Abstract
A captive juvenile Bornean orangutan (Pongo pygmaeus) died from an unknown disseminated parasitic infection. Deep sequencing of DNA from infected tissues, followed by gene-specific PCR and sequencing, revealed a divergent species within the newly proposed genus Versteria (Cestoda: Taeniidae). Versteria may represent a previously unrecognized risk to primate health.
Keywords: Cestoda, Taeniidae, Versteria, metacestode, primate, orangutan, Pongo pygmaeus, deep sequencing, parasites
We describe the identification of a previously genetically uncharacterized species within the newly proposed Taeniid (Cestoda) genus Versteria (1), which caused fatal metacestode infection in a captive juvenile Bornean orangutan (Pongo pygmaeus). The orangutan was born in Colorado, USA, on April 4, 2007, and, after maternal rejection, was transported to the Milwaukee County Zoo in Milwaukee, Wisconsin, USA, for adoption by a surrogate mother on February 7, 2008. On December 27, 2012, keepers noted that the orangutan was exhibiting loss of appetite and an intermittent, moist cough. The animal became increasingly lethargic and was found dead 2 days later.
The Study
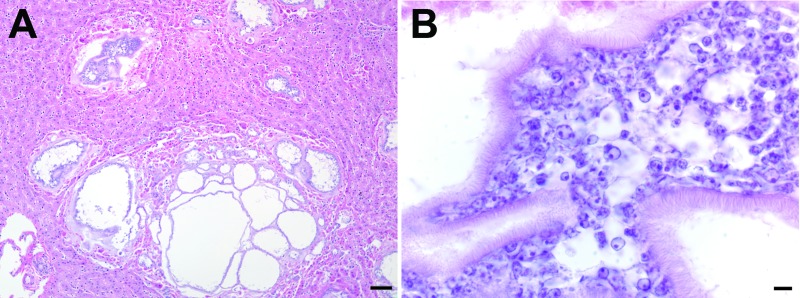
Postmortem examination revealed diffuse hemorrhages in the lungs (which did not collapse), splenomegaly, a pale mottled liver, and thoracic and pericardial effusions. Diagnostic microbiologic examination of tracheal washes and lung tissue identified only common environmental bacteria, and tests for viruses and fecal examination for parasites were all negative. Histopathologic examination of the liver revealed cystic structures containing eukaryotic parasite cells between ≈4 and 5 μm in diameter (Figure 1). Similar cells were observed in the parenchyma and blood vessels of lung and spleen (not shown). On the basis of these results and clinical observations, the cause of death was determined to be acute respiratory distress due to disseminated infection with an unknown parasite.
Figure 1.
Microscopic images of liver sections from a Bornean orangutan fatally infected with Versteria metacestodes. Images of liver sections stained with hematoxylin and eosin (H&E) stain were captured at 10× magnification (A; scale bar = 30 μm) and 100× magnification; B; scale bar = 5 μm). Large numbers of parasite cells can be seen within well-defined cystic structures separated from the surrounding host tissue by clearly visible membranes.
Because attempts to identify the parasite by morphologic features were inconclusive, total DNA extracted from infected organs was subjected to deep sequencing to detect molecular sequences of pathogens. DNA was isolated from liver, lung, and spleen by using the QIAGEN DNeasy Blood and Tissue Kit (QIAGEN Inc., Valencia, CA, USA), followed by treatment with RNase (Epicenter Biotechnologies, Madison, WI, USA) to remove RNA. DNA libraries were then generated by using the Nextera DNA Sample Prep Kit (Illumina, San Diego, CA, USA) and sequenced on an Illumina MiSeq instrument as described (2). Resulting sequence data were analyzed by using CLC Genomics Workbench 5.5 (CLC bio, Aarhus, Denmark). Briefly, low quality (<q30) and short (<100-bp) sequences were removed, sequences were aligned against an orangutan (P. abelii) genome (3), and nonmapped sequences were subjected to de novo assembly.
Deep sequencing of total DNA from infected tissues resulted in ≈2,400,000 sequences after quality trimming. Subtractive mapping against the orangutan genome removed ≈97% of these sequences. De novo assembly of the remaining ≈50,000 sequences resulted in 293 contiguous sequences, 7 of which had high similarity to GenBank sequences corresponding to Taenia spp. (expected values <1 × 10−18). Subsequent mapping of nonhost sequences against the T. solium genome (4) resulted in 8,494 matches.
On the basis of deep-sequencing results, PCR primers were used to amplify 3 mitochondrial genes informative for resolving relationships within the Taeniidae (Table). For 12s ribosomal RNA (12s rRNA), primers CES12sF (5′-AGGGGATAGGACACAGTGCCAGC-3′) and CES12sR (5′-CGGTGTGTACMTGAGYTAAAC-3′) were modified from GenBank accession nos. KC344674–KC344701. For cytochrome c oxidase subunit I (cox1), published primers JB3 (5′-TTTTTTGGGCATCCTGAGGTTTAT-3′) and JB4.5 (5′-TAAAGAAAGAACATAATGAAAATG-3′) were used, and for NADH dehydrogenase subunit 1 (nad1), published primers JB11 (5′-AGATTCGTAAGGGGCCTAATA- 3′) and JB12 (5′-ACCACTAACTAATTCACTTTC-3′) were used (5). PCRs were conducted in 20-μL volumes with 1-μL DNA template by using the Phusion kit (New England Biolabs Inc., Ipswich, MA, USA), cycled as follows: 98°C, 30 s; 35 cycles of 94°C, 10 s, annealing, 30 s, 72°C, 30 s; and final extension at 72°C for 10 min (annealing temperatures for 12s rRNA, cox1, and nad1 were 60°C, 50°C, and 55°C, respectively).
Table. GenBank accession numbers of taeniid DNA sequences used in phylogenetic analyses.
| Taxon | Origin | GenBank accession nos.* |
||
|---|---|---|---|---|
| 12s rRNA | cox1 | nad1 | ||
| Echinococcus | ||||
| E. canadensis | Kazakhstan | NC_011121 | NC_011121 | NC_011121 |
| E. equinus | United Kingdom | AF346403 | AF346403 | AF346403 |
| E. felidis | Uganda | AB732958 | AB732958 | AB732958 |
| E. granulosus | United Kingdom | NC_008075 | NC_008075 | NC_008075 |
| E. multilocularis | Japan | NC_000928 | NC_000928 | NC_000928 |
| E. oligarthrus | Panama | NC_009461 | NC_009461 | NC_009461 |
| E. ortleppi | Argentina | NC_011122 | NC_011122 | NC_011122 |
| E. shiquicus | Tibet | NC_009460 | NC_009460 | NC_009460 |
|
E. vogeli
|
Colombia |
NC_009462 |
NC_009462 |
NC_009462 |
| Hydatigera | ||||
| H. krepkogorski | China | AB731762 | AB731762 | |
| H. parva | Spain | AB731760 | AB731760 | |
| H. taeniaeformis (A) | China | NC_014768 | NC_014768 | NC_014768 |
|
H. taeniaeformis (B) |
Finland |
|
AB731761 |
AB731761 |
| Taenia | ||||
| T. arctos | Finland | GU252130 | GU252132 | |
| T. asiatica | Korea | NC_004826 | NC_004826 | NC_004826 |
| T. crassiceps | Canada | NC_002547 | NC_002547 | NC_002547 |
| T. hydatigena | China | NC_012896 | NC_012896 | NC_012896 |
| T. krabbei | Norway | EU544578 | EU544631 | |
| T. laticollis | Finland | AB731727 | AB731727 | |
| T. madoquae | Kenya | AB731726 | AB731726 | |
| T. martis | Croatia | NC_020153 | NC_020153 | NC_020153 |
| T. multiceps | China | NC_012894 | NC_012894 | NC_012894 |
| T. multiceps gaigeri | Iran | HM101469 | HM101470 | |
| T. omissa | Canada | JX860631 | JX860632 | |
| T. ovis | New Zealand | AB731675 | AB731675 | AB731675 |
| T. pisiformis | China | NC_013844 | NC_013844 | NC_013844 |
| T. polyacantha arctica | Greenland | EU544594 | EU544646 | |
| T. polyacantha polyacantha | Denmark | EU544583 | EU544636 | |
| T. regis (A) | Kenya | AM503328 | AM503346 | |
| T. regis (B) | Kenya | AM503329 | AM503347 | |
| T. saginata | Africa | NC_009938 | NC_009938 | NC_009938 |
| T. serialis (A) | Kenya | AM503319 | AM503336, | |
| T. serialis (B) | Kenya | AM503322 | AM503339 | |
| T. solium | China | NC_004022 | NC_004022 | NC_004022 |
| T. sp. AL-2012 | Finland | JX860629 | JX860630 | |
|
T. twitchelli
|
Russia |
|
AB731759 |
AB731759 |
| Versteria | ||||
| V. mustelae (A) | Finland | EU544567 | EU544620 | |
| V. mustelae (B) | Russia | EU544571 | EU544624 | |
| V. sp.† | United States | KF303339 | KF303340 | KF303341 |
*Multiple sequences were chosen to capture the maximum extent of intraspecific genetic divergence within highly diverse taxa (variants arbitrarily labeled A or B). †Sequences generated in this study.
Amplicons underwent electrophoresis on 1% agarose gels stained with ethidium bromide, were purified with the Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA, USA), and were Sanger-sequenced in both directions by using PCR primers on ABI 3730xl DNA Analyzers (Applied Biosystems, Carlsbad, CA, USA) at the University of Wisconsin Biotechnology Center. Sequence chromatograms were edited and assembled by using Sequencher version 4.9 (Gene Codes Corporation, Ann Arbor, MI, USA). Sequences were aligned with homologous sequences from all taeniid species in GenBank as of April 7, 2013 (Table). To construct phylogenetic trees, we used the maximum-likelihood method in MEGA5.2 software (6).
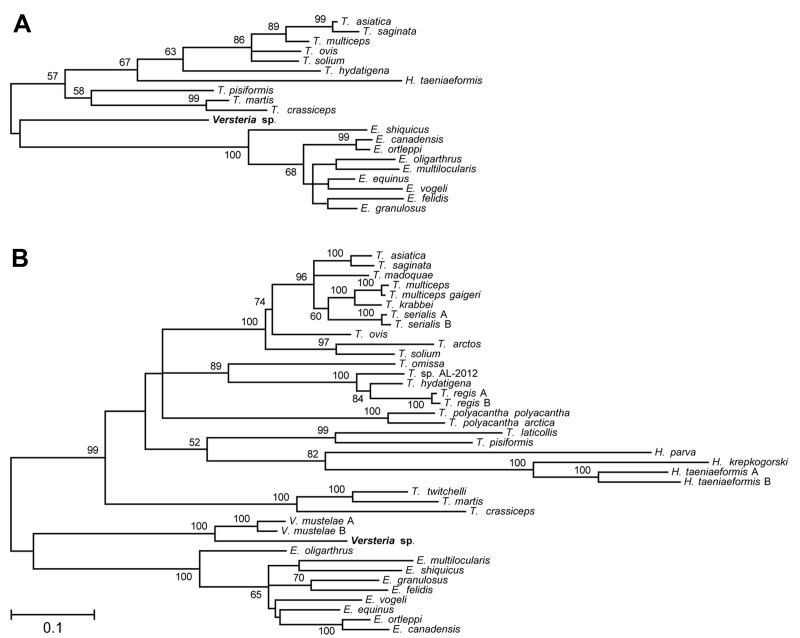
Figure 2 shows phylogenetic trees of newly generated 12s rRNA (panel A) and concatenated cox1/nad1 (panel B) sequences and representative taeniid sequences. The trees closely agree with recently published taeniid phylogenies (1). Concatenated cox1/nad1 sequences from the orangutan cluster with V. mustelae (formerly, T. mustelae) with 100% bootstrap support, placing the organism within the newly proposed genus Versteria (1) with confidence. However, the new cox1 and nad1 sequences are ≈12% different from those of published V. mustelae sequences. This degree of divergence is equal to or greater than that separating established Echinococcus and Taenia spp. (7) (Figure 2).
Figure 2.
Phylogenetic trees of the Taeniidae, including newly generated sequences derived from tissues of a fatally infected Bornean orangutan. Trees were constructed from DNA sequence alignments of 12s rRNA (A) and concatenated cox1/nad1 (B) sequences from the orangutan (Versteria sp.; bold; accession nos. KF303339–303341) and representative Echinococcus, Hydatigera, Taenia, and Versteria sequences from GenBank (see Table). The maximum likelihood method was used, with the likeliest model of molecular evolution chosen for both datasets by using MEGA5.2 software (6). Models of molecular evolution and tree likelihood values are HKY+G, -lnL = 2279.42 for 12s rRNA, and GTR+G+I, -lnL = 11582.71 for cox1/nad1. Numbers next to branches indicate bootstrap values (%), estimated from 1,000 resamplings of the data (only bootstrap values ≥50% are shown). Scale bar indicates nucleotide substitutions per site.
Members of the newly proposed genus Versteria have morphologic features that distinguish them from members of the other taeniid genera, such as miniature rostellar hooks, small scolex, rostellum, and suckers; a short strobili; and a small number of testes (1). However, no such distinguishing morphologic features could be identified by microscopy in the case described here. V. mustelae tapeworms infect multiple small animal intermediate host species and have been found in the upper midwestern United States in a hunter-killed fox squirrel (Sciurus niger rufeventer) with hepatic cysts (8). The definitive hosts of V. mustelae tapeworms are small carnivores of the family Mustelidae, such as weasels and martens (9). The genus Versteria also contains V. brachyacantha (10) tapeworms, which infect the African striped weasel (Poecilogale albinucha), but sequences of this species are not represented in GenBank. North American V. mustelae tapeworms are capable of asexual multiplication in the intermediate host (11); however, sequence data are only available for Eurasian specimens (7). The parasite described herein could thus represent a novel species or a previously genetically uncharacterized North American V. mustelae variant.
Conclusions
This study illustrates the utility of deep sequencing for diagnosing and characterizing enigmatic parasites. Similar methods have aided in the discovery of RNA viruses (12), but their application to eukaryotic pathogens has lagged, presumably because of technical challenges associated with distinguishing host from parasite DNA. In this light, it is noteworthy that our efforts were greatly facilitated by the availability of an orangutan genome against which to perform in silico subtractive mapping (3). As more host genomes become available, and as costs of equipment, reagents, and bioinformatics software decline, such methods promise to enter the diagnostic mainstream, as a complement to traditional morphologic and molecular approaches.
Encysted taeniid metacestodes can remain dormant for years before asexual multiplication (13); thus, this animal could have become infected at virtually any point in its life. Rapid progression to fatal disease could indicate an underlying condition, such as immune deficiency. Alternatively, this particular Versteria species may be inherently virulent.
Regarding source of infection, orangutans engage in geophagy (14), a behavior that this animal frequently practiced, suggesting that the infectious agent could have been obtained from contaminated soil. However, other sources (e.g., food, water, fomites) cannot be excluded. Infectious eggs could have entered the orangutan’s environment through direct deposition by a definitive host or through complex pathways of environmental transport. To date, no other animals in the zoologic collections in Colorado or Wisconsin, where the orangutan was housed, have experienced similar disease, nor have similar infections been reported in persons, to our knowledge.
In any case, this animal’s rapid and severe disease progression raises concerns about the health of captive apes in similar settings. Moreover, the close evolutionary relationship between orangutans and humans (3) raises concerns about the parasite’s zoonotic potential.
Acknowledgments
We are grateful to the staff and administration of the Milwaukee County Zoo for their dedication and support. We thank S. Sibley, A. Bailey, T. Friedrich, and B. Beehler for helpful discussions.
This work was supported in part by the National Institutes of Health, USA, grants TW009237, R01AI084787, P51OD011106, and P51RR000167, and by the Wisconsin Partnership Program through the Wisconsin Center for Infectious Disease. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Biography
Dr Goldberg is professor of epidemiology at the University of Wisconsin-Madison. He is also associate director for research in the University of Wisconsin-Madison Global Health Institute. His research interests include the ecology, epidemiology, and evolution of emerging infections in nonhuman primates and other animals.
Footnotes
Suggested citation: Goldberg TL, Gendron-Fitzpatrick A, Deering KM, Wallace RS, Clyde VL, Lauck M, et al. Fatal metacestode infection in Bornean orangutan caused by unknown Versteria species. Emerg Infect Dis [Internet]. 2014 Jan [date cited]. http://dx.doi.org/10.3201/eid2014.131191
References
- 1.Nakao M, Lavikainen A, Iwaki T, Haukisalmi V, Konyaev S, Oku Y, et al. Molecular phylogeny of the genus Taenia (Cestoda: Taeniidae): proposals for the resurrection of Hydatigera Lamarck, 1816 and the creation of a new genus Versteria. Int J Parasitol. 2013;43:427–37. 10.1016/j.ijpara.2012.11.014 [DOI] [PubMed] [Google Scholar]
- 2.Lauck M, Hyeroba D, Tumukunde A, Weny G, Lank SM, Chapman CA, et al. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS ONE. 2011;6:e19056. 10.1371/journal.pone.0019056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–33. 10.1038/nature09687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, Brooks KL, et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 2013;496:57–63 . 10.1038/nature12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowles J, Blair D, McManus DP. A molecular phylogeny of the genus Echinococcus. Parasitology. 1995;110:317–28. 10.1017/S0031182000080902 [DOI] [PubMed] [Google Scholar]
- 6.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavikainen A, Haukisalmi V, Lehtinen MJ, Henttonen H, Oksanen A, Meri S. A phylogeny of members of the family Taeniidae based on the mitochondrial cox1 and nad1 gene data. Parasitology. 2008;135:1457–67. 10.1017/S003118200800499X [DOI] [PubMed] [Google Scholar]
- 8.Langham RF, Rausch RL, Williams JF. Cysticerci of Taenia mustelae in the fox squirrel. J Wildl Dis. 1990;26:295–6. 10.7589/0090-3558-26.2.295 [DOI] [PubMed] [Google Scholar]
- 9.Kinsella JM. Comparison of helminth parasites of the cotton rat, Sigmodon hispidus, from several habitats in Florida. Am Mus Novit. 1974;2540:1–12. [Google Scholar]
- 10.Baer JG, Fain A. Cestodes nouveaux du Congo Belge. Acta Trop. 1951;8:59–63. [PubMed] [Google Scholar]
- 11.Loos-Frank B. An up-date of Verster's (1969) 'Taxonomic revision of the genus Taenia Linnaeus' (Cestoda) in table format. Syst Parasitol. 2000;45:155–84. 10.1023/A:1006219625792 [DOI] [PubMed] [Google Scholar]
- 12.Lipkin WI, Firth C. Viral surveillance and discovery. Curr Opin Virol. 2013;3:199–204.http:// [DOI] [PMC free article] [PubMed]
- 13.Whitfield PJ, Evans NA. Parthenogenesis and asexual multiplication among parasitic platyhelminths. Parasitology. 1983;86:121–60. 10.1017/S0031182000050873 [DOI] [PubMed] [Google Scholar]
- 14.Mackinnon J. The behaviour and ecology of wild orang-utans (Pongo pygmaeus). Anim Behav. 1974;22:3–74. 10.1016/S0003-3472(74)80054-0 [DOI] [Google Scholar]