M stage. All clinicopathologic variables except for lymphatic or venous invasion were significantly associated with MUC13m expression
Figure 2.
Univariate survival analysis of membranous type MUC13; MUC13m (2a), cytoplasmic type MUC13;MUC13c (2b), CDH17 (2c), LGALS4 (2d), REG4 (2e), OLFM4 (2f), MUC5AC (2g) and KRT20 (2h).
Table 3.
Correlation analysis between the combined expression status of MUC13m, CDH17, LGALS4, REG4 and clinicopathologic parameters.
| Biomarkers | CDH17 | REG4 | LGALS4 | MUC13m | |||||
|---|---|---|---|---|---|---|---|---|---|
| Expression | + | − | + | − | + | − | + | − | |
| Age | 58.0±12.8 | 56.8±12.6 | 59.2±12.1* | 56.6±13.2* | 57.6±12.4 | 58.3±13.1 | 61.4±11.5* | 54.6±12.8* | |
| Sex | M:F | 2.3:1 | 3.3:1 | 2.4:1 | 2.8:1 | 2.3:1 | 3.1:1 | 3.7:1* | 2.1:1* |
| Differentiation | Undifferentiated | 139 (55.4%)* | 112 (44.6%)* | 106 (47.5%) | 117 (52.5%) | 131 (58.0%) | 95 (42.0%) | 43 (17.4%)* | 204 (82.6%)* |
| Lauren | diffuse | 98 (54.1%)* | 83 (45.9%)* | 75 (46.6%)* | 86 (53.4%)* | 95 (58.3%) | 68 (41.7%) | 15 (8.4%)* | 164 (91.6%)* |
| Lymphatic invasion | 151 (58.5%) | 107 (41.5%) | 117 (48.1%) | 126 (51.9%) | 123 (50.2%)* | 122 (49.8%)* | 105 (41.2%) | 150 (58.8%) | |
| Venous invasion | 47 (62.7%) | 28 (37.3%) | 33 (45.2%) | 40 (54.8%) | 35 (47.9%) | 38 (52.1%) | 35 (47.3%) | 39 (52.7%) | |
| Perineural invasion | 124 (58.5%) | 88 (41.5%) | 88 (44.0%)* | 112 (56.0%)* | 113 (56.2%) | 88 (43.8%) | 67 (31.9%)* | 143 (68.1%)* | |
| T stage | |||||||||
| T1 | 45 (36.3%) | 79 (63.7%) | 39 (39.4%)† | 60 (60.6%)† | 38 (36.5%) | 66 (63.5%) | 51 (41.8%)†† | 71 (58.2%)†† | |
| T2 | 23 (33.3%) | 46 (66.7%) | 35 (53.0%)† | 31 (47.0%)† | 34 (50.7%) | 33 (49.3%) | 39 (58.2%)†† | 28 (41.8%)†† | |
| T3 | 57 (37.7%) | 94 (62.3%) | 74 (52.5%)† | 67 (47.5%)† | 60 (42.6%) | 81 (57.4%) | 85 (57.0%)†† | 64 (43.0%)†† | |
| T4 | 46 (47.9%) | 50 (52.1%) | 52 (56.5%)† | 40 (43.5%)† | 45 (48.9%) | 47 (51.1%) | 67 (70.5%)†† | 28 (29.5%)†† | |
| N stage | |||||||||
| N0 | 68 (36.6%) | 118 (63.4%) | 72 (45.6%) | 86 (54.4%) | 59 (36.2%)†† | 104 (63.8%)†† | 93 (51.1%)† | 89 (48.9%)† | |
| N1 | 21 (33.3%) | 42 (66.7%) | 32 (54.2%) | 27 (45.8%) | 28 (46.7%)†† | 32 (53.3%)†† | 32 (52.5%)† | 29 (47.5%)† | |
| N2 | 20 (33.9%) | 39 (66.1%) | 24 (41.4%) | 34 (58.6%) | 24 (41.4%)†† | 34 (58.6%)†† | 32 (54.2%)† | 27 (45.8%)† | |
| N3 | 62 (47.0%) | 70 (53.0%) | 72 (58.5%) | 51 (41.5%) | 66 (53.7%)†† | 57 (46.3%)†† | 85 (64.9%)† | 46 (35.1%)† | |
| M stage | M0 | 149 (38.5%) | 238 (61.5%) | 167 (48.1%)† | 180 (51.9%)† | 146 (41.4%)†† | 207 (58.6%)†† | 206 (54.1%)† | 175 (45.9%)† |
| M1 | 22 (41.5%) | 31 (58.5%) | 33 (64.7%)† | 18 (35.3%)† | 31 (60.8%)†† | 20 (39.2%)†† | 36 (69.2%)† | 16 (30.8%)† | |
MUC13m indicates membranous type MUC13.
P<0.05
Pearson's correlation is significant at the 0.05 level
Pearson's correlation is significant at the 0.01 level
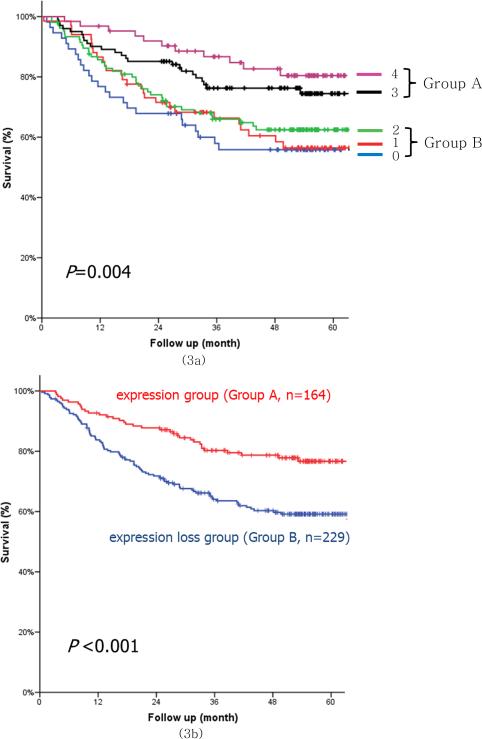
The combined expression of CDH17, REG4, LGALS4 and MUC13m was divided into five groups according to expression patterns. No expression of the four protein markers was found in 56 cases (14.2%); one marker was seen in 67 cases (17.0%), two in 106 cases (27.0%), three in 101 cases (25.7%), and four in 63 cases (16.0%). Among CDH17, REG4 and LGALS4 and MUC13m, the combined expression of 3 or 4 protein biomarkers was classified as the positive expression group (Group A) and that of 2 or fewer protein markers was classified as the expression loss group (Group B). Group B showed a younger age population, undifferentiated and diffuse type tumors, larger tumor size, a larger number of metastatic lymph nodes, more frequent lymphatic/perineural invasion, less frequent R0 resection and more advanced stage (Table 4). Regarding disease-specific survival analysis, the combined expression loss of protein biomarkers was significantly correlated with the prognosis, and Group B showed a significantly poorer survival rate than Group A (Fig. 3).
Table 4.
Clinicopathologic characteristics between Groups A and B
| Group A (n=164) | Group B (n=229) | P value | ||
|---|---|---|---|---|
| Age | 59.9±12.1 | 56.5±13.0 | 0.009 | |
| Sex | Male:Female | 2.5:1 (117/47) | 2.6:1 (166/63) | 0.820 |
| Differentiation | Differentiated | 90 (54.9%) | 73 (31.9%) | <0.001 |
| Undifferentiated | 74 (45.1%) | 148 (64.6%) | ||
| Unknown | 0 | 8 (3.5%) | ||
| Lauren | Intestinal | 91 (55.5%) | 74 (32.3%) | <0.001 |
| Diffuse | 53 (32.3%) | 107 (46.7%) | ||
| Mixed | 20 (12.2%) | 45 (19.7%) | ||
| Unknown | 0 | 3 (1.3%) | ||
| Lymphatic invasion | Invasion (+) | 90 (54.9%) | 152 (66.4%) | 0.027 |
| Invasion (−) | 74 (45.1%) | 77 (33.6%) | ||
| Venous invasion | Invasion (+) | 28 (17.1%) | 44 (19.2%) | 0.692 |
| Invasion (−) | 136 (82.9%) | 185 (80.8%) | ||
| Perineural invasion | Invasion (+) | 65 (39.6%) | 13 (58.1%) | <0.001 |
| Invasion (−) | 99 (60.4%) | 96 (41.9%) | ||
| Size (cm) | 5.1±2.4 | 6.1±3.5 | 0.001 | |
| Metastatic LN | 4.5±8.2 | 6.8±9.2 | 0.008 | |
| Retrieved LN | 31.1±13.0 | 31.7±12.9 | 0.635 | |
| Radicality | R0 | 153 (93.3%) | 196 (85.6%) | 0.022 |
| R1/R2 | 11 (6.7%) | 33 (14.4%) | ||
| T stage | T1 | 54 (32.9%) | 43 (18.8%) | 0.008 |
| T2 | 25 (15.2%) | 39 (17.0%) | ||
| T3 | 56 (34.1%) | 85 (37.1%) | ||
| T4 | 29 (17.7%) | 62 (27.1%) | ||
| N stage | N0 | 76 (46.3%) | 79 (34.5%) | 0.003 |
| N1 | 26 (15.9%) | 32 (14.0%) | ||
| N2 | 28 (17.1%) | 30 (13.1%) | ||
| N3 | 34 (20.7%) | 88 (38.4%) | ||
| M stage | M0 | 151 (92.1%) | 192 (83.8%) | 0.016 |
| M1 | 13 (7.9%) | 37 (16.2%) | ||
| TNM stage | Stage I | 64 (39.0%) | 60 (26.2%) | 0.009 |
| Stage II | 40 (24.4%) | 51 (22.3%) | ||
| Stage III | 47 (28.7%) | 81 (35.4%) | ||
| Stage IV | 13 (7.9%) | 37 (16.2%) |
Group A indicates the positive expression group in which ≥ 3 proteins among membranous type MUC13m, CDH17, LGALS4 and REG4 were expressed. Group B indicates the expression loss group in which ≤ 2 proteins among membranous type MUC13, CDH17, LGALS4 and REG4 were expressed.
Figure 3.
Survival curves for the combined expression of membranous type MUC13, CDH17, LGALS4, REG4 (3a) and the combination of the positive expression (Group A) and expression loss (Group B) groups (3b). Group A indicates the positive expression group in which ≥ 3 proteins among membranous type MUC13, CDH17, LGALS4 and REG4 were expressed. Group B indicates the expression loss group in which ≤ 2 proteins among membranous type MUC13, CDH17, LGALS4 and REG4 were expressed.
By subgroup analysis, Group B showed a significantly poorer prognosis than Group A for undifferentiated cancer, ≥ T2 advanced gastric cancer (AGC), and node-positive (N+) as well as node-negative cancer (Fig. 4). For TNM stage, even though there was no significant difference between the two groups in both stage I and stage IV cancers, Group B showed a significantly poorer prognosis than Group A in stages II/III (Fig. 5). In multivariate analysis, the combined loss of expression for MUC13m, CDH17, REG4 and LGALS4 proved to be an independent prognostic factor in undifferentiated cancer, AGC, N+ cancer and stage II/III cancer (Table 5).
Figure 4.
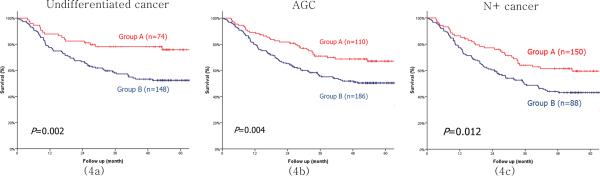
Survival curves between groups A and B in undifferentiated gastric cancer (4a), AGC (4b), and N+ cancer (4c). Group A indicates the positive expression group in which ≥ 3 proteins among membranous type MUC13m, CDH17, LGALS4 and REG4 were expressed. Group B indicates the expression loss group in which ≤ 2 proteins among membranous type MUC13, CDH17, LGALS4 and REG4 were expressed. AGC indicates gastric cancer with ≥ T2 stage. N+ indicates gastric cancer with ≥ N1 stage.
Figure 5.
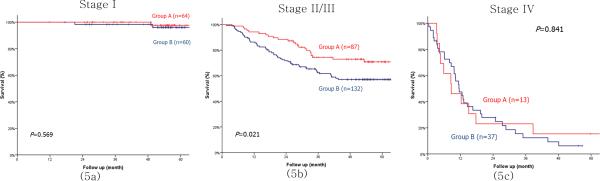
Survival curves between groups A and B in stages I (5a), II/III (5b) and IV (5c). Group A indicates the positive expression group in which ≥ 3 proteins among membranous type MUC13m, CDH17, LGALS4 and REG4 were expressed. Group B indicates the expression loss group in which ≤ 2 proteins among membranous type MUC13, CDH17, LGALS4 and REG4 were expressed.
Table 5.
Cox multivariate analysis for disease-specific survival in gastric cancer subgroups.
| Subgroups | Prognostic factor | Univariate | Multivariate | Hazard Ratio (95.0% CI) |
|---|---|---|---|---|
| Undifferentiated cancer | TNM stage (stage I) | <0.001 | <0.001 | |
| stage II | 0.093 | 3.952 (0.797-19.602) | ||
| stage III | <0.001 | 22.756 (5.045-102.636) | ||
| stage IV | <0.001 | 66.992 (13.865-323.682) | ||
| Lymphatic invasion | <0.001 | 0.695 | 1.138 (0.596-2.176) | |
| Venous invasion | <0.001 | 0.551 | 1.168 (0.7-1.949) | |
| Perineural invasion | <0.001 | 0.450 | 0.799 (0.447-1.43) | |
| Protein expression (expression/loss) | 0.002 | 0.020 | 1.927 (1.11-3.345) | |
| AGC | N stage (N0) | <0.001 | <0.001 | |
| N1 | 0.120 | 2.273 (0.808-6.389) | ||
| N2 | <0.001 | 5.390 (2.156-13.474) | ||
| N3 | <0.001 | 11.861 (5.11-27.527) | ||
| Differentiation | 0.030 | 0.129 | 1.355 (0.915-2.005) | |
| Lymphatic invasion | <0.001 | 0.471 | 0.816 (0.469-1.419) | |
| Venous invasion | <0.001 | 0.014 | 1.617 (1.102-2.372) | |
| Perineural invasion | <0.001 | 0.330 | 1.242 (0.803-1.922) | |
| Protein expression (expression/loss) | 0.004 | 0.019 | 1.625 (1.084-2.437) | |
| N+ cancer | T stage (T1) | <0.001 | <0.001 | |
| T2 | 0.777 | 1.415 (0.128-15.624) | ||
| T3 | 0.017 | 11.136 (1.539-80.597) | ||
| T4 | 0.004 | 18.573 (2.569-134.283) | ||
| Differentiation | 0.034 | 0.151 | 1.342 (0.898-2.004) | |
| Lymphatic invasion | 0.023 | 0.781 | 1.085 (0.609-1.934) | |
| Venous invasion | <0.001 | 0.006 | 1.714 (1.171-2.509) | |
| Perineural invasion | <0.001 | 0.469 | 0.833 (0.507-1.367) | |
| Protein expression (expression/loss) | 0.012 | 0.023 | 1.602 (1.068-2.402) | |
| Stage II /III | Age(≤70 vs. >70 yr) | 0.019 | 0.050 | 1.646 (0.999-2.712) |
| TNM stage (stage II vs. III) | <0.001 | <0.001 | 6.416 (3.181-12.941) | |
| Venous invasion | <0.001 | 0.041 | 1.683 (1.021-2.773) | |
| Protein expression (expression/loss) | 0.021 | 0.010 | 1.916 (1.166-3.149) |
AGC indicates gastric cancer with ≥ T2 stage. N+ indicates gastric cancer with ≥ N1 stage.
Discussion
The current study revealed that the profiling of the combined expression of four metaplasia biomarkers could be used as an independent prognostic marker in undifferentiated type gastric cancer or in AGC. Little is known about the genetic pathway or the mechanism for carcinogenesis or tumor progression in undifferentiated gastric cancer, especially compared with that of differentiated gastric cancer.[20, 21] During tumor progression, gastric adenocarcinomas tend to lose differentiation markers present in metaplasia, leading to less differentiated phenotypes.[14] Other investigations have also suggested that the predominant histologic type may be altered from the differentiated to the undifferentiated type with increasing tumor size and the progression of the tumor.[22, 23] Certain undifferentiated-type carcinomas may derive from the differentiated type tumors through progressive loss of cell-to-cell adhesion by the loss of cell-adhesion molecules.[14] The results of these previous reports and our present findings suggest that the cumulative loss of the expression of differentiation-related proteins during the progression of the tumor may cause differentiated gastric cancer to evolve into the undifferentiated type, resulting in a poorer prognosis.
The proteins biomarkers utilized in our investigations have similar functions related to cell to cell adhesion or differentiation. CDH17 (cadherin-17, liver-intestine cadherin) is a member of the cadherin family and works as a functional calcium-dependent cell adhesion molecule.[11, 24] We have previously reported that CDH 17 is a promising prognostic marker for early stage gastric cancer.[11]
The human MUC13 gene encodes an epithelial and hemopoietic transmembrane mucin, and it is most highly expressed in epithelial tissues, especially those of the gastrointestinal and respiratory tracts.[25] In gastric cancer, MUC13 is a good differentiation marker for the gastrointestinal mucosa and has distinct roles in the carcinogenesis of diffuse type gastric cancer.[26]
LGALS4, a novel biomarker for gastric cancer, belongs to a subfamily of galectins which is composed of two carbohydrate recognition domains within the same peptide and is involved in a number of biological steps including cell to cell adhesion, growth regulation and signaling pathways.[27, 28] Within human epithelial cancer cell lines, LGALS4 is overexpressed in highly differentiated cell lines, but is absent in less differentiated lines.[28] Balan et al. also reported LGALS4 as a useful tumor marker for gastric cancer progression and the potential for lymph node metastasis[29] Based on these previous studies and the results of our study, it appears that LGALS4 is expressed in more differentiated cancers, particularly Group A patients in our study, and loss of protein expression is associated with tumor progression or dedifferentiation in Group B patients.
The regenerating gene (REG) was originally isolated from regenerating rat pancreatic islets and is a trophic factor for both islet cells and gastric epithelial cells.[30, 31] Human REG4 is expressed in gastrointestinal cancers, pancreas cancer and carcinoid tumors, and it is associated with intestinal metaplasia or increased cellular differentiation in the gastric cancer.[32] The biological function of REG4 is poorly understood. REG4 has been known to enhances peritoneal metastasis in gastric cancer or to be involved in apoptosis resistance because of its 5-FU resistance.[15, 33] On the other hand REG4 expression has often been considered as a good differentiation biomarker for gastric precancerous lesions.[32] These results suggest that REG4 may promote differentiation in metaplasia while enhancing carcinogenesis or tumor progression in gastric cancer with more advanced stage, which highlights the difficulties of using a single marker in prognostic studies.
In this study, we found that the cumulative expression of cell-to-cell adhesion or differentiation related proteins is an independent prognostic factor in not only undifferentiated gastric cancer but also AGC or N+ gastric cancer. Cell-to-cell adhesion molecule expression may be influenced by maturation as well as malignant transformation of cells, and down-regulation of cell adhesion molecules may be a common event in the carcinogenesis of gastric cancer.[34, 35] Previous studies have demonstrated that the loss of cell adhesion molecule expression was significantly related to the loss of contact inhibition of growth, cellular dedifferentiation and the disorganization of tissue architecture, and is potentially important in the formation of metastases from adenocarcinoma.[35-38]. In addition to the proteins assessed in our study, a number of cell-adhesion proteins, including E-cadherin, dysadherin, CD44 focal adhesion kinase, syndecan-1 and integrins, have been associated with the prognosis of gastric cancer.[17, 37, 39-41] Unlike these previous studies, we have postulated that the cumulative loss of expression of adhesion or differentiation protein biomarkers, which were selectively expressed in premalignant metaplasia, could induce the disorganization of tissue architecture as well as cellular dedifferentiation, elevate the invasive potential into adjacent tissue including lymphatics, and influence the behavior of AGC or N+ cancer.
In the present study, protein expressions in each gastric cancer case are based on the immunohistochemical analysis of one core on TMAs. In terms of the possible diversity of histological components or molecular abnormality of advanced gastric cancer, we have already shown excellent agreement in the staining results of the different intratumoral areas of gastric carcinomas including advanced gastric cancer, and the effects of intratumoral heterogeneity can be averaged out in such a large scale analysis as our study.[7, 42, 43]
Unfortunately, we could not demonstrate an independent prognostic factor for each TNM stage. However, stage I may be too early to show the differences in protein expression levels of various cell-to-cell adhesion molecules or differentiation markers, while stage IV in the current UICC/AJCC TNM staging system is already too advanced and incurable to reflect a prognostic difference.[19] According to the current treatment strategies, stage II/III are curable advanced stages which are continuously being investigated and challenged. Therefore, compared with stage I or IV, an independent prognostic factor in advanced stage II/III can offer much more important clinical value.
In conclusion, the loss of the combined expression of metaplasia biomarkers including CDH17, MUC13m, REG4, and LGALS4, is an independent prognostic indicator in the undifferentiated type gastric cancer and stage II/III gastric cancer.
Synopsis.
Immunohistochemical staining using tissue microarrays indicates that the combined loss of expression of MUC13, CDH17, LGALS4 and REG4 is an independent prognostic indicator in undifferentiated or stage II/III gastric cancer.
Table 1.
A detailed description of 7 selected primary antibodies used in immunohistochemistry.
| Antigen | Antibody (dilution) |
|---|---|
| MUC13 | Mouse IgG1 (1/500) |
| CDH17 | Mouse IgG1 (1/250) |
| OLFM4 | Rabbit polyclonal (1/200) |
| KRT20 | Mouse IgG2a (Prediluted) |
| LGLAS4 | Mouse IgG1 (1/50) |
| MUC5AC | Mouse IgG1 (1/100) |
| REG4 | Goat polyclonal (1/100) |
Acknowledgements
This study was supported by funding to HJL from the SNUH Research Fund (grant number 05-2009-002), and funding to JRG from NIH RO1 DK071590, RO1 DK071590-S1 and a VA Merit Review Award.
Footnotes
Financial disclosure: The authors declare no conflict of interest.
References
- 1.Van De Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 2.Beer DG, Kardia SLR, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–24. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 3.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–47. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Jatkoe T, Zhang Y, Mutch MG, Talantov D, Jiang J, McLeod HL, et al. Gene expression profiles and molecular markers to predict recurrence of Dukes’ B colon cancer. J Clin Oncol. 2004;22:1564–71. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 5.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–60. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 6.Bubendorf L, Nocito A, Moch H, Sauter G. Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J Pathol. 2001;195:72–9. doi: 10.1002/path.893. [DOI] [PubMed] [Google Scholar]
- 7.Lee HS, Cho SB, Lee HE, Kim MA, Kim JH, Park DJ, Yang HK, et al. Protein Expression Profiling and Molecular Classification of Gastric Cancer by the Tissue Array Method. Clin Cancer Res. 2007;13:4154–63. doi: 10.1158/1078-0432.CCR-07-0173. [DOI] [PubMed] [Google Scholar]
- 8.Yasui W, Oue N, Ito R, Kuraoka K, Nakayama H. Search for new biomarkers of gastric cancer through serial analysis of gene expression and its clinical implications. Cancer Sci. 2004;95:385–92. doi: 10.1111/j.1349-7006.2004.tb03220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oue N, Sentani K, Noguchi T, Ohara S, Sakamoto N, Hayashi T, Anami K, et al. Serum olfactomedin 4 (GW112, hGC-1) in combination with Reg IV is a highly sensitive biomarker for gastric cancer patients. Int J Cancer. 2009;125:2383–92. doi: 10.1002/ijc.24624. [DOI] [PubMed] [Google Scholar]
- 10.Nam KT, Lee HJ, Mok H, Romero-Gallo J, Crowe JE, Jr, Peek RM, Jr, Goldenring JR. Amphiregulin-deficient mice develop spasmolytic polypeptide expressing metaplasia and intestinal metaplasia. Gastroenterology. 2009;136:1288–96. doi: 10.1053/j.gastro.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HJ, Nam KT, Park HS, Kim MA, LaFleur BJ, Aburatani H, Yang HK, et al. Gene Expression Profiling of Metaplastic Lineages Identifies CDH17 as a Prognostic Marker in Early Stage Gastric Cancer. Gastroenterology. 2010;139:213–25. doi: 10.1053/j.gastro.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nozaki K, Ogawa M, Williams JA, Lafleur BJ, Ng V, Drapkin RI, Mills JC, et al. A molecular signature of gastric metaplasia arising in response to acute parietal cell loss. Gastroenterology. 2008;134:511–22. doi: 10.1053/j.gastro.2007.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng H, Xu X, Miao YU, Takahashi H, Masuda S, Takano Y. The role of Reg IV gene and its encoding product in gastric carcinogenesis. Hum Pathol. 2010;41:59–69. doi: 10.1016/j.humpath.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura T, Yao T, Kabashima A, Nishiyama K, Maehara Y, Tsuneyoshi M. Loss of phenotypic expression is related to tumour progression in early gastric differentiated adenocarcinoma. Histopathology. 2005;47:357–67. doi: 10.1111/j.1365-2559.2005.02242.x. [DOI] [PubMed] [Google Scholar]
- 15.Kuniyasu H, Oue N, Sasahira T, Yi L, Moriwaka Y, Shimomoto T, Fujii K, et al. Reg IV enhances peritoneal metastasis in gastric carcinomas. Cell Prolif. 2009;42:110–21. doi: 10.1111/j.1365-2184.2008.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahara E. Molecular mechanism of stomach carcinogenesis. J Cancer Res Clin Oncol. 1993;119:265–72. doi: 10.1007/BF01212724. [DOI] [PubMed] [Google Scholar]
- 17.Hippo Y, Yashiro M, Ishii M, Taniguchi H, Tsutsumi S, Hirakawa K, Kodama T, et al. Differential gene expression profiles of scirrhous gastric cancer cells with high metastatic potential to peritoneum or lymph nodes. Cancer Res. 2001;61:889–95. [PubMed] [Google Scholar]
- 18.Japanese Gastric Cancer Association Japanese Classification of Gastric Carcinoma-2nd English Edition. Gastric Cancer. 1998;1:10–24. doi: 10.1007/s101209800016. [DOI] [PubMed] [Google Scholar]
- 19.Edge SB, Byrd DR, Compton CC. AJCC cancer staging manual. 7th ed. NY Springer; New York: 2009. [Google Scholar]
- 20.Yamamoto E, Suzuki H, Takamaru H, Yamamoto H, Toyota M, Shinomura Y. Role of DNA Methylation in the Development of Diffuse-Type Gastric Cancer. Digestion. 2011;83:241–9. doi: 10.1159/000320453. [DOI] [PubMed] [Google Scholar]
- 21.Tamura G, Sato K, Akiyama S, Tsuchiya T, Endoh Y, Usuba O, Kimura W, et al. Molecular characterization of undifferentiated-type gastric carcinoma. Lab Invest. 2001;81:593–8. doi: 10.1038/labinvest.3780268. [DOI] [PubMed] [Google Scholar]
- 22.Saito A, Shimoda T, Nakanishi Y, Ochiai A, Toda G. Histologic heterogeneity and mucin phenotypic expression in early gastric cancer. Pathol Int. 2001;51:165–71. doi: 10.1046/j.1440-1827.2001.01179.x. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda Y, Mori M, Kamakura T, Haraguchi Y, Saku M, Sugimachi K. Increased incidence of undifferentiated type of gastric cancer with tumor progression in 912 patients with early gastric cancer and 1245 with advanced gastric cancer. Cancer. 1994;73:2459–63. doi: 10.1002/1097-0142(19940515)73:10<2459::aid-cncr2820731003>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 24.Gessner R, Tauber R. Intestinal Cell Adhesion Molecules: Liver Intestine Cadherin. Ann N Y Acad Sci. 2000;915:136–43. doi: 10.1111/j.1749-6632.2000.tb05236.x. [DOI] [PubMed] [Google Scholar]
- 25.Williams SJ, Wreschner DH, Tran M, Eyre HJ, Sutherland GR, McGuckin MA. Muc13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J Biol Chem. 2001;276:18327–36. doi: 10.1074/jbc.M008850200. [DOI] [PubMed] [Google Scholar]
- 26.Shimamura T, Ito H, Shibahara J, Watanabe A, Hippo Y, Taniguchi H, Chen Y, et al. Overexpression of MUC13 is associated with intestinal-type gastric cancer. Cancer Sci. 2005;96:265–73. doi: 10.1111/j.1349-7006.2005.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demetter P, Nagy N, Martin B, Mathieu A, Dumont P, Decaestecker C, Salmon I. The galectin family and digestive disease. J Pathol. 2008;215:1–12. doi: 10.1002/path.2334. [DOI] [PubMed] [Google Scholar]
- 28.Huflejt ME, Leffler H. Galectin-4 in normal tissues and cancer. Glycoconj J. 2004;20:247–55. doi: 10.1023/B:GLYC.0000025819.54723.a0. [DOI] [PubMed] [Google Scholar]
- 29.Balan V, Nangia-Makker P, Raz A. Galectins as Cancer Biomarkers. Cancers. 2010;2:592–610. doi: 10.3390/cancers2020592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terazono K, Yamamoto H, Takasawa S, Shiga K, Yonemura Y, Tochino Y, Okamoto H. A novel gene activated in regenerating islets. J Biol Chem. 1988;263:2111–4. [PubMed] [Google Scholar]
- 31.Yamagishi H, Fukui H, Sekikawa A, Kono T, Fujii S, Ichikawa K, Tomita S, et al. Expression profile of REG family proteins REG Iα and REG IV in advanced gastric cancer: comparison with mucin phenotype and prognostic markers. Mod Pathol. 2009;22:906–13. doi: 10.1038/modpathol.2009.41. [DOI] [PubMed] [Google Scholar]
- 32.Oue N, Mitani Y, Aung PP, Sakakura C, Takeshima Y, Kaneko M, Noguchi T, et al. Expression and localization of Reg IV in human neoplastic and non-neoplastic tissues: Reg IV expression is associated with intestinal and neuroendocrine differentiation in gastric adenocarcinoma. J Pathol. 2005;207:185–98. doi: 10.1002/path.1827. [DOI] [PubMed] [Google Scholar]
- 33.Mitani Y, Oue N, Matsumura S, Yoshida K, Noguchi T, Ito M, Tanaka S, et al. Reg IV is a serum biomarker for gastric cancer patients and predicts response to 5-fluorouracil-based chemotherapy. Oncogene. 2007;26:4383–93. doi: 10.1038/sj.onc.1210215. [DOI] [PubMed] [Google Scholar]
- 34.Eidelman S, Damsky CH, Wheelock MJ, Damjanov I. Expression of the cell-cell adhesion glycoprotein cell-CAM 120/80 in normal human tissues and tumors. Am J Pathol. 1989;135:101–10. [PMC free article] [PubMed] [Google Scholar]
- 35.Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 1998;1:333–9. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayer B, Johnson JP, Leitl F, Jauch KW, Heiss MM, Schildberg FW, Birchmeier W, et al. E-cadherin expression in primary and metastatic gastric cancer: down-regulation correlates with cellular dedifferentiation and glandular disintegration. Cancer Res. 1993;53:1690–5. [PubMed] [Google Scholar]
- 37.Mayer B, Jauch KW, Schildberg FW, Funke I, Gunthert U, Figdor CG, Johnson JP. De-novo expression of CD44 and survival in gastric cancer. Lancet. 1993;342:1019–22. doi: 10.1016/0140-6736(93)92879-x. [DOI] [PubMed] [Google Scholar]
- 38.Brackenbury R. Molecular mechanisms of cell adhesion in normal and transformed cells. Cancer Metastasis Rev. 1985;4:41–58. doi: 10.1007/BF00047736. [DOI] [PubMed] [Google Scholar]
- 39.Shimada Y, Yamasaki S, Hashimoto Y, Ito T, Kawamura J, Soma T, Ino Y, et al. Clinical significance of dysadherin expression in gastric cancer patients. Clin Cancer Res. 2004;10:2818–23. doi: 10.1158/1078-0432.ccr-0633-03. [DOI] [PubMed] [Google Scholar]
- 40.Park JH, Lee BL, Yoon J, Kim J, Kim MA, Yang HK, Kim WH. Focal adhesion kinase (FAK) gene amplification and its clinical implications in gastric cancer. Hum Pathol. 2010;41:1664–73. doi: 10.1016/j.humpath.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 41.Wiksten JP, Lundin J, Nordling S, Lundin M, Kokkola A, von Boguslawski K, Haglund C. Epithelial and stromal syndecan 1 expression as predictor of outcome in patients with gastric cancer. Int J Cancer. 2001;95:1–6. doi: 10.1002/1097-0215(20010120)95:1<1::aid-ijc1000>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 42.Lee HS, Lee HK, Kim HS, Yang HK, Kim YI, Kim WH. MUC1, MUC2, MUC5AC, and MUC6 expressions in gastric carcinomas. Cancer. 2001;92:1427–34. doi: 10.1002/1097-0142(20010915)92:6<1427::aid-cncr1466>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 43.Lee HS, Lee HK, Kim HS, Yang HK, Kim WH. Tumour suppressor gene expression correlates with gastric cancer prognosis. J Pathol. 2003;200:39–46. doi: 10.1002/path.1288. [DOI] [PubMed] [Google Scholar]