Abstract
Background & Aims
We studied the relationship between IL28B gene-related SNP rs12979860 and early viral kinetics (day 0–28) during peginterferon and ribavirin treatment, in 173 African Americans (AA) and 188 Caucasian Americans (CA) with HCV genotype 1.
Methods
We studied the relationship between IL28B 16 gene-related SNP rs12979860 and early viral kinetics (day 0–28) 17 during peginterferon and ribavirin treatment, in 171 African 18 Americans (AA) and 188 Caucasian Americans (CA) with HCV 19 genotype 1.
Results
Compared to non-C/C genotypes, C/C was associated with greater declines in serum HCV RNA during phase 1 (day 0–2), phase 2 (day 7–28), and day 0–28 and higher response (undetected HCV RNA) rates at weeks 4 and 12 in AA and CA. A static phase and increases in HCV RNA from day 2 to 7 were more common in patients with non-C/C genotypes. C/C was also associated with higher week 24, 48, and 72 response rates in CA (p <0.01) but not in AA. At baseline, SNP genotype was the only independent predictor of phase 1; SNP genotype and phase 1 were independent predictors of phase 2 (p<0.001). There were no racial differences in HCV RNA declines during phase 1, day 2–7, phase 2, and day 0–28 with the same SNP genotype. AA with C/C and C/T genotypes had lower week 24, 48, and 72 (SVR) rates than did CA (p = 0.03). SNP C/C predicted higher SVR rates in AA and CA with high baseline HCV RNA (≥ 600,000 IU/ml), and in CA with ≥ 1 log10 IU/ml decrease in HCV RNA from day 0 to 28.
Conclusions
SNP rs12979860 is strongly associated with both phase 1 and phase 2 HCV RNA kinetics in AA and CA with HCV genotype 1.
Keywords: IL28B gene, Hepatitis C virus, Viral kinetics, Single nucleotide polymorphism
Introduction
A sustained virologic response (SVR) following antiviral treatment can delay liver fibrosis progression and decrease the incidence of HCC due to chronic HCV infections [1,2]. However, less than 50% of HCV genotype 1 infected patients experience a SVR after peginterferon plus ribavirin therapy [3,4]. SVR rates are lower in AA infected with HCV genotype 1 compared to CA [5]. Several studies have shown a strong association between single nucleotide polymorphisms (SNP) near the IL28B gene on chromosome 19 and SVR following peginterferon and ribavirin treatment in patients with HCV genotype 1 [6]. The SNP rs12979860 C/C genotype was associated with a 2–3-fold higher SVR compared to T/T and C/T genotypes. The C-allele was much less common in AA compared to CA, and the difference in allele frequencies was felt to explain up to 50% of the racial disparity in SVR [6].
How genetic variation near the IL28B gene regulates treatment response to peginterferon and ribavirin has not been completely defined. Yet, it is known that a faster decline in serum HCV RNA during the first 28 days of therapy predicts a higher SVR [7]. Serum HCV RNA dynamics from day 0 to 28 peginterferon, alone or combined with ribavirin show at least 2 distinct phases [8–11]. Phase 1 is characterized by a decline in serum HCV RNA from day 0 to 2, hypothesized to represent IFN effectiveness at blocking HCV production. HCV RNA drops at a slower rate during phase 2 (day 7–28), which is thought to indicate clearance of infected cells. The magnitude of phase 2 is a strong predictor of SVR [8,9]. In previous studies, AA patients with HCV genotype 1 exhibited weaker phase 1 and phase 2 responses [9,12]. Thompson et al. observed a faster decline in HCV RNA from day 0 to 14 and day 0 to 28, and higher week 4 and 12 response rates during peginterferon combination treatment in HCV genotype 1 patients with the SNP (rs12979860) C/C compared to T/T and C/T genotypes [13]. The relationship between IL28B SNP and phase 1 and phase 2 kinetics has not been studied in AA and CA. However, the C/C genotype was an independent predictor of phase 1 in European patients [14]. Based on superior efficacy to standard therapy, two HCV NS34a protease inhibitors combined with peginterferon and ribavirin were recently approved for treatment of HCV genotype 1 in the US [15,16]. Yet, both drugs were less efficacious in AA compared to CA, probably related to greater resistance to peginterferon and ribavirin in AA patients. In the present study, we defined the relationships between SNP rs12979860 and early viral kinetics in AA and CA infected with HCV genotype 1 during peginterferon and ribavirin treatment.
Materials and methods
Study population
The VIRAHEP-C study was described previously [5]. Briefly, 401 treatment-naïve adults (196AA and 205 CA) with HCV genotype 1 infections received peginterferon alfa-2a 180 μg weekly and ribavirin 1000–1200 mg per day for up to 48 weeks. Adherence to peginterferon and RBV was monitored using the electronic Medication Event Management System (Aardex, Zug, Switzerland). Informed consent was obtained from each patient; the current cohort includes 361 subjects who consented to genetics studies. The study protocol conformed to ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Institutional Review Board at each site.
HCV RNA kinetics day 0–28
Serum HCV RNA concentrations on day 0, 1, 2, 7, 14, and 28 were measured with the COBAS Amplicor Hepatitis C Virus Monitor Test, 2.0™ assay (sensitivity 600 IU/ml; Roche Molecular Diagnostics, Alameda, CA) [7]. Levels below the detection limit were tested in duplicate with the Roche Amplicor™ assay (sensitivity >50 IU/ml). Samples negative by Monitor Test and positive with the Amplicor assay were assigned an HCV RNA concentration of 599 IU/ml. Phase 1 was defined as the difference in HCV RNA concentration (log10 IU/ml) from day 0 to 1 or day 0 to 2, whichever was greater [7]. Phase 2 was defined as the best fit slope of serum HCV RNA (log10 IU/ml/wk) concentrations from day 7 to 28.
Study endpoints
Response rates at weeks 4 (rapid virologic response or RVR), 12 (early virologic response or EVR), 24, 48, and 72 (SVR) were defined as an undetectable serum HCV RNA (<50 IU/ml). Viremia breakthrough was characterized by an undetectable serum HCV RNA at week 24, but detectable at week48. Relapse was defined as an undetectable HCV RNA at week 48, but detectable at week 72. A null response was defined as < 1 log10 IU/ml decrease in HCV RNA from day 0 to 28.
SNP rs12979860 genotyping
Samples were genotyped for SNP rs12979860 using the allele specific discrimination kit (ABI TaqMan) and an ABI7900HT Sequence Detection System (Applied Biosystems) as previously described [17].
Statistical analysis
Patient demographic and clinical characteristics were compared using t-tests and Chi-square tests. The effect of SNP genotype on virologic response was evaluated under the additive genetic model, with the genotype variable coded as the number of C alleles (0, 1,2). Changes in serum HCV RNA on days 1, 2, 7, 14, and 28 relative to day 0 were compared using nonparametric tests. Phase 1 (day 0–2], phase 2 (day 7–28) and declines in serum HCV RNA from day 2 to 7 and day 0 to 28 were compared using analysis of variance (ANOVA). Multivariate logistic regression was used to define variables associated with SVR and early viral kinetics phase 1 and 2. Bivariate logistic regression was used to evaluate associations between phase 1 and phase 2 and week 4, 12, 24, 48, and 72 treatment responses. All analyses were performed using SAS 9.2 (SAS Institute, Cary, North Carolina).
Results
Patient characteristics
Table 1 shows patient characteristics by SNP rs12979860 genotype in AA and CA. T/T and C/T genotypes were more frequent in AA; the C/C genotype was more common in CA (p<0.001). Baseline serum HCV RNA levels were higher in AA with C/T compared to T/T and C/C. Baseline serum HCV RNA concentration did not vary by SNP genotype in CA. The liver fat score was higher in CA with T/T compared to C/T and C/C. Patient age, gender distribution, BMI, HOMA-IR score, liver fibrosis stage and adherence to peginterferon and ribavirin did not differ by SNP genotype in either AA or CA. Compared to CA, AA were older, and had greater BMI (Supplementary Table 1A) and lower pretreatment serum ALT as reported previously [5]. Also, AA patients received a lower percentage of the maximum ribavirin dose during the first 4 and the first 24 weeks. Furthermore, AA patients had significantly lower week 4 (RVR) (9% vs. 23%), 12 (EVR) (44% vs. 71%), 24 (52% vs. 83%), 48 (ETR) (46% vs. 79%), and 72 (SVR) (26% vs. 54%) (p <0.001) rates. Baseline characteristics as a function of SNP rs12979860 genotype are shown in Supplementary Table 1B. Patients with T/T and C/T had greater BMI and insulin resistance (HOMA-IR). Patients with C/C and C/T had higher pretreatment serum HCV RNA, while those with C/C had higher and ALT concentrations relative to non-C/C genotypes.
Table 1.
Patient characteristics.
| African-Americans (n = 173)
|
Caucasians (n = 188)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| TT n = 58; 33% |
CT n = 95; 55% |
CC n = 20; 12% |
pa | TT n = 19; 10% |
CT n = 83; 44% |
CC n = 86; 46% |
pa | |
| Age, yr (SD) | 49.2 (7.5) | 48.9 (7.1) | 48.9 (4.5) | 0.86 | 49.3 (7.0) | 47.6 (8.5) | 46.3 (8.9) | 0.42 |
|
| ||||||||
| Male, n (%) | 36 (62.1) | 60 (63.2) | 17 (85.0) | 0.14 | 16 (84.2) | 52 (62.7) | 55 (64.0) | 0.19 |
|
| ||||||||
| Body Mass Index, kg/m2 (SD) | 31.2 (6.3) | 30.4 (6.2) | 31.3 (6.6) | 0.62 | 27.7 (4.9) | 28.8 (4.9) | 28.0 (6.0) | 0.32 |
|
| ||||||||
| HOMA IR, (SD) | 5.4 (4.4) | 5.7 (6.5) | 6.3 (8.4) | 0.56 | 3.4 (2.0) | 5.1 (9.4) | 3.6 (5.2) | 0.36 |
|
| ||||||||
| Serum HCV RNA, log10 IU/ml [median (IQR)] | 6.3 (0.9) | 6.5 (1.0) | 6.3 (1.1) | 0.02 | 6.5 (1.1) | 6.5 (1.1) | 6.6 (1.2) | 0.88 |
|
| ||||||||
| Serum ALT, IU/L (SD) | 68.3 (42.9) | 64.5 (29.9) | 110.8 (84.7) | 0.07 | 109.7 (81.0) | 97.3 (79.5) | 118.3 (106.6) | 0.84 |
|
| ||||||||
| Ishak Fibrosis score [median (IQR)] | 2.0 (2.0) | 2.0 (2.0) | 2.0 (2.0) | 0.71 | 2.0 (3.0) | 2.0 (2.0) | 2.0 (2.0) | 0.82 |
|
| ||||||||
| Liver fat score [median (IQR)] | 0.0 (1.0) | 0.0 (1.0) | 0.0 (0.5) | 0.16 | 1.0 (1.0) | 0.0 (1.0) | 0.0 (1.0) | 0.05 |
|
| ||||||||
| Mean peginterferon adherence, % (SD) | ||||||||
| Day 0-wk 4 | 94 (10.5) | 95 (14.0) | 93 (14.0) | 0.47 | 97 (8.1) | 95 (10.7) | 96 (9.1) | 0.72 |
| Day 0-wk 24 | 87 (24.3) | 92 (19.5) | 91 (22.7) | 0.25 | 98 (3.2) | 94 (20.1) | 92 (20.9) | 0.77 |
|
| ||||||||
| Mean ribavirin adherence, % (SD) | ||||||||
| Day 0-wk 4 | 94 (12.2) | 96 (8.0) | 95 (9.7) | 0.88 | 94 (14.9) | 97 (6.4) | 98 (4.3) | 0.75 |
| Day 0-wk 24 | 77 (28.7) | 83 (21.5) | 81 (24.8) | 0.72 | 93 (13.2) | 89 (19.6) | 87 (22.7) | 0.29 |
Continuous variables expressed as mean (SD) or median (IQR). Results compared using Wilcoxon rank-sum test.
HCV RNA kinetics day 0–28
Compared to CA, AA patients had smaller declines in serum HCV RNA on days 1, 2, 7, 14, and 28 (Supplementary Fig. 1), and less vigorous phase 1 (day 0–2) [0.69 + 0.63 vs. 0.98 + 0.76 log10 IU/ml; p <0.001 ANOVA], phase 2 (day 7–28) [0.38 + 0.25 vs. 0.45 + 0.28 log10 IU/ml/wk; p = 0.03], day 2–7 [0.11 + 0.5 vs. 0.1 + 0.5; p <0.001] and day 0–28 [1.5 + 1.0 vs. 2.25 + 1.23; p <0.0001] responses. A null response (<1 log10 IU/ml HCV RNA decline day 0–28) was more common in AA (37% vs. 20%; p = 0.003) and in patients with SNP non-CC genotypes (T/T-44.2%; C/T-32.0%; C/C-9.4%; p <0.0001). Yet, AA (Fig. 1A) and CA (Fig. 1B) with the SNP C/C genotype had larger declines in serum HCV RNA on days 1, 2, 7, 14, and 28 than those with T/T and C/T. Phase 1 (Fig. 2A), phase 2 (Fig. 2C), and day 0–28 (Fig. 2C) declines were also more robust in AA and CA with C/C relative to T/T and C/T genotypes. Hermann et al. described a second phase starting on day 2–4 and lasting up to day 28 in which HCV RNA was either static or increased (i.e. shoulder) followed by a third phase of decay thought to reflect a delay in the pharmacologic activity of ribavirin [10,11]. In the present study, patients with T/T and C/T genotypes were more likely to have either a static phase or an increase (rebound) in HCV RNA from day 2 to 7 (Fig. 1A and B). Among patients with >1 log10 IU/ml decrease in HCV RNA from day 0 to 28, 40% of CA with CC exhibited a static or rebound phase compared to 61% and 60% of CA with C/T and T/T (p = 0.02). The corresponding proportions in AA were 47%, 70%, and 76% (p = 0.35). However, changes in HCV RNA from day 2 to 7 varied by SNP genotype only in the CA (Fig. 2B). Resumption in HCV RNA decay was noted between day 7 and 28. As reported previously, we did not observe an acceleration in HCV RNA between day 7 and 56 [7]. With one exception, phase 1, phase 2, day 2–7, and day 0–28 declines in HCV RNA did not differ significantly between AA and CA with the same SNP genotype. The change in HCV RNA from day 2 to 7 was greater in CA compared to AA with C/T (p = 0.04).
Fig. 1. Serum HCV RNA day 0–28 AA (A) and CA (B).
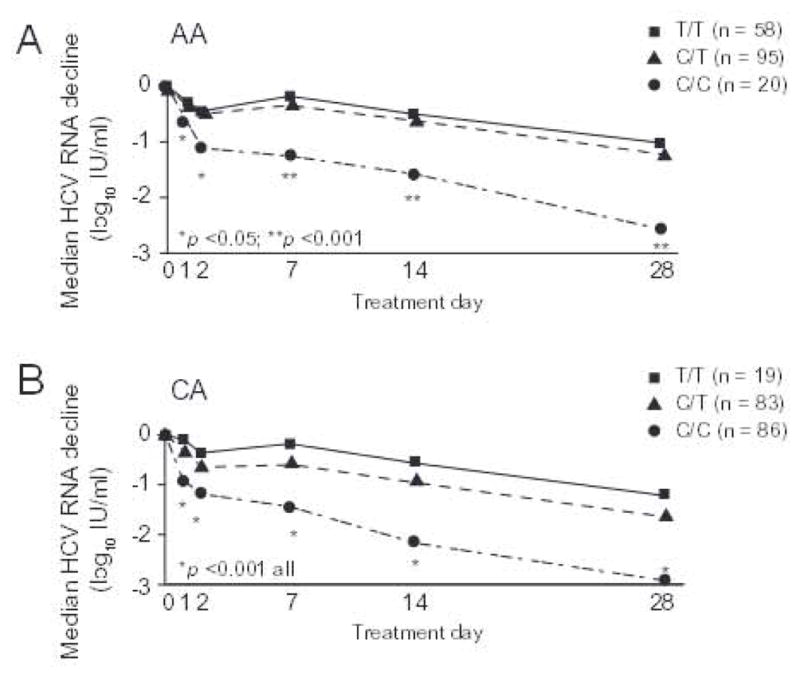
Serum HCV RNA concentrations were measured as per Materials and methods. Results compared using Kruskal-Wallis tests.
Fig. 2. Early HCV RNA kinetics phases (day 0–28).
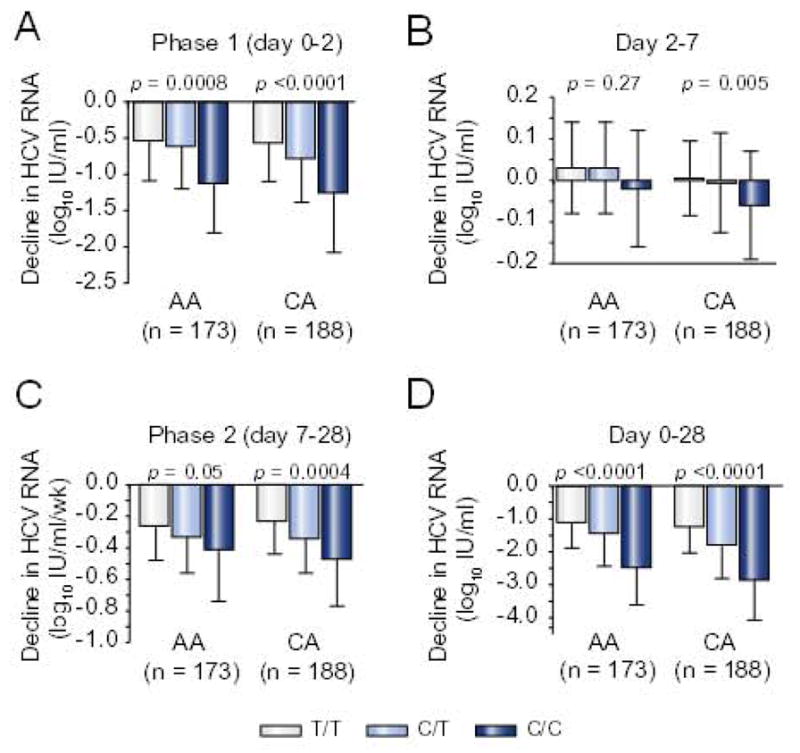
Changes in serum HCV RNA concentrations, (A) phase 1 (day 0–2), (B) day 2–7; (C) phase 2 (day 7–28); (D) day 0–28. Results compared by ANOVA.
Variables associated with phase 1 and phase 2 responses
In univariate analyses, CA race (p <0.0001), SNP C/C genotype (p <0.0001), Ishak liver fibrosis score <3 (p = 0.01), greater portal tract inflammation (HAI >2; p = 0.004), and less lobular inflammation (HAI <4; p = 0.03) at baseline were associated with a larger phase 1 decline in HCV RNA (Supplementary Table 2). CA race (p = 0.01), SNP C/C genotype (p <0.0001) and less hepatic steatosis (p = 0.004) were associated with a faster phase 2 decline. These 6 variables, plus body weight <75 kg (p = 0.01) and less insulin resistance (HOMA score <2) (p = 0.03) were associated with the drop in HCV RNA from day 0 to 28 (Supplementary Table 2). SNP genotype, race and body weight were also associated with day 2–7 change in HCV RNA. In a multivariable regression model, only SNP genotype (C/C vs. non-C/C) was associated with phase 1 (p <0.0001). Phase 1 (stratified by <1 log10 IU/ml vs. >1 log10 IU/ml) was added to the multivariate model for phase 2 [8,9,13]. SNP genotype was associated with phase 2 in both groups (phase 1 <1 log10; p =0.0008; >1 log10, p =0.011). Phase 1 (p = 0.013) predicted phase 2 only in those with <1 log10 IU/ml HCV RNA decline in the total study sample. C/C was associated with a faster phase 2 decline rate in CA with either a phase 1 <1 (p = 0.003) or >1.0 (p = 0.02) log10 IU/ml, but not in AA stratified by phase 1 (Supplementary Fig. 4).
Week 4–72 treatment responses
Compared to T/T and C/T, the SNP C/C genotype was associated with higher week 4 (RVR), 12 (EVR), 24, 48, and 72 (SVR) response rates overall (Supplementary Fig. 2). These differences were highly significant at each time in CA (Fig. 3B), but only at weeks 4 and 12 in AA (Fig. 3A). Breakthrough rates did not vary by SNP genotype in AA patients (Fig. 3A). In contrast, breakthrough was less frequent in CA with C/C and C/T relative to T/T (p = 0.003) (Fig. 3B). Relapse rates did not differ by SNP genotype in either race. Similar results were obtained when 262 patients (112 AA and 250 CA) with optimum medication adherence (i.e. >80% of the maximum peginterferon and ribavirin doses) from day 0 to week 24 were studied (Supplementary Fig. 3A and B). There were no racial differences in response rates from week 4 to 72 among patients with the T/T genotype (Supplementary Fig. 5A). Among patients with C/T (Supplementary Fig. 5B) and C/C genotypes (Supplementary Fig. 5C), CA had higher response rates at weeks 24 (p <0.01), 48 (p <0.005) and 72 (p <0.03). Breakthrough and relapse rates did not differ between AA and CA matched for SNP genotype. Racial differences in week 24–72 responses among patients with C/T and C/C persisted when only patients with optimum medication adherence were analyzed.
Fig. 3. SVR by SNP rs12979860 genotype in AA (A) and CA (B).
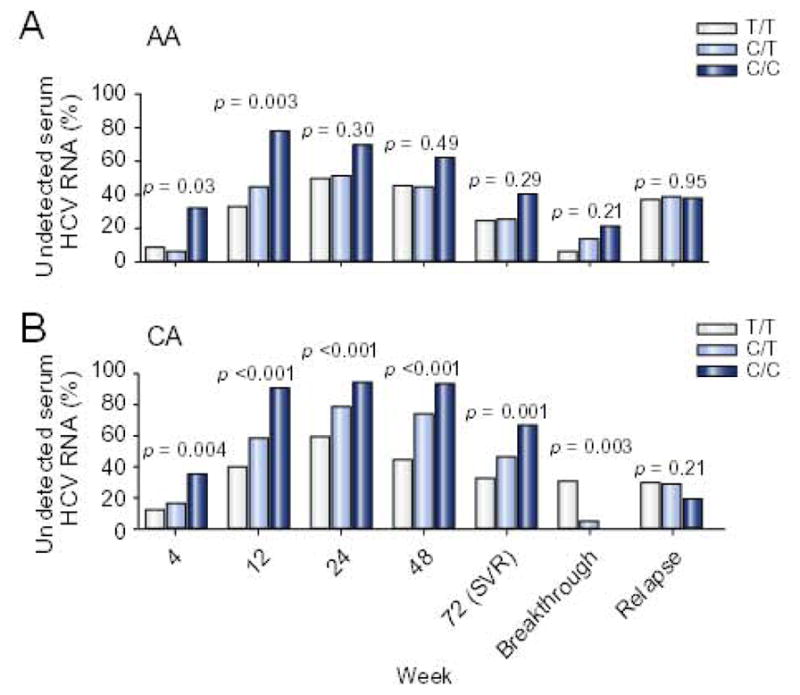
Results compared by regression analysis and additive genetic model.
Variables associated with SVR
Variables associated with SVR in univariate analyses in the total sample were patient race (p <0.0001), SNP genotype (p <0.0001), baseline HCV RNA (p = 0.04), Ishak fibrosis score (p = 0.001), and peginterferon (p = 0.005) and ribavirin adherence (p = 0.006) (Supplementary Tables 3A–C). In a multivariable regression model for the total group, CA race, C/C genotype (additive genetic model), and higher peginterferon and ribavirin adherence were associated with greater odds for SVR (Table 2, model 1); higher baseline serum HCV RNA and a higher Ishak fibrosis score were associated with a lower SVR. SNP genotype, Ishak score, and peginterferon adherence were associated with SVR in CA; only baseline serum HCV RNA was associated with SVR in AA (Supplementary Table 4A). SNP genotype also predicted SVR in a recessive genetic model [OR-3.05 (1.62–5.73), p = 0.0005], but not in a dominant model [OR-1.50 (0.74–3.03), p = 0.26] (Supplementary Tables 4B and C, respectively).
Table 2.
Variables associated with SVR in multivariable regression model.a
| Model 1 | Model 2 | |
|---|---|---|
| Race CA vs. AA | 2.3 (1.3–4.1) p = 0.005 |
1.9 (0.95–3.8) p = 0.07 |
| SNP rs12979860 C/C vs. T/T + C/T |
1.9 (1.3–2.9) p = 0.003 |
0.84 (0.47–1.5) p = 0.56 |
| Baseline HCV RNA | 0.51 (0.34–0.77) p = 0.001 |
0.42 (0.24–0.73) p = 0.002 |
| Ishak Fibrosis | 0.79 (0.66–0.96) p = 0.01 |
0.88 (0.71–1.1) p = 0.24 |
| HOMA-IR | 0.96 (0.91–1.01) p = 0.10 |
0.97 (0.92–1.01) p = 0.17 |
| Peginterferon adherence Day 0-wk 24 |
1.03 (1.00–1.06) p = 0.03 |
1.04 (1.004–1.08) p = 0.03 |
| Ribavirin adherence Day 0-wk 24 |
1.02 (1.00–1.04) p = 0.05 |
1.02 (0.99–1.04) p = 0.09 |
| Phase 1 | - | 0.69 (0.31–1.52) p = 0.36 |
| Phase 2 | - | 3.6 (0.30–42.6) p = 0.31 |
| Day 0–28 log10 HCV RNA decline |
- | 2.66 (1.23–5.75) p = 0.01 |
| RVR | - | 1.8 (0.46–6.98) p = 0.4 |
Results show odds ratio and 95% confidence intervals (CI); n = 361.
Declines in HCV RNA during phase 1, phase 2, and both day 2–7 and day 0–28 were greater in patients with a RVR or a SVR compared to nonresponders at weeks 4 and 72 (Supplementary Table 2). In bivariate regression analyses, RVR was more strongly associated with phase 1 [odds ratio (OR)-4.2, 95% CI (2.4–7.3)] than with phase 2 [OR-0.1 (0.01–0.30)]. In contrast, week 24, 48, and 72 responses were more strongly associated with phase 2 than with phase 1 [SVR: phase 1, OR-2.0 (1.4–3.1); phase 2, OR-21.8 (7.1–67.5)]. To determine the relative importance of early HCV kinetics and baseline variables to SVR, on-treatment variables during the first 4 weeks were combined to variables in model 1 (Table 2, model 2). Baseline serum HCV RNA level (OR 0.46) and the extent of HCV RNA decline from day 0 to 28 (OR 2.66) were strong predictors of SVR; peginterferon adherence (OR 1.04) was weakly associated with SVR. If the day 2–7 change in serum HCV RNA was also included in model 2, only baseline viral load was associated with SVR. Patients with a low baseline HCV RNA (<600,000 IU/ml) had a higher SVR than those with a high baseline RNA (>600,000 IU/ml) (AA-44% vs. 17%, p <0.001; CA-62% vs. 48%; p = 0.02) (Table 3). SVR did not differ by SNP genotype in patients with a low baseline viral load. However, in patients with a high baseline HCV RNA, SVR rates were 2–4 times higher in those with C/C compared to non-C/C genotypes. In patients with C/C, SVR rates did not differ significantly between low and high baseline HCV RNA; in contrast, SVR rates were 2–5 times higher in C/T and T/T patients with a low viral load.
Table 3.
SVR by baseline HCV RNA and day 0–28 response.a
| AA (n = 173)
|
CA (n = 188)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| T/T (%) | C/T (%) | C/C (%) | p | T/T (%) | C/T (%) | C/C (%) | p | |
| HCV RNA (IU/ml) | ||||||||
|
| ||||||||
| <600,000 | 11/24 (46) | 11/27 (41) | 3/6 (50) | 0.97 | 4/6 (67) | 17/24 (71) | 16/26 (61) | 0.61 |
|
| ||||||||
| ≥600,000 | 3/34 (9) | 12/68 (18) | 5/14 (36) | 0.03 | 2/13 (15) | 21/59 (36) | 41/60 (62) | <0.001 |
|
| ||||||||
| Day 0–28 HCV RNA declineb | ||||||||
|
| ||||||||
| <1 | 5/26 (19) | 2/36 (6) | 0/2 (0) | 0.10 | 0/8 (0) | 7/21 (33) | 0/8 (0) | 1.00 |
|
| ||||||||
| ≥1 (inclusive) | 9/27 (33) | 20/55 (36) | 7/17 (41) | 0.60 | 6/10 (60) | 29/58 (50) | 55/71 (77) | 0.008 |
|
| ||||||||
| ≥1 and <2 | 3/17 (18) | 10/30 (33) | 1/4 (25) | 0.41 | 3/7 (43) | 9/25 (36) | 5/8 (62) | 0.41 |
|
| ||||||||
| ≥2 and <3 | 6/10 (60) | 7/20 (35) | 2/7 (29) | 0.17 | 3/3 (100) | 14/23 (61) | 17/25 (68) | 0.74 |
|
| ||||||||
| ≥3 | - | 3/5 (60) | 4/6 (67) | 0.82 | - | 6/10 (60) | 33/38 (87) | 0.07 |
Effect of SNP genotype evaluated under additive genetic model. Numerator shows number with SVR; denominator shows number with a specified baseline HCV RNA or day 0–28 decline in HCV RNA.
log10 IU/ml; SVR-low vs. high viral load (AA-44%vs. 17%, p <0.001; CA-62% vs. 48%; p = 0.02); >1 log10 vs. null (AA-36% vs. 5%, p <0.001; CA-65% vs. 19%; p = 0.001).
Compared to null responders (<1 log10 IU/ml decline in HCV RNA from day 0 to 28), those with a > 1 log10 decrease in RNA had higher SVR rates, overall and stratified by SNP genotype (Table 3). SVR did not vary by SNP genotype in null responders. Among patients with >1 log10 RNA decline from day 0 to 28, in contrast, CA (but not AA) with C/C had a higher SVR than those with non-C/C genotypes. SVR increased gradually with the extent of decline in HCV RNA from day 0 to 28 (Table 3; rows 5–7). Within each day 0–28 decline category, however, SVR rates did not differ by SNP genotype in either AA or CA. Yet, among patients with the same SNP genotype and either baseline viral load or day 0–28 decline category, CA tended to have higher SVR rates. In patients without an RVR, AA had a lower SVR than CA (22% vs. 49%; p <0.001) (Supplementary Table 5). However, SVR did not vary by SNP genotype in AA and CA with an RVR. The C/C genotype was associated with a higher SVR in CA without an RVR (p = 0.004). SVR did not vary by SNP genotype in patients with and without an EVR.
Discussion
SNP rs12979860 was associated with greater declines in serum HCV RNA during phase 1 (day 0–2), phase 2 (day 7–28), day 2–7, and day 0–28 and higher week 4 and 12 response rates in both AA and CA receiving peginterferon and ribavirin therapy for HCV genotype 1. AA had less vigorous phase 1 and 2, day 2–7, and day 0–28 declines and lower week 4 and 12 responses relative to CA that were explained largely by differences in SNP genotype frequencies. Phase 1 and phase 2 predicted SVR, and SNP C/C (vs. non-C/C) was strongly associated with both phases in multivariable analyses. Week 24, 48, and 72 (SVR) responses were 2–3 times higher in CA patients with SNP C/C relative to non-C/C genotypes. Compared to CA with C/T and C/C, week 24, 48 and 72 response rates were lower in AA, and SVR did not vary by SNP genotype in AA.
Bochud et al. reported that SNP rs12979860 genotype is an independent predictor of the phase 1 decline as well as higher RVR and SVR rates during peginterferon α-2a and ribavirin therapy for HCV genotype 1 in European patients [14]. However, this SNP was not associated with the magnitude of phase 2. Consistent with their results, SNP rs12979860 was the sole predictor of phase 1 in a multivariable regression model in the present study [14]. In contrast to Bochud, SNP genotype and phase 1 <1 log10IU/ml were also independently associated with phase 2. The discrepancy in results may be explained by a difference in the definitions for phase 1- day 0–2 in present study and day 0–4 in Bochud’s [14]. Others have reported that a phase 1 >0.5 log10IU/ml is needed for an optimum phase 2 response to peginterferon ± ribavirin [18]. The finding that phase 1 predicts phase 2 in patients with phase <1 log10 IU/ml is in accord with this observation, and suggests there was no phase 1 threshold for the second phase response above a 1 log10 IU/ml decrease. However, the SNP C/C genotype predicted a faster decline in phase 2 regardless of phase 1 category, especially in CA.
In keeping with previous reports in HCV genotype 1 patients, more robust phase 1 (day 0–2) and phase 2 (day 7–28) declines in serum HCV RNA predicted a higher SVR rate during peginterferon and ribavirin [9]. Also, phase 2 was the better predictor of SVR than phase 1. Furthermore, AA patients exhibited weaker phase 1, phase 2, day 2–7, and day 0–28 declines and lower response rates from week 4 to 72 (SVR) compared to CA [9,12]. The racial disparity in early viral kinetics (day 0–28) and week 4 and 12 response rates was strongly related to variation in SNP allele frequencies [6,13,19]. There were no racial differences in phase 1, phase 2, and day 0–28 declines in HCV RNA and in week 4 and week 12 responses in patients grouped by SNP genotype. The lower RVR in AA with the C/T genotype was an exception that might be explained by the small number of AA with a RVR. We also observed an association between SNP rs12979860 genotype and the extent of HCV RNA decay from day 2 to 7, the transition between phase 1 and phase 2. Compared to C/C, patient with non-C/C genotypes were more likely to have either a static phase or rebound in HCV RNA. This variability is likely related to differences in IFN effectiveness (i.e. phase 1). Indeed, the change in HCV RNA from day 2 to 7 was also more strongly correlated with the extent of phase 1 than the phase 2 slope (data not shown). Furthermore, the rebound in HCV RNA from day 2 to 7 was related to a decline in serum peginterferon levels between the first 2 peginterferon injections and was inversely proportional to the ribavirin plasma concentration in previous studies [18,20]. The importance of ribavirin to the phase 2 decline rate in HCV genotype 1 patients is known to vary inversely with IFN effectiveness (i.e. phase 1) [21]. Ribavirin adherence did not differ by SNP genotype in either race. Assuming similar ribavirin exposure, lower IFN effectiveness could explain why a flat or rebound phase from day 2 to 7 and a weaker phase 2 responses were more prevalent in patients with T/T and C/T genotypes [21].
Thompson et al. previously studied the relationship between SNP rs12979860 and treatment response to initial peginterferon and ribavirin therapy for HCV genotype 1 in US patients [13]. In keeping with their results, we observed higher week 4 (RVR), 12, 24, 48, and 72 (SVR) response rates in CA patients with C/C compared to non-C/C genotypes [13]. Likewise, the C/C genotype predicted a higher SVR in CA without a RVR. Furthermore, SNP genotype did not influence the SVR rate in either race based on week 12 (EVR). In contrast, the C/C genotype was only associated with higher week 4 (RVR) and 12 (EVR) responses in AA in the current study. Week 24–72 response rates did not vary by SNP genotype in AA. McCarthy also found no association between this SNP and SVR in 58 AA patients infected with HCV genotype 1 [19]. These inconsistencies may be related to the smaller sample size and a lower percentage of AA with C/C in the present study. Nevertheless, SVR was lower in AA compared to CA with C/C and race, C/C genotype, serum HCV RNA, and Ishak fibrosis score were independently associated with SVR in this and other studies in American patients [6,13,19]. Higher peginterferon adherence was also weakly associated with SVR, but this did not account for differences in treatment response based on race and SNP genetics [5,7].
A multivariable model was used to explore the link between baseline, drug adherence, and early viral kinetics variables associated with SVR (Table 2, model 2 compared to model 1). Only baseline serum HCV RNA, day 0–28 HCV RNA decline, and peginterferon adherence were associated with SVR in this model. Other baseline variables (SNP genotype, race, and Ishak’s fibrosis score) were no longer significant. The day 0–28 decrease in HCV RNA overall was a better predictor of SVR than phase 1 and 2 and the week 4 response (i.e. RVR). The C/C genotype was associated with a higher SVR in AA and CA with a high baseline serum HCV RNA (>600,000 IU/ml) and in CA with >1 log10IU/ml fall in HCV RNA from day 0 to 28. SNP genotype did not influence SVR in patients with a baseline HCV RNA <600,000 IU/ml or a null response. Thus, the greatest clinical utility of SNP genotype for predicting SVR to initial peginterferon and ribavirin for HCV genotype 1 might be in patients with high baseline viral loads. Yet, grouped by SNP genotype and either baseline HCV RNA or day 0–28 decline in RNA, AA had lower SVR rates than CA. Presumably these racial differences reflect biological processes that are not linked to SNP rs12979860.
In conclusion, SNP 12979860 is strongly associated with the strength of phase 1 (day 0–2), phase 2 (day 7–28), and the day 2–7 response during early HCV RNA kinetics and week 4 and 12 response rates during peginterferon and ribavirin treatment in both AA and CA patients infected with HCV genotype 1. In pharmacodynamic terms, these findings suggest that this SNP predicts IFN effectiveness at blocking HCV production and clearance of infected hepatocytes, respectively. SNP 12979860 explained more than 50% of the disparity in early viral kinetics and viral clearance during the first 12 weeks between AA and CA. This SNP was strongly associated with week 24, 48, and 72 (SVR) responses in CA patients, but did not predict treatment responses beyond week 12 in AA patients. A better understanding of the biological basis for the effect of IL28B gene-related SNPs on treatment response and of racial disparities in week 24–72 response to peginterferon and ribavirin treatment might identify novel therapeutic targets for HCV genotype 1.
Supplementary Material
Acknowledgments
Financial support
Supported by NIDDK 1 R01 DK066920-01, 1 K24 DK072036-01. The VIRAHEP-C study was a cooperative agreement funded by the NIDDK with a Cooperative Research and Development Agreement (CRADA) with Roche Laboratories, Inc. Grant Nos.: U01 DK60329, U01 DK60340, U01 DK60324, U01 DK60344, U01 DK60327, U01 DK60335, U01 DK60352, U01 DK60342, U01 DK60345, U01 DK60309, U01 DK60346, U01 DK60349, U01 DK60341. Other support: National Center for Research Resources (NCRR) General Clinical Research Centers Program grants: M01 RR00645 (New York Presbyterian), M02 RR000079 (University of California, San Francisco), M01 RR16500 (University of Maryland), M01 RR000042 (University of Michigan), M01 RR00046 (University of North Carolina).
Abbreviations
- AA
African American(s)
- CA
Caucasian American(s)
- EVR
early virologic response
- CI
confidence interval
- D28
day 28
- HCV
hepatitis C virus
- IFN
interferon
- BMI
body mass index
- HOMA-IR
homeostasis model assessment for insulin resistance
- NR
nonresponders
- OR
odds ratio
- ln
natural log
- R
response or responder[s]
- RVR
rapid virologic response
- SNP
single nucleotide polymorphism
- SVR
sustained virologic response
Footnotes
Conflict of interest
C.H. serves on the Roche-Genentech Pegasys Advisory Board.
Supplementary data associated with this article can be found, in the online version, at doi: 10.1016/j.jhep.2011.10.004.
References
- 1.Maylin S, Martinot-Peignoux M, Moucari R, Boyer N, Ripault MP, Cazals-Hatem D, et al. Eradication of hepatitis C virus in patients successfully treated for chronic hepatitis C. Gastroenterology. 2008;3:821–829. doi: 10.1053/j.gastro.2008.05.044. [DOI] [PubMed] [Google Scholar]
- 2.Pradat P, Tillmann HL, Sauleda S, Braconier JH, Saracco G, Thursz M, et al. Long-term follow-up of the hepatitis C HENCORE cohort: response to therapy and occurrence of liver-related complications. J Viral Hepat. 2007;8:556–563. doi: 10.1111/j.1365-2893.2006.00829.x. [DOI] [PubMed] [Google Scholar]
- 3.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 4.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C. N Engl J Med. 2002:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 5.Conjeevaram HS, Fried MW, Jeffers LJ, Terrault N, Wiley-Lucas T, Afdhal N, et al. Peginterferon and Ribavirin Treatment in African American and Caucasian American Patients with Chronic Hepatitis C Genotype 1. Gastroenterology. 2006:470–477. doi: 10.1053/j.gastro.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;7262:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 7.Hoofnagle JH, Wahed AS, Brown RS, Jr, Howell CD, Belle SH Virahep-C Study Group. Early Changes in Hepatitis C Virus (HCV) Levels in Response to Peginterferon and Ribavirin Treatment in Patients with Chronic HCV Genotype 1 Infection. J Infect Dis. 2009;8:1112–1120. doi: 10.1086/597384. [DOI] [PubMed] [Google Scholar]
- 8.Layden JE, Layden TJ, Reddy KR, Levy-Drummer RS, Poulakos J, Neumann AU. First phase viral kinetic parameters as predictors of treatment response and their influence on the second phase viral decline. J Viral Hepat. 2002:340–345. doi: 10.1046/j.1365-2893.2002.00377.x. [DOI] [PubMed] [Google Scholar]
- 9.Girard C, Ravallec M, Mariller M, Bossy JP, Cahour A, Lopez-Ferber M, et al. Effect of the 5′ non-translated region on self-assembly of hepatitis C virus genotype 1 a structural proteins produced in insect cells. J Gen Virol. 2004;0:3659–3670. doi: 10.1099/vir.0.79909-0. [DOI] [PubMed] [Google Scholar]
- 10.Herrmann E, Lee JH, Marinos G, Modi M, Zeuzem S. Effect of ribavirin on hepatitis C viral kinetics in patients treated with pegylated interferon. Hepatology. 2003:1351–1358. doi: 10.1053/jhep.2003.50218. [DOI] [PubMed] [Google Scholar]
- 11.Dahari H, Ribeiro RM, Perelson AS. Triphasic decline of hepatitis C virus RNA during antiviral therapy. Hepatology. 2007;1:16–21. doi: 10.1002/hep.21657. [DOI] [PubMed] [Google Scholar]
- 12.Layden-Almer JE, Ribeiro RM, Wiley T, Perelson AS, Layden TJ. Viral dynamics and response differences in HCV-infected African American and white patients treated with IFN and ribavirin. Hepatology. 2003:1343–1350. doi: 10.1053/jhep.2003.50217. [DOI] [PubMed] [Google Scholar]
- 13.Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;1:120–129. doi: 10.1053/j.gastro.2010.04.013. [e18] [DOI] [PubMed] [Google Scholar]
- 14.Bochud PY, Bibert S, Negro F, Haagmans B, Soulier A, Ferrari C, et al. IL28B polymorphisms predict reduction of HCV RNA from the first day of therapy in chronic hepatitis C. J Hepatol. 2011 doi: 10.1016/j.jhep.2011.01.050. [DOI] [PubMed] [Google Scholar]
- 15.McHutchison JG, Everson GT, Gordon SC, Jacobson IM, Sulkowski M, Kauffman R, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;18:1827–1838. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 16.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;13:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009 doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feld JJ, Lutchman GA, Heller T, Hara K, Pfeiffer JK, Leff RD, et al. Ribavirin improves early responses to peginterferon through improved interferon signaling. Gastroenterology. 2010;1:154–162. doi: 10.1053/j.gastro.2010.03.037. [e4] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarthy JJ, Li JH, Thompson A, Suchindran S, Lao XQ, Patel K, et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;7:2307–2314. doi: 10.1053/j.gastro.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howell CD, Dowling TC, Paul M, Wahed AS, Terrault NA, Taylor M, et al. Peginterferon pharmacokinetics in African American and Caucasian American patients with hepatitis C virus genotype 1 infection. Clin Gastroenterol Hepatol. 2008;5:575–583. doi: 10.1016/j.cgh.2008.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixit NM, Layden-Almer JE, Layden TJ, Perelson AS. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature. 2004:922–924. doi: 10.1038/nature03153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


