Abstract
Tissue resident memory (Trm) represent a newly described memory T cell population. We have previously characterized a population of Trm that persists within the brain following acute virus infection. Although capable of providing marked protection against a subsequent local challenge, brain Trm do not undergo recall expansion following dissociation from the tissue. Furthermore, these Trm do not depend on the same survival factors as the circulating memory T cell pool as assessed either in vivo or in vitro. To gain greater insight into this population of cells we compared the gene-expression profiles of Trm isolated from the brain to circulating memory T cells isolated from the spleen following an acute virus infection. Trm displayed altered expression of genes involved in chemotaxis, expressed a distinct set of transcription factors and overexpressed several inhibitory receptors. Cumulatively, these data indicates that Trm are a distinct memory T cell population disconnected from the circulating memory T cell pool and displaying a unique molecular signature which likely results in optimal survival and function within their local environment.
Introduction
Classically, memory CD8 T cells are broadly divided into two subsets termed central memory (Tcm) and effector memory (Tem). Tcm CD8 T cells express the adhesion molecule L- selectin (CD62L) and the chemokine receptor CCR7, traffic through lymph nodes, spleen and blood, and represent a long-lived population of memory cells with high proliferative capacity upon re-challenge. Effector memory CD8 T cells (Tem) lack CD62L and CCR7, are present in spleen and blood, re-circulate through the peripheral tissues and maintain immediate cytotoxic potential (1, 2). Non-lymphoid tissues house a large proportion of the memory T cell pool (3). Previously, it was thought that these were simply effector memory T cells trafficking through the tissue as part of their immunological surveillance. Recent studies have shown that a proportion of these memory T cells reside in the tissue and represent a distinct memory T cell population (4). These peripherally deposited memory T cells, termed resident memory (Trm) are a self-sustaining population that persist long term within non-lymphoid tissue, commonly at sites of prior infection. Trm have been identified in a variety of peripheral tissues including the skin and sensory ganglia of mice latently infected with herpes simplex virus (HSV) (4–6), and in the gut (7), brain (8), lung (9, 10) and salivary glands (11). These cells rapidly acquire effector function upon secondary pathogenic encounter (6) and those in the skin are highly protective against subsequent local infection (4, 12).
We have recently characterized the memory CD8+ T cell population that persists within the brain following an acute systemic vesicular stomatitis virus (VSV) infection (8). We showed that memory T cells persisting within the brain survive without replenishment from the circulation. These cells selectively expressed the integrin CD103, the expression of which was dependent on antigen recognition within the tissue. Antigen persistence was not required for memory T cell retention within the brain. The memory CD8 T cells isolated from the brain died rapidly upon isolation from the tissue and failed to undergo recall expansion after adoptive transfer into the bloodstream of antigen-challenged recipients. Cumulatively, these data showed that memory CD103+ CD8 T cells that persist within the brain following an acute virus infection are bona fide tissue resident memory T cells.
Recent work by several groups in both mice and human studies provides compelling evidence to support the presence of a locally confined memory T cell population that is deposited at former sites of antigen encounter (13). However, to date there was no molecular genetic profile defining these peripherally deposited memory T cells. Using a previously characterized model of intranasal infection with VSV we genetically profiled Trm CD8 T cells that develop within the brain and compared them to memory T cells that are part of the circulating memory T cell pool. Our data clearly indicates that Trm are a distinct memory T cell population with a unique molecular signature.
Materials and Methods
Mice
C57BL/6, B6.SJL-PtprcaPep3b/BoyJ (CD45.1) mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and housed in specific pathogen-free conditions in the animal facilities at the University of Washington (Seattle, WA). OT-I TCR transgenic mice congenic for Ly5.1 or deficient in bim and MHC II deficient mice were bred and maintained in the same facilities. All experiments were done in accordance with the Institutional Animal Care and Use Committee guidelines of the University of Washington.
T cell adoptive transfer and infections
Mice received 104 naïve OT-I.CD45.1 CD8+ T cells via i.v. injection prior to infection. Cells were sorted from the brain and spleens of mice at the indicated times post infection using a FACS Aria II and 3×103 cells were transferred into mice either i.c. or i.v. prior to infection.
Mice were infected intranasally (i.n.) with 5×104 PFU or intraperitoneally (i.p) with 2×107 PFU of a recombinant vesicular stomatitis virus that expresses GFP and a secreted form of OVA (14). Mice were infected intracranially with 10 CFU of either wild type or a recombinant Listeria monocytogenes that expresses a secreted form of OVA (LM-OVA). Growth and quantification of LM-OVA was performed as described previously (15).
For in vitro stimulation experiments, 20,000 memory OT-I T cells were cultured at a 1:1 ratio with SIINFEKL pulsed (1µM) or un-pulsed irradiated splenocytes. The absolute number of live cells was determined 60 hours later.
Viral titer determination
The presence of infectious VSV in tissue samples was determined using standard PFU assays on confluent Vero cell monolayers as described previously (8).
Flow cytometry
Single cell suspensions were prepared from spleens and LN by mechanical disruption. Brains were enzymatically digested for 1 hour at 37°C in 3 ml of collagenase type 3 (Worthington) (3mg/ml in RPMI 1640 supplemented with 2% FCS) and lymphocytes were separated on a percoll gradient. Cells were stained for 25 min on ice with the appropriate cocktail of monoclonal antibodies and washed with PBS with 1% BSA. The following conjugated monoclonal antibodies were obtained from BD or eBioscience: anti CD8α, CD45.1, CD103. Cells were analysed on a FACSCanto II using Flowjo software (Tree Star).
Survival assay
Cells were cultured for 18 hours in RPMI 1640 supplemented with 2% FCS, 2 mM glutamine, 5×10−5 M 2-ME, and antibiotics and with 10ng/ml of either recombinant IL-7 or IL-15. Annexin V staining was performed using an Annexin V staining kit (BD) following manufacturers instructions.
RNA samples
Mice received 104 naïve OT-I.CD45.1 CD8+ T cells via i.v. injection prior to i.n. infection with VSV-OVA. CD103+ and CD103− OT-I cells were sorted from the brain and spleens of mice on day 20 p.i. using a FACS Aria II. Cells were subjected to two successive rounds of sorting. All samples were maintained at 4°C for the duration of the sort, and purity was 95%–99% for all populations. The treatments and cell sorts were repeated for three independent pools of mice. One or two samples were extracted of each cell population from each mouse pool, making thirteen samples in total. RNA was isolated and hybridized to Affymetrix Mouse Gene 1.0ST GeneChips by the Immunological Genome Project (www.immgen.org), following standard operating procedures.
Microarray analysis
The microarray intensities were background corrected, normalized and summarized according to the Robust Multichip Average algorithm (16) using the aroma.affymetrix software package (http://www.aroma-project.org). The chip definition file MoGene-1.0-st-v1,r3.cdf was used to define a core set of probe-sets with reliable gene annotation.
Differential expression analysis was undertaken using the limma software package for R (17). Genes were filtered as non-expressed if they failed to exceed the bottom 25% of expression values on at least three arrays. Differential expression was assessed using empirical Bayes moderated t-statistics and F-statistics (18). The analysis was adjusted for correlation between samples from the same mouse pool using the duplicate Correlation function. P-values were adjusted to control the false discovery rate (FDR) using the method Benjamini and Hochberg (19)
Results
Brain Trm do not undergo recall expansion following dissociation from the tissue in which they reside
Intranasal infection with VSV results in a transient systemic infection which is cleared from all organs including lung, spleen and brain by day 8 post infection. Using a recombinant virus that expresses the model antigen OVA (VSV-OVA) in combination with OT-I TCR transgenic cells we have previously demonstrated that long after the resolution of the acute VSV-OVA infection, memory OT-I CD8 T cells persist within the brain (8). The memory CD8 T cells persisting within the brain can be sub-divided into two populations based on CD103 expression although by day 20 p.i. the vast majority of memory OT-I cells persisting within the brain express this integrin (Fig 1A). Further characterization of these brain residing CD103+ memory T cells revealed that they are resident memory T cells (Trm) (8).
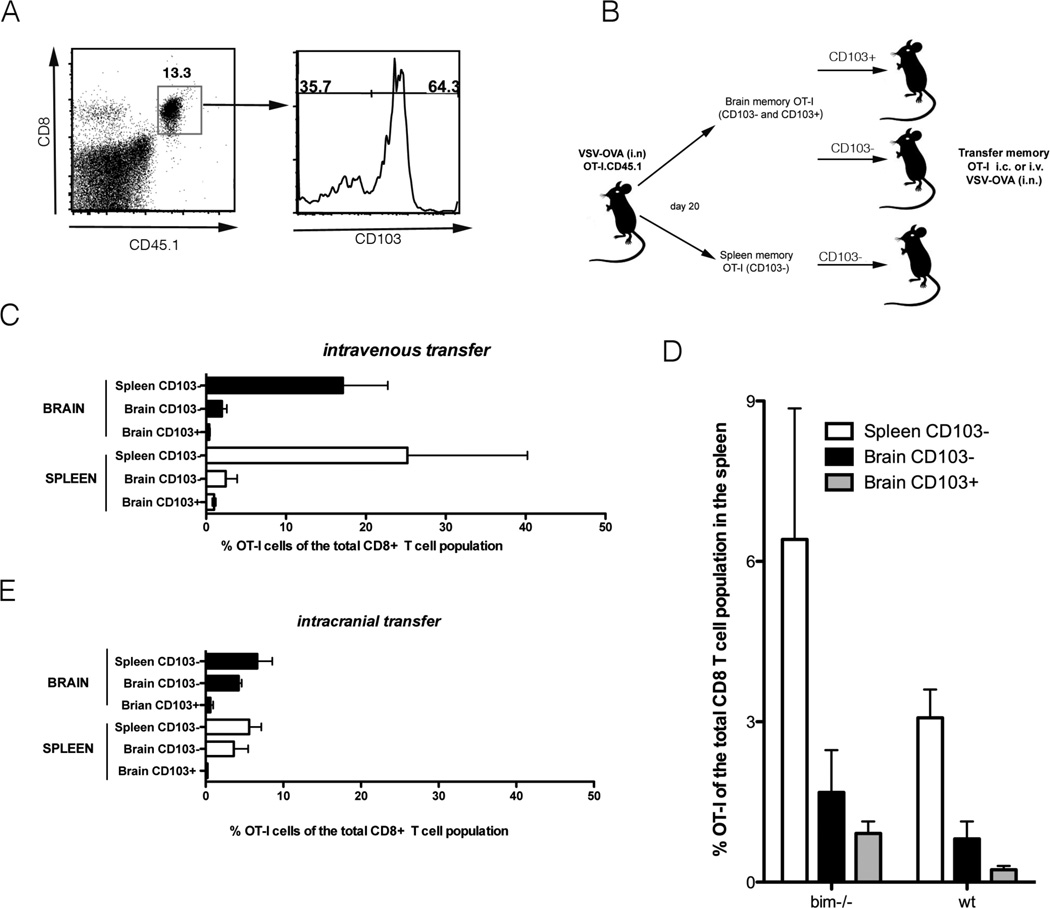
Figure 1. Resident memory T cells do not undergo recall expansion following dissociation from the tissue in which they reside.
Mice were seeded with OT-I.CD45.1 T cells before i.n. infection with VSV-OVA. On day 20 p.i., OT-I cells were recovered from the brain and spleen of mice, sorted into CD103+ and CD103−, and adoptively transferred into naïve recipient mice that were challenged with VSV-OVA (i.n.) (A) Representative flow cytometry profile of brain on day 20 p.i. demonstrating the OT-I.CD103+ and OT-I.CD103− subsets. (B) Schematic diagram representing the experimental setup (C) The proportion of OT-I of the total CD8+ T cell population in the brain and spleen on day 8 p.i. following intravenous transfer of memory T cells. Data represents the mean + SEM (n = 3–5) and data is pooled from 2 independent experiments. (D) Mice were seeded with either bim−/− OT-I.CD45.1 or wild type (wt) OT-I T cells before i.n. infection with VSV-OVA. On day 20 p.i., OT-I cells were recovered from the brain and spleen of mice, sorted into CD103+ and CD103−, and adoptively transferred (i.v.) into naïve recipient mice that were challenged with VSV-OVA (i.n.). Depicted is the proportion of OT-I of the total CD8+ T cell population in the spleen on day 8 p.i. Data represents the mean + SEM (n = 4–8) and data is pooled from 2 independent experiments. (E) As described in part C except memory T cells were transferred intracranially prior to i.n. challenge with VSV-OVA.
We have previously shown that brain CD103+ Trm undergo poor recall expansion following dissociation from the brain tissue. Specifically, when we sorted CD103+ and CD103− OT-I CD8+ T cells from the brains and spleens of mice on day 20 post intranasal infection with VSV-OVA and adoptively transferred these T cells i.v. into naïve recipient mice and subsequently challenged these animals with VSV-OVA we observed that all memory populations expanded in response to secondary challenge except the brain resident CD103+ memory T cells ((8), Fig 1 B, C). Delaying isolation of the spleen and lung memory T cells until a later memory time point (day 50 p.i.) to allow further maturation of the memory cells failed to restore the proliferation potential of brain CD103+ Trm (Supplementary Fig 1A). The lack of recall expansion by the CD103+ memory T cell population after isolation and adoptive transfer may reflect a defect in survival following tissue dissociation. It is noteworthy that we could not rescue the inability of brain CD103+ memory T cells to expand when we repeated the experiments described above using OT-I cells deficient in the pro-apoptotic Bcl-2 family member Bim (OT-I.bim−/−) (Fig 1D) which is known to be critical for controlling normal homeostasis of memory T cells (20). This suggests that if the inability of brain CD103+ memory T cells to mount a secondary recall response is due to a defect in survival following dissociation from the tissue then this is a bim independent death.
We next sought to determine if we could enhance the recall response of brain CD103+ memory T cells by re-introducing them back into the microenvironment of the brain. To do this, we sorted CD103+ and CD103− OT-I CD8+ T cells from the brains and spleens of mice on day 20 post infection with VSV-OVA and adoptively transferred low numbers of these memory T cells directly into the brain (i.c.) of naïve recipient mice. We subsequently challenged these animals with VSV-OVA. Even when reseeded back into the brain tissue, brain CD103+ memory T cells failed to mount a recall response (Fig 1E). This is in contrast to both CD103− brain and splenic memory T cells which underwent expansion. Brain CD103+ Trm also displayed impaired recall expansion when stimulated ex vivo. We sorted CD103+ and CD103− OT-I CD8+ T cells from the brains and spleens of mice on day 20 post intranasal infection with VSV-OVA. These cells were cultured in vitro with SIINFEKL peptide loaded splenocytes. We observed that all memory populations expanded in response to ex vivo stimulation except the brain resident CD103+ memory T cells (Supplementary Fig 1B). Cumulatively, these data demonstrate that brain CD103+ memory T cells are functionally distinct from brain CD103− and splenic CD103− memory T cell populations.
Brain memory T cells do not depend on the same survival factors as circulating memory T cells both in vitro and in vivo
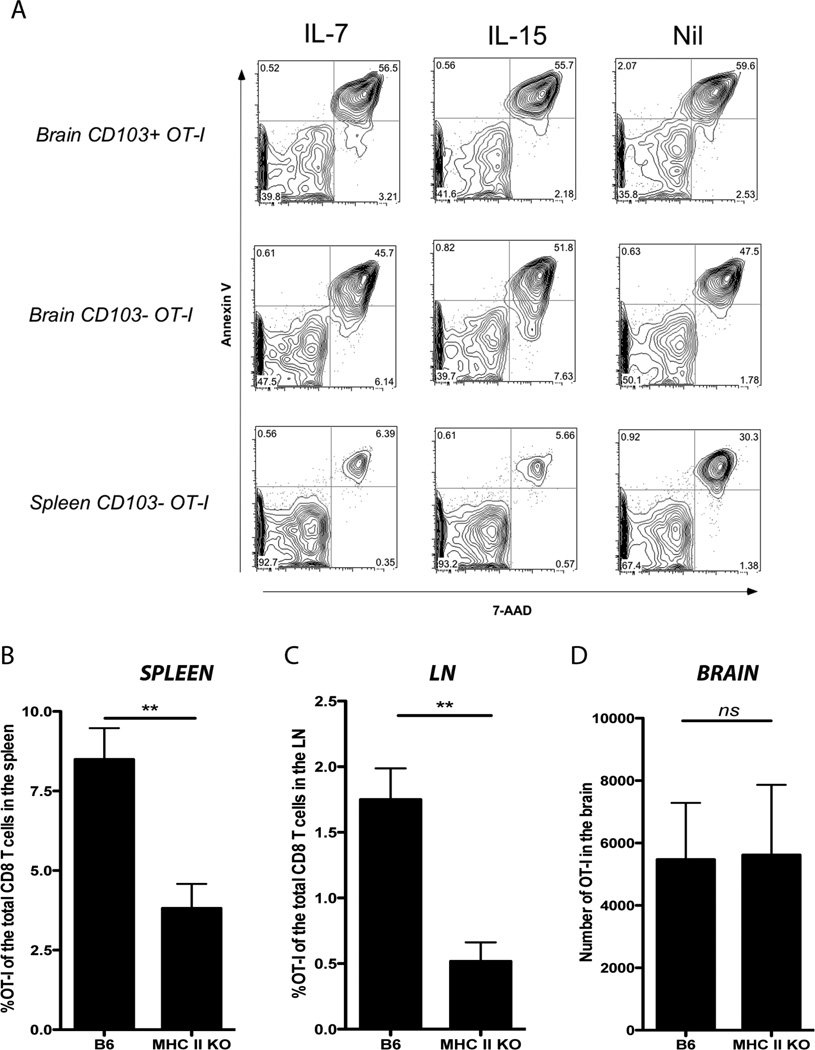
We previously reported that memory CD8 T cells isolated from the brain die rapidly in vitro (8). We wished to determine whether the addition of cytokines known to be important in memory T cell survival could improve ex vivo survival of these tissue residing memory T cells. CD103+ and CD103− OT-I cells were sorted from the brain and spleen of mice on day 20 post infection with VSV-OVA. The cells were cultured in vitro in the presence of IL-7 or IL-15 and survival was assessed 18 hours later by annexin V and 7-amino-actinomycin D (7-AAD) staining. Although IL-7 and IL-15 greatly enhanced the survival of splenic memory T cells, neither cytokine rescued either brain memory T cell population (Fig 2 A).
Figure 2. Brain memory T cells do not depend on the same survival factors as circulating memory T cells in vitro or in vivo.
(A) Mice were seeded with OT-I.CD45.1 T cells before i.n. infection with VSV-OVA. On day 20 p.i., memory OT-I.CD103+ and CD103− cells were sorted from the brain and spleen of mice and cultured in vitro overnight in the presence of IL-7 or IL-15. Cells were stained with Annexin V and 7-AAD, and the percentage of dead cells (Annexin V+/− and 7-AAD+) was determined by flow cytometry. (B) Mice (either B6 or MHC class II KO) were seeded with OT-I.CD45.1 T cells before i.n. infection with VSV-OVA. Graphs depict the percentage or number of OT-I cells of the total CD8 T cell population in the spleen, lymph node (LN) and brain on day 60 p.i. Bars represent the mean + SEM (n= 9–17). Data is pooled from 4 independent experiments [P < 0.05, student t test].
Memory T cells within the brain also do not appear to rely on the same survival factors as splenic memory T cells in vivo. We demonstrated this by monitoring the persistence of memory CD8 T cells within the brain of MHC class II deficient mice. It has been previously reported that memory CD8 T cells undergo a gradual decay in MHC class II deficient mice (21). To assess whether brain resident memory T cells were vulnerable to a similar decay, we adoptively transferred naïve OT-I cells into either B6 or MHC class II deficient mice prior to i.n. infection with VSV-OVA. Analysis of the brain, LN and spleen of these mice on day 60 p.i. revealed that although memory OT-I within the LN and spleen from MHC class II KO mice underwent significant decay (Fig 2B, C), consistent with the original studies (21) we observed no difference in the number of memory T cells within the brain when comparing B6 to MHC class II deficient mice (Fig 2D). Thus, brain memory T cells and circulating memory T cells do not depend on the same survival factors for their long term maintenance. Furthermore, these data further support the notion that the memory T cell population within the brain exists independently of the circulating memory T cell pool because if this population was continually supplemented by circulating memory T cells then we would also expect a decay in brain memory T cell numbers mirroring the events within the circulation.
Brain Trm provide protection during a localized infection
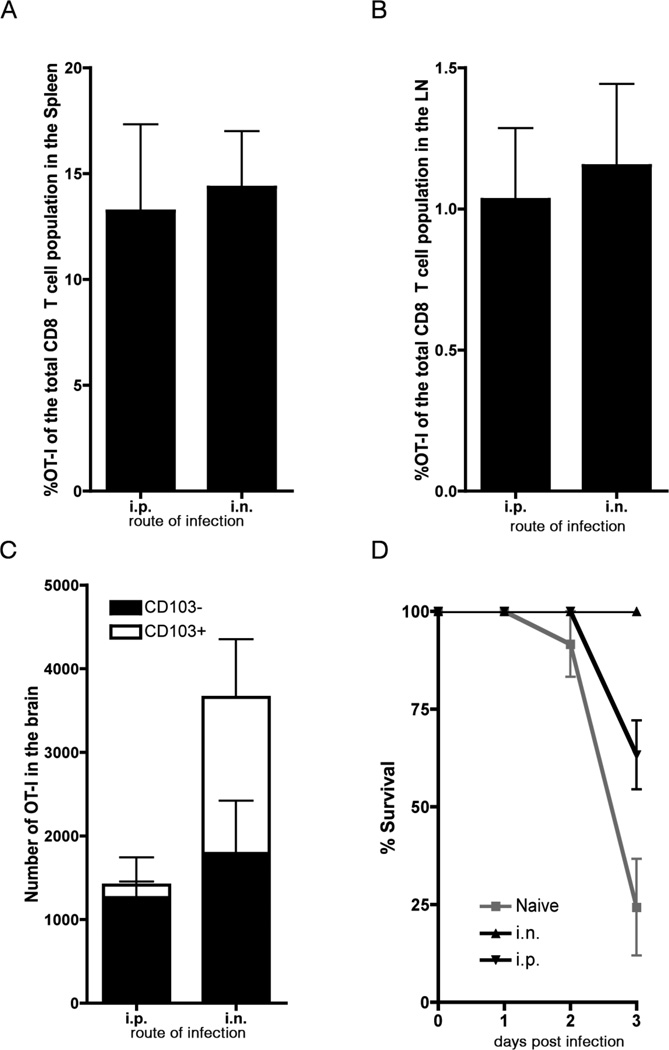
Brain CD103+ Trm exhibit survival defects ex vivo although when left in situ, they do provide an overt advantage during a localized secondary infection. To demonstrate this we took advantage of the fact that only local virus infection of the brain results in the generation of CD103+ Trm. Mice seeded with low numbers of OT-I.CD45.1 T cells were infected either i.n. or i.p. within VSV-OVA. Irrespective of the route of infection, we observed a similar sized memory OT-I response in the LN and spleen (Fig 3A, B). Unlike i.n. administration of VSV, i.p. infection does not result in virus infection of the brain (Supplementary Fig 2). Analysis of the OT-I cell population within the brains of mice infected with VSV-OVA either i.p or i.n. 20 days earlier shows that in both cases there are approximately equivalent numbers of CD103− OT-I cells. However, only following i.n. immunization, which results in a virus infection of the brain, did we observe the development and persistence of CD103+ memory OT-I cells. (Fig 3C) In summary this model provides us with a system where we have equivalent circulating memory T cell populations in the LN and spleen and similar numbers of brain CD103− memory T cells. The only difference between the two cohorts of mice is the presence of CD103+ memory OT-I in the animals that had been primed intranasally.
Figure 3. Resident memory T cells within the brain can provide protection against local infection.
Mice were seeded with OT-I.CD45.1 T cells before intranasal (i.n.) or intraperitoneal (i.p) infection with VSV-OVA. On day 20 p.i. mice were challenged intracranially with LM-OVA. The proportion of OT-I T cells of the total CD8+ T cell population in the (A) spleen and (B) lymph node and (C) the number of OT-I CD8 T cells in the brain on day 20 p.i. Bars represent the mean + SEM (n = 6) data is representative of 2 independent experiments. (D) Survival (measured as a loss of > 20% of starting weight) of animals primed either i.p or i.n. with VSV-OVA and challenged i.c. with LM-OVA. Shown is the mean + SEM (n = 15). Data is pooled from 3 independent experiments.
Mice primed 20 days earlier with VSV-OVA via either the i.p or i.n route were challenged intrancranially with a recombinant Listeria that expresses OVA (Lm-OVA) and were monitored for survival (Fig 3D). We observed 100% survival amongst animals primed with VSV-OVA i.n. and therefore containing brain CD103+ memory T cells. In contrast, by day 3 post challenge with Lm-OVA 40% of mice primed via the i.p route, and therefore lacking brain CD103+ memory OT-I T cells had succumbed to infection. This protection is not merely due to presence of innate immune mechanisms present in the brain of mice primed i.n. with VSV-OVA as when we intracranially challenge these mice with a wild type Listeria (not expressing OVA) we failed to observe any protection (Supplementary Fig 3). Thus, brain CD103+ memory T cells are functional in situ and provide enhanced protection during a localized infection.
Gene expression profile analysis of brain CD8+ Trm
To further characterize brain CD103+ Trm we performed gene array analysis. To define the molecular signature of CD103+ Trm we compared the transcriptomes of CD103+ and CD103− OT-I T cells sorted from the brain to CD103− memory OT-T cells isolated from the spleen of mice 20 days following i.n. infection with VSV-OVA. Note that there were too few CD103+ memory cells in spleen to include in the analysis. An example of pre-sort and post-sort analysis of purified cell subsets is shown in Supplementary Figure 4.
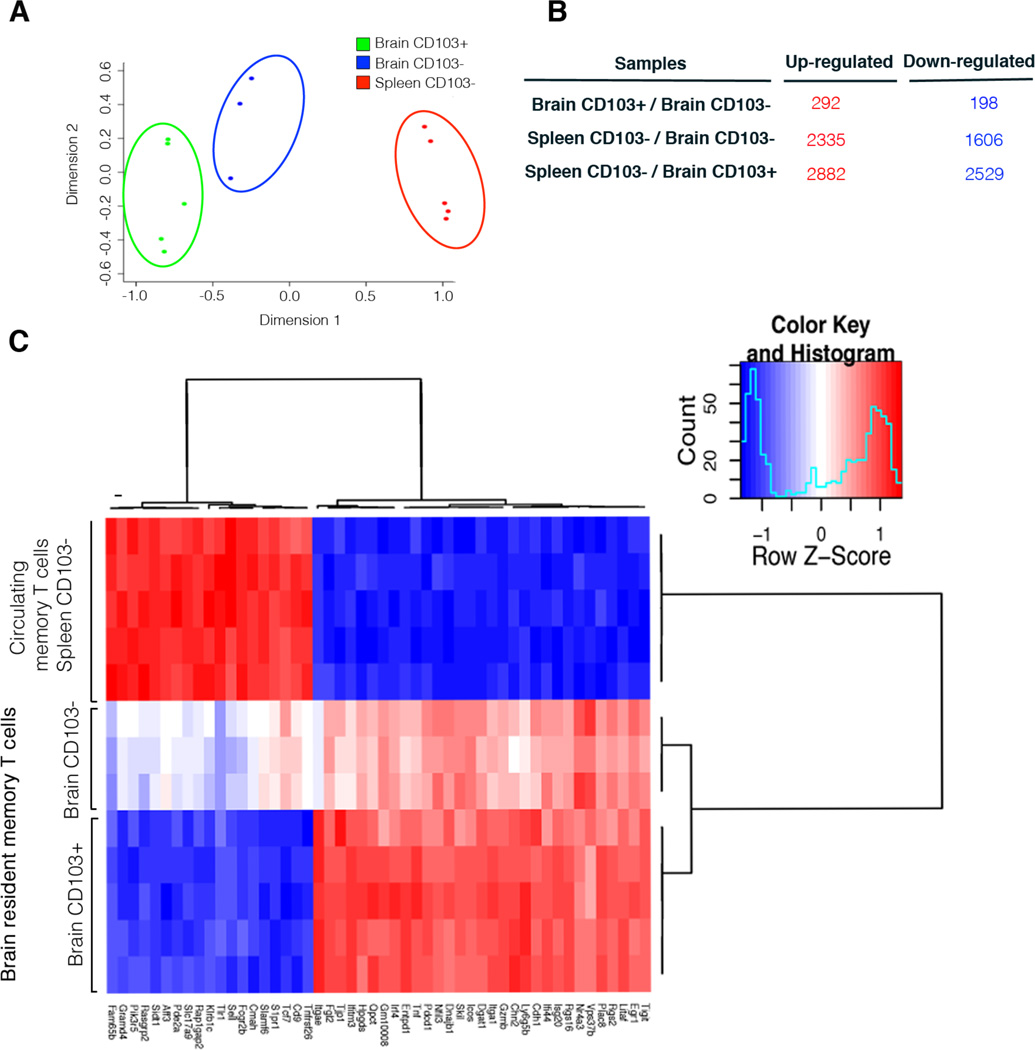
Multidimensional scaling (MDS) was used to display the leading fold-changes between the expression profiles, demonstrating that the three cell populations have distinct profiles and that brain CD103+ OT-I cells are most similar to brain CD103− cells (Fig 4A). Furthermore, when comparing the brain populations to the splenic memory T cell pool it is the brain CD103− OT-I cells that share more genetic similarity to the splenic memory T cells population.
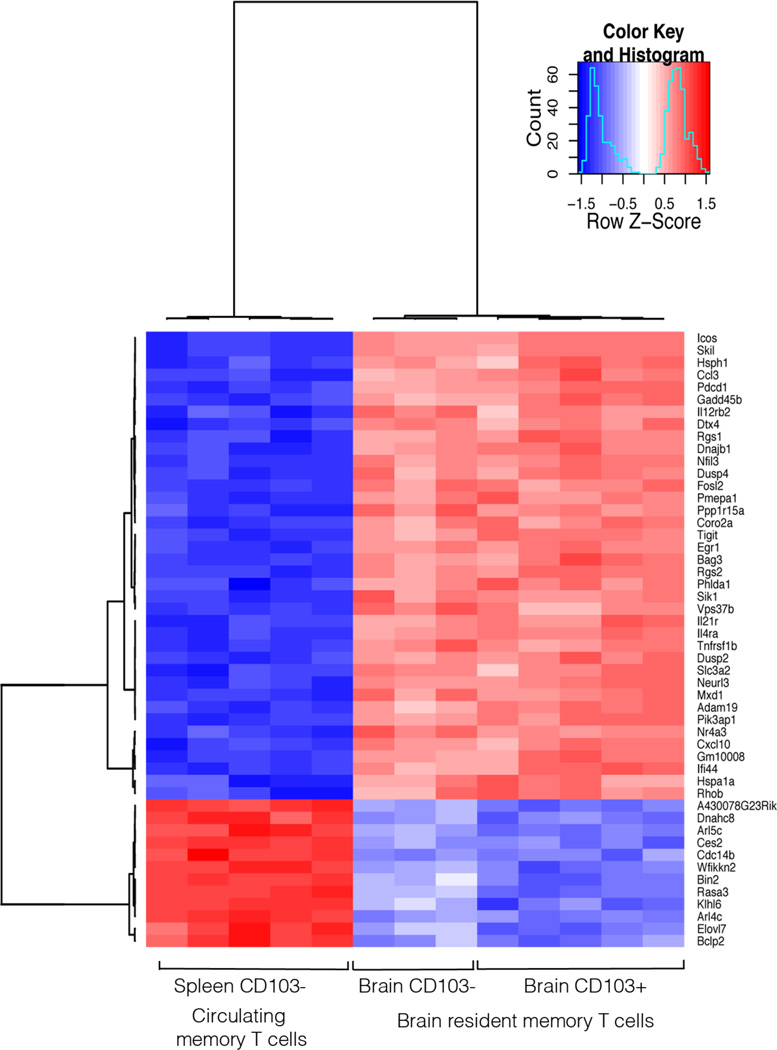
Figure 4. Brain resident memory T cells have a different molecular signature compared to circulating memory T cells.
(A) A MDS plot depicting the relationship between Brain CD103+, Brain CD103− and Spleen CD103− OT-I T cell populations. Distances between samples represent leading fold change, the average log2-fold change between the 500 genes with largest differences. (B) Number of genes differentially expressed between brain resident and splenic memory T cells (FDR < 0.05). (C) Heatmap of expression profiles (normalized log2-intensities) of Brain OT-I.CD103+, Brain OT-I CD103− and Spleen OT-I.CD103− memory T cells. Heatmap shows the 50 genes with the most significant difference between the cell populations.
Although the two memory OT-I populations isolated from the brain are closely related their genetic profiles indicate that they do represent distinct cell populations. We find that there are a total of 490 genes that are significantly differentially expressed (FDR < 0.05) when comparing the brain CD103+ to brain CD103− memory OT-I cells (Fig 4B). There were more differences in gene expression comparing either CD103− or CD103+ brain OT-I to the splenic memory T cell population with 3941 and 5411 genes being upregulated or downregulated, respectively (Fig 4B).
Figure 4C shows a heat map analysis of the 50 most differentially expressed genes when comparing brain CD103+, brain CD103− and spleen CD103− memory OT-I cells. These data further highlights that the gene expression profiles of brain CD103+ and CD103− memory T cells are closely related. However these two brain-derived subsets were markedly different from splenic memory T cells in terms of their gene expression profile. A list of the top 10 most differentially expressed genes when comparing the 3 subsets is presented in Table 1. While Table 2 is a list of genes that are significantly differentially expressed between the 3 cell subsets and grouped into immune system related categories of interest.
Table 1.
| Brain CD103+ vs Brain CD103− | |||
|---|---|---|---|
| ID | SYMBOL | logFC | P value |
| 10378286 | Itgae | 3.01 | 1.26E-18 |
| 10522051 | Klf3 | −2.23 | 7.71E-16 |
| 10385776 | Tcf7 | −2.47 | 7.21E-15 |
| 10439583 | Sidt1 | −1.32 | 8.58E-14 |
| 10351691 | Slamf6 | −2.11 | 1.10E-13 |
| 10407940 | Naip3 | 2.52 | 9.30E-13 |
| 10501586 | Slpr1 | −2.42 | 9.74E-13 |
| 10388234 | Gsg2 | 2.03 | 2.26E-12 |
| 10403821 | Tcrg-V3 | 2.01 | 4.69E-12 |
| 10589994 | Eomes | −1.81 | 5.72E-12 |
| Spleen CD103− vs Brain CD103+ | |||
| ID | SYMBOL | logFC | P value |
| 10378286 | Itgae | −5.08 | 5.72E-24 |
| 10439583 | Sidt1 | 3.17 | 8.14E-22 |
| 10522051 | Klf3 | 3.81 | 3.01E-21 |
| 10569017 | Ifitm3 | −3.53 | 1.04E-19 |
| 10351691 | Slamf6 | 3.62 | 4.55E-19 |
| 10554240 | Isg20 | −3.01 | 6.38E-19 |
| 10350733 | Rgs16 | −3.02 | 9.13E-19 |
| 10437687 | Litaf | −2.47 | 1.77E-18 |
| 10360028 | Fcgr2b | 3.15 | 1.87E-18 |
| 10385776 | Tcf7 | 3.31 | 2.69E-18 |
| Spleen CD103− vs Brain CD103− | |||
| ID | SYMBOL | logFC | P value |
| 10439583 | Sidt1 | 1.85 | −2.26E-16 |
| 10346799 | Icos | −2.09 | 2.84E-16 |
| 10554240 | Isg20 | −2.41 | 5.40E-16 |
| 10350733 | Rgs16 | −2.45 | 5.59E-16 |
| 10437687 | Litaf | −2.05 | 7.70E-16 |
| 10378286 | Itgae | −2.07 | 1.06E-15 |
| 10533729 | Vps37b | −2.72 | 3.56E-15 |
| 10358389 | Rgs2 | −2.83 | 3.70E-15 |
| 10454782 | Egr1 | −2.22 | 3.95E-15 |
| 10530145 | Tlr1 | 2.15 | 5.57E-15 |
Table 2.
| Cytokine and chemokine receptor activity | |||
|---|---|---|---|
| Brain CD103+/ Brain CD103− |
Spleen CD103−/ Brain CD103+ |
Spleen CD103−/ Brain CD103− |
|
| IL6ra | −0.5 | 1.6 | 1.1 |
| IL21r | 0.2 | −1.4 | −1.2 |
| Cx3Cr1 | −0.7 | 2.7 | 2 |
| IL2ra | 1.3 | −2.2 | −0.9 |
| IL12rb2 | −0.2 | −1.5 | −1.6 |
| IL4ra | 0.1 | −1.4 | −1.3 |
| Ccr7 | −1.3 | 2.1 | 0.8 |
| Cytokine and Chemokine activity | |||
|
Brain CD103+/ Brain CD103− |
Spleen CD103−/ Brain CD103+ |
Spleen CD103−/ Brain CD103− |
|
| Ccl4 | 0.6 | −2.2 | −1.6 |
| Xcl1 | −0.5 | −2.2 | −2.7 |
| Cxcl10 | 0.1 | −3.2 | −3.2 |
| Ccl3 | 0.4 | −2.4 | −2 |
| Tnfsf8 | −1.1 | 2.8 | 1.7 |
| Ccl9 | −0.7 | 2 | 1.3 |
| Tnfsf10 | 0.7 | −1.9 | −1.1 |
| Tnf | 0.7 | −2.4 | −1.7 |
| Regulation of transcription, DNA dependent | |||
|
Brain CD103+/ Brain CD103− |
Spleen CD103−/ Brain CD103+ |
Spleen CD103−/ Brain CD103− |
|
| Klf3 | −2.2 | 3.8 | 1.6 |
| Adam8 | 0.9 | −1.7 | −0.8 |
| Aff3 | −1.1 | 2.2 | 1 |
| Atf3 | 0.9 | −3.1 | −2.2 |
| Bach2 | −0.7 | 1.4 | 0.7 |
| Cd38 | 0.7 | −2 | −1.3 |
| Cdh1 | 0.7 | −2.9 | −2.2 |
| Dmrta1 | −1.2 | 1.4 | 0.3 |
| Egr1 | 0.1 | −2.3 | −2.2 |
| Eomes | −1.8 | 2.1 | 0.3 |
| Fosl2 | .01 | −2.2 | −2.2 |
| Irf4 | 0.7 | −3.4 | −2.7 |
| Jun | 0.2 | −2 | −1.7 |
| Lefl | −1.1 | 1.46 | 0.4 |
| Litaf | 0.4 | −2.5 | −2 |
| Nr4a3 | −0.3 | −3.1 | −3.5 |
| Tcf7 | −2.5 | 3.3 | 0.8 |
| Programmed cells death | |||
|
Brain CD103+/ Brain CD103− |
Spleen CD103−/ Brain CD103+ |
Spleen CD103−/ Brain CD103− |
|
| Bag3 | 0.2 | −2.3 | −2.1 |
| Cdh1 | 0.7 | −2.9 | −2.2 |
| Dapl1 | −1.1 | 3 | 1.8 |
| Gadd45b | 0.3 | −2 | −1.7 |
| Gramd4 | −0.8 | 1.9 | 1.1 |
| Traf4 | 0.2 | −1.2 | −1.1 |
| Tnfsf10 | 0.7 | −1.9 | −1.1 |
| Rhob | 0.3 | −3.2 | −2.9 |
| Pdcd1 | 0.3 | −2.3 | −2 |
| Gzmb | 0.5 | −2.2 | −1.7 |
| Negative regulation of immune system process | |||
|
Brain CD103+/ Brain CD103− |
Spleen CD103−/ Brain CD103+ |
Spleen CD103−/ Brain CD103− |
|
| Btla | −1.4 | 1.9 | −1.4 |
| Cd55 | −0.8 | 2.36 | 1.5 |
| Cd86 | 0.5 | −1.9 | −1.4 |
| Ctla4 | 0.6 | −2.87 | −2.2 |
| Dtx1 | −0.5 | 2 | 1.5 |
| Fcgr2b | −1.1 | 3.15 | 2.1 |
| Tigit | 0.3 | −2.7 | −2.3 |
Brain CD103+ T cells express genes characteristic of activated effector cells, including granzyme B and IL-2Ra at higher levels than spleen memory cells. Interestingly, 2 inhibitory receptors in the CD28:CTLA-4 family (22) (CTLA-4 and PD-1 ) are up-regulated in brain CD103+ Trm compared to brain CD103− and splenic CD103− memory T cells. Furthermore, transcripts encoding key regulators of T cell differentiation such as eomesodermin (eomes), t cell factor 1 (tcf1), lymphoid enhancer binding factor (lef1) and T-bet are down regulated in CD103+ Trm compared to the other cell subsets. Brain CD103+ Trm also express the highest levels amongst all the subsets of several interferon stimulated genes including, interferon induced transmembrane proteins 3 (IFITM3) in addition to Irf4 and Isg20.
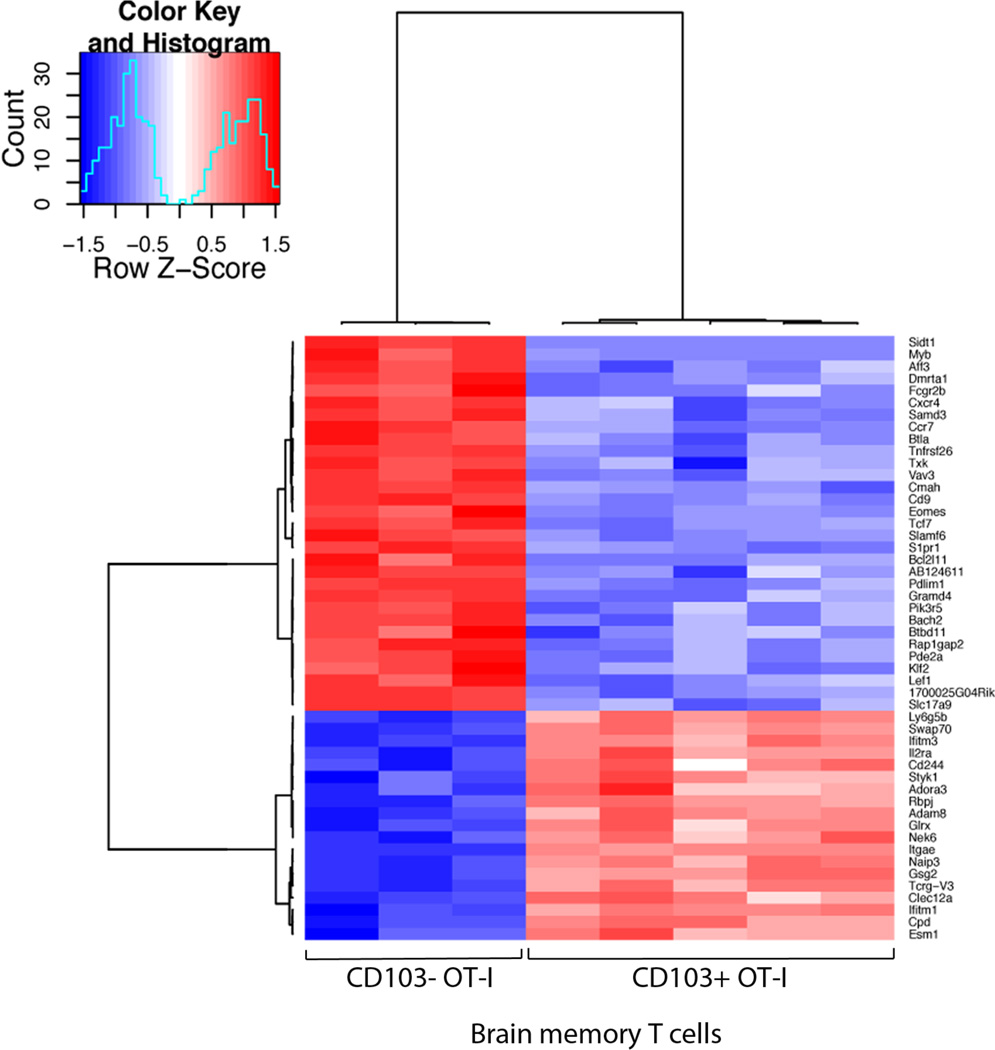
Both CD103+ and CD103− memory T cell populations isolated from the brain share similar expression of several chemokine and cytokine receptors. Figure 5 shows a heat map analysis of genes that are similarly expressed between the memory T cells isolated from the brain but which differ from the splenic memory T cells. In comparison to the splenic memory T cell pool the memory T cell isolated from the brain displayed up-regulation of several chemokine genes including, CCL3, CXCL10, CCL4 and downregulation of others including CX3Cr1 and CCL9. It is noteworthy both CXCL10 (23) and CCL3 (24) have been previously linked to the recruitment of T cells into the brain. There are also several migration molecules that are differentially expressed between the two brain populations (Fig 6). These include Sphingosine-1-Phosphate receptor 1 (S1P1R), CCR7 and CXCR4. Interestingly, S1P1R is involved in regulating T cell egress from lymph nodes. Expression of CD69 has been shown to suppress the function of the S1P1R and thereby reduces T cell migration out of the lymph node in response to a S1P gradient (25). CD69 expression has been documented on Trm isolated from different tissues (4, 13). Thus in addition to a role in circulating T cell egress, S1P1R also may be involved in tissue egress as suggested by earlier reports (26).
Figure 5. Heatmap of Brain memory OT-I signature genes.
Microarray analysis of Brain OT-I.CD103+, Brain OT-I CD103− and Spleen OT-I.CD103− memory T cells presented as a heatmap of the 50 genes that have the greatest difference in expression when comparing Brain (both CD103+ and CD103−) to Spleen populations (presented as normalized log2-intensities).
Figure 6. Gene expression profile comparing Brain CD103+ and Brain CD103− resident memory T cells.
Microarray analysis of Brain OT-I.CD103+ and Brain OT-I CD103− memory T cells presented as a heatmap of the 50 genes with the greatest difference in expression (presented as normalized log2-intensities)
These array data indicate that brain CD103+ Trm are a distinct memory T cell population displaying a unique molecular signature.
Discussion
Memory CD103+ CD8 T cells that persist within the brain following an acute virus infection are bona fide tissue resident memory T cells. We base this classification on several functional and phenotypical characteristics including i) they are a self sustaining T cell population that persists independently of the circulating memory T cell pool ii) they are resident within the tissue and do not recirculate iii) they express the integrin CD103; and iv) they do not survive well following dissociation from the tissue (8). However, to date there was no molecular genetic profile defining these peripherally deposited memory T cells. For the first time we determine the molecular signature of a Trm population.
Genetic profiling revealed that CD103+ Trm express low levels of the transcription factors tcf1 and eomes in comparison to the other memory T cell subsets analyzed. Both transcription factors have been demonstrated to be important in differentiation and persistence of memory CD8+ T cells (27, 28). Zhou et al., (27) show that tcf-1 deficiency impaired central memory T cell differentiation and tcf-1 deficient memory T cells were progressively lost over time. This memory T cell decay was most striking when analyzing memory T cells within the spleen and interestingly, when other organs were assessed (ie. liver and lung) the decay was not as pronounced. This implies that tcf1 may only be important in the persistence of circulating memory T cells and not memory T cells lodged within peripheral tissues. It is noteworthy that Zhou et al., (27) also performed a transcriptome analysis of Tcf1-deficient memory T cells. They show that Tcf1-deficient T cells express lower levels of eomes, CCR7 and SELL and higher levels of granzyme A and B compared to wild type memory T cells. Interestingly, brain CD103+ memory T cells express low levels of Tcf1 and the subsequent genetic profile associated with tcf1 deficiency including low levels of eomes, CCR7, SELL, and high levels of granzyme A and B. Furthermore, studies by Jeannet et al., demonstrate that tcf1 deficient memory T cells are impaired in their ability to expand upon secondary challenge (29). Brain CD103+ T cells fail to undergo recall expansion following dissociation from the tissue in which they reside and this lack of expansion may be associated with the tcf1 deficiency these cells display.
Our microarray analysis reveals that brain CD103+ Trm express high levels of several inhibitory receptors including CTLA-4 and PD-1 (30). It is interesting to speculate that expression of these inhibitory receptors by CD103+ Trm is in an effort to maintain peripheral tolerance. Trm are a highly sensitive, activated T cell population that resides within a variety of peripheral tissues. Expression of these inhibitory receptors may serve as a means to prevent this memory T cell population from accidentally being activated and unnecessarily attacking self. Brain CD103+ Trm are not exhausted or senescent and here we show that during a local bacterial infection these cells are able to provide substantial protection. Furthermore, it has been shown that brain CD103+ Trm can make cytokines and are cytotoxic (8). Hence, during a localized infection Trm must be released from this inhibitory state as these cells are clearly functional during a secondary bout of infection.
Brain CD103+ Trm express elevated levels of the antiviral protein IFITM3 (31). The constitutive expression of antiviral proteins may protect Trm from pathogens that they will undoubtedly encounter as a consequence of being situated within the peripheral tissue. We have show that Trm in the lung also maintain high levels of expression of IFITM3 and this expression appears to render these cells more resistant to influenza virus infection (Wakim et al., "submitted for publication"). Resistance to virus infection is an extremely beneficial attribute for a memory T cell population localized to common portals of pathogen entry to possess. This finding further supports the notion that Trm develop characteristics that allow them to better survive and function within their local environment. Furthermore the detection of IFITM3 in a lung Trm population (Wakim et al., "submitted for publication") implies that there are universal traits expressed among Trm in different organs. It remains to be determined whether Trm in other tissues express a similar signature to brain Trm or whether Trm in different tissue microenvironments express unique genetic profiles.
We have previously reported that the brain CD103+ Trm represent a T cell pool that has encountered cognate antigen locally within the brain microenvironment, most likely during the acute stages of the infection. This local antigen presentation promoted CD103 expression and the retention of these memory T cells within the brain tissue (8). This is in contrast to the CD103− memory T cell population which we suspect represent a T cell population that was recruited to the brain during the acute infection but fail to attain local antigen stimulation. These CD103− memory T cells do persist within the brain tissue, albeit poorly compared to their CD103+ expressing counterparts. Functional assays highlight further differences between these two cell populations with brain CD103− memory T cells undergoing better ex vivo recall expansion in comparison to brain CD103+ Trm (8). We have now identified several hundred genes that are differentially expressed between brain CD103+ and brain CD103− memory T cells. Hence this local ‘programming’ of Trm not only influences CD103 expression and thus T cell retention but initiates the expression of a series of genes that likely influence T cell survival, function and maintenance within the tissue microenvironment. These data highlights that there is vast complexity within the memory T cell pool and even memory T cells occupying the same niche have strikingly different genetic profiles.
Gene expression profile analysis indicates that CD103+ Trm are a distinct memory T cell population displaying a unique molecular signature which likely results in optimal survival and function within their local environment. In summary, these studies provide a base work with which to begin dissection of the factors that influence tissue resident memory T cell function, survival and maintenance. This knowledge will ultimately open the way for novel vaccination strategies that promote Trm formation.
Supplementary Material
Acknowledgments
This work benefited from data assembled by the ImmGen consortium (32
L.M.W is supported by an Overseas Biomedical Fellowship from the National Health and Medical Research Council of Australia. This work was supported by the Howard Hughes Medical Institute and National Institutes of Health Grant U19 AI083019 (to M.J.B.).
Footnotes
The datasets presented in this publication have been deposited in NCBI's Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE39152
References
- 1.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 2.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 3.Marshall DR, Turner SJ, Belz GT, Wingo S, Andreansky S, Sangster MY, Riberdy JM, Liu T, Tan M, Doherty PC. Measuring the diaspora for virus-specific CD8+ T cells. Proc Natl Acad Sci U S A. 2001;98:6313–6318. doi: 10.1073/pnas.101132698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- 5.Khanna KM, Bonneau RH, Kinchington PR, Hendricks RL. Herpes simplex virus-specific memory CD8+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity. 2003;18:593–603. doi: 10.1016/s1074-7613(03)00112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakim LM, Waithman J, van Rooijen N, Heath WR, Carbone FR. Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science. 2008;319:198–202. doi: 10.1126/science.1151869. [DOI] [PubMed] [Google Scholar]
- 7.Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, Akondy R, Wang J, Casey KA, Barber DL, Kawamura KS, Fraser KA, Webby RJ, Brinkmann V, Butcher EC, Newell KA, Ahmed R. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207:553–564. doi: 10.1084/jem.20090858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakim LM, Woodward-Davis A, Bevan MJ. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc Natl Acad Sci U S A. 2010;107:17872–17879. doi: 10.1073/pnas.1010201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purwar R, Campbell J, Murphy G, Richards WG, Clark RA, Kupper TS. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS One. 2011;6:e16245. doi: 10.1371/journal.pone.0016245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teijaro JR, Turner D, Pham Q, Wherry EJ, Lefrancois L, Farber DL. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J Immunol. 2011;187:5510–5514. doi: 10.4049/jimmunol.1102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmann M, Pircher H. E-cadherin promotes accumulation of a unique memory CD8 T-cell population in murine salivary glands. Proc Natl Acad Sci U S A. 2011;108:16741–16746. doi: 10.1073/pnas.1107200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackay LK, Stock AT, Ma JZ, Jones CM, Kent SJ, Mueller SN, Heath WR, Carbone FR, Gebhardt T. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc Natl Acad Sci U S A. 2012;109(18):7037–7042. doi: 10.1073/pnas.1202288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ariotti S, Haanen JB, Schumacher TN. Behavior and Function of Tissue-Resident Memory T cells. Adv Immunol. 2012;114:203–216. doi: 10.1016/B978-0-12-396548-6.00008-1. [DOI] [PubMed] [Google Scholar]
- 14.Turner MJ, Jellison ER, Lingenheld EG, Puddington L, Lefrancois L. Avidity maturation of memory CD8 T cells is limited by self-antigen expression. J Exp Med. 2008;205:1859–1868. doi: 10.1084/jem.20072390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sacks JA, Bevan MJ. TRAIL deficiency does not rescue impaired CD8+ T cell memory generated in the absence of CD4+ T cell help. J Immunol. 2008;180:4570–4576. doi: 10.4049/jimmunol.180.7.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smyth GK. Limma: linear models for microarray data. New York: Springer; 2005. [Google Scholar]
- 18.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 20.Wojciechowski S, Tripathi P, Bourdeau T, Acero L, Grimes HL, Katz JD, Finkelman FD, Hildeman DA. Bim/Bcl-2 balance is critical for maintaining naive and memory T cell homeostasis. J Exp Med. 2007;204:1665–1675. doi: 10.1084/jem.20070618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun JC, Williams MA, Bevan MJ. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat Immunol. 2004;5:927–933. doi: 10.1038/ni1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- 23.Klein RS, Lin E, Zhang B, Luster AD, Tollett J, Samuel MA, Engle M, Diamond MS. Neuronal CXCL10 directs CD8+ T-cell recruitment and control of West Nile virus encephalitis. J Virol. 2005;79:11457–11466. doi: 10.1128/JVI.79.17.11457-11466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trifilo MJ, Bergmann CC, Kuziel WA, Lane TE. CC chemokine ligand 3 (CCL3) regulates CD8(+)-T-cell effector function and migration following viral infection. J Virol. 2003;77:4004–4014. doi: 10.1128/JVI.77.7.4004-4014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, Cyster JG, Matloubian M. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 26.Ledgerwood LG, Lal G, Zhang N, Garin A, Esses SJ, Ginhoux F, Merad M, Peche H, Lira SA, Ding Y, Yang Y, He X, Schuchman EH, Allende ML, Ochando JC, Bromberg JS. The sphingosine 1-phosphate receptor 1 causes tissue retention by inhibiting the entry of peripheral tissue T lymphocytes into afferent lymphatics. Nat Immunol. 2008;9:42–53. doi: 10.1038/ni1534. [DOI] [PubMed] [Google Scholar]
- 27.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 29.Jeannet G, Boudousquie C, Gardiol N, Kang J, Huelsken J, Held W. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci U S A. 2010;107:9777–9782. doi: 10.1073/pnas.0914127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe N, Gavrieli M, Sedy JR, Yang J, Fallarino F, Loftin SK, Hurchla MA, Zimmerman N, Sim J, Zang X, Murphy TL, Russell JH, Allison JP, Murphy KM. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat Immunol. 2003;4:670–679. doi: 10.1038/ni944. [DOI] [PubMed] [Google Scholar]
- 31.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.