Abstract
Short interfering RNAs (siRNAs) are promising drug candidates for a wide range of targets including those previously considered “undruggable”. However, properties associated with the native RNA structure limit drug development and chemical modifications are necessary. Here we describe the structure-guided discovery of functional modifications for the guide strand 5’ end using computational screening with the high resolution structure of human Ago2, the key nuclease on the RNA interference pathway. Our results indicate the guide strand 5’-end nucleotide need not engage in Watson-Crick (W/C) H-bonding but must fit the general shape of the 5’-end binding site in MID/PIWI domains of hAgo2 for efficient knockdown. 1,2,3-Triazol-4-yl bases formed from the CuAAC reaction of azides and 1-ethynylribose, which is readily incorporated into RNA via the phosphoramidite, perform well at the guide strand 5’-end. In contrast, purine derivatives with modified Hoogsteen faces or N2 substituents are poor choices for 5’-end modifications. Finally, we identified a 1,2,3-triazol-4-yl base incapable of W/C H-bonding that performs well at guide strand position 12, where base pairing to target was expected to be important. This work expands the repertoire of functional nucleotide analogs for siRNAs.
Introduction
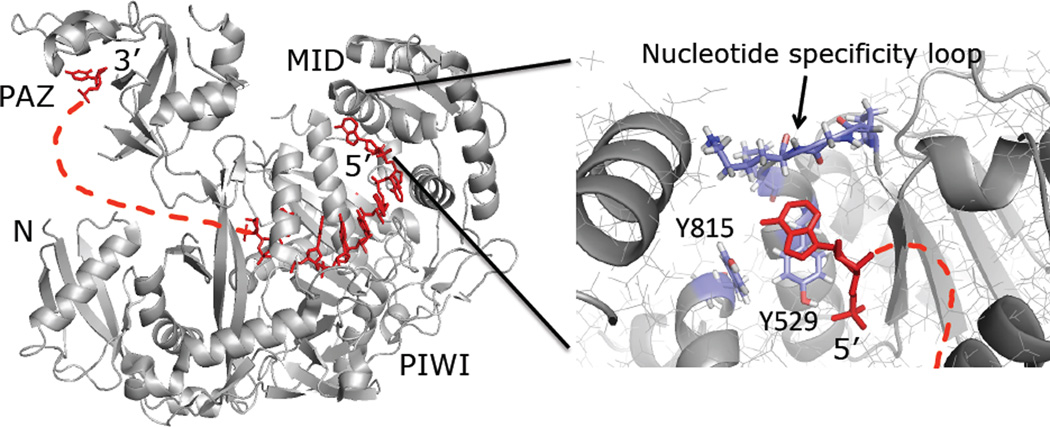
Short interfering RNA (siRNA)-triggered gene knockdown via the RNA interference (RNAi) pathway has revolutionized the study of gene function and spurred the development of new oligonucleotide-based therapeutics.1,2 However, the natural RNA structure poses limits to drug development and modifications are necessary to improve properties (e.g. increase potency, reduce nuclease sensitivity, facilitate delivery, reduce off-target effects, etc.).3–6 Thus, it is important to identify modifications at specific positions in the siRNA strands where changes in the natural RNA structure improve siRNA performance.7,8 The key step in the RNAi pathway is cleavage of target RNA by the RNaseH-like enzyme Ago2 in complex with the siRNA guide strand. Recent mechanistic9 and structural studies10,11 have shown that the four domains of the human Ago2 protein (N-terminal, PAZ, MID and PIWI) each have a distinct role in guide strand binding and the target RNA cleavage reaction (Figure 1). For instance, the hAgo2 PAZ domain binds the 3’ end of the guide strand with direct contacts made to the last three nucleotides.11,12 On the other hand, nucleotides2–6 of the guide strand are bound to the PIWI domain in an A-form helical geometry with their Watson-Crick (W/C) edges available for binding target RNA. The PIWI domain is also responsible for the target RNA cleavage reaction. Importantly, the 5’-most nucleotide (i.e. guide nucleotide 1) is tucked into a MID domain pocket where extensive contacts are made to both the 5’-phosphate and the nucleobase (Figure 1). Interactions with the edge of the base via a rigid loop in the protein (i.e. the nucleotide specificity loop) explain the MID domain’s preference for binding pU and pA over pG and pC (Figure 1).13 This nucleotide does not W/C pair with target RNA, but modifications to its structure that reduce MID domain binding affinity reduce siRNA efficacy.14 For instance, an abasic site at this location results in weaker MID domain binding and lower target cleavage activity.9 This is most likely because the nucleobase at guide position 1 stacks onto Tyr 529 of the MID domain and an abasic residue lacks this interaction (Figure 1). Since this guide strand nucleotide is not involved in target nucleotide recognition yet influences siRNA activity, modifications at this position have the potential to enhance RNAi potency and could be generally useful for all siRNAs. Furthermore, two recent reports of crystal structures of human Ago2-guide strand complexes clearly define 5’-end/hAgo2 interactions and suggest a structure-guided approach may be possible for the discovery of functional 5’ end modifications.10,11 We report here a combined computational screening and synthesis strategy to identify new 5’-end modifications for siRNA guide strands. We used the structure of a hAgo2/guide strand complex and computational screening of nucleobase analogs accessible by copper-catalyzed azide/alkyne cycloaddition (CuAAC) reactions. The screening identified several high scoring candidates for 5’-end modifiers including triazole derivatives that appeared capable of accessing a cleft in hAgo2 adjacent to the 5’ nucleotide-binding site. Synthesis and testing of a group of guide strand 5’-end modifications showed a good correlation between siRNA potency and predicted hAgo2 interactions.
Figure 1.
Results and Discussion
Computational screening for 5’ end modifiers
To access the computationally predicted hAgo2 binding affinity of guide strand 5’ terminal nucleobases, we constructed a library of synthetically accessible structures (see below), expanded this library to conformers of each molecule, then docked the conformational library into a rigid receptor created from the published crystal structure of hAgo2 bound to guide strand RNA (4EI1).10 This was achieved using the OpenEye Suite of programs (see Experimental Section).15 The receptor was created in such a way as to ensure that even the largest analogs investigated could be accommodated and with constraints imposed on the 5’-phosphate group and the 3’-hydroxyl to simulate how the 5’-end nucleotide of the guide strand is bound by hAgo2. Each base was assigned a binding score measuring the quality of the fit of the analog into the binding pocket (e.g. π stacking, sterics, complementarity of H-bonding donors and acceptors, etc). These scores were generated from the utilization of several different computational docking scoring methods as well as analysis of the top pose overlay with the guide strand 5’-end adenosine from the hAgo2-RNA crystal structure. A full description of the docking and scoring method can be found in the Experimental Section.
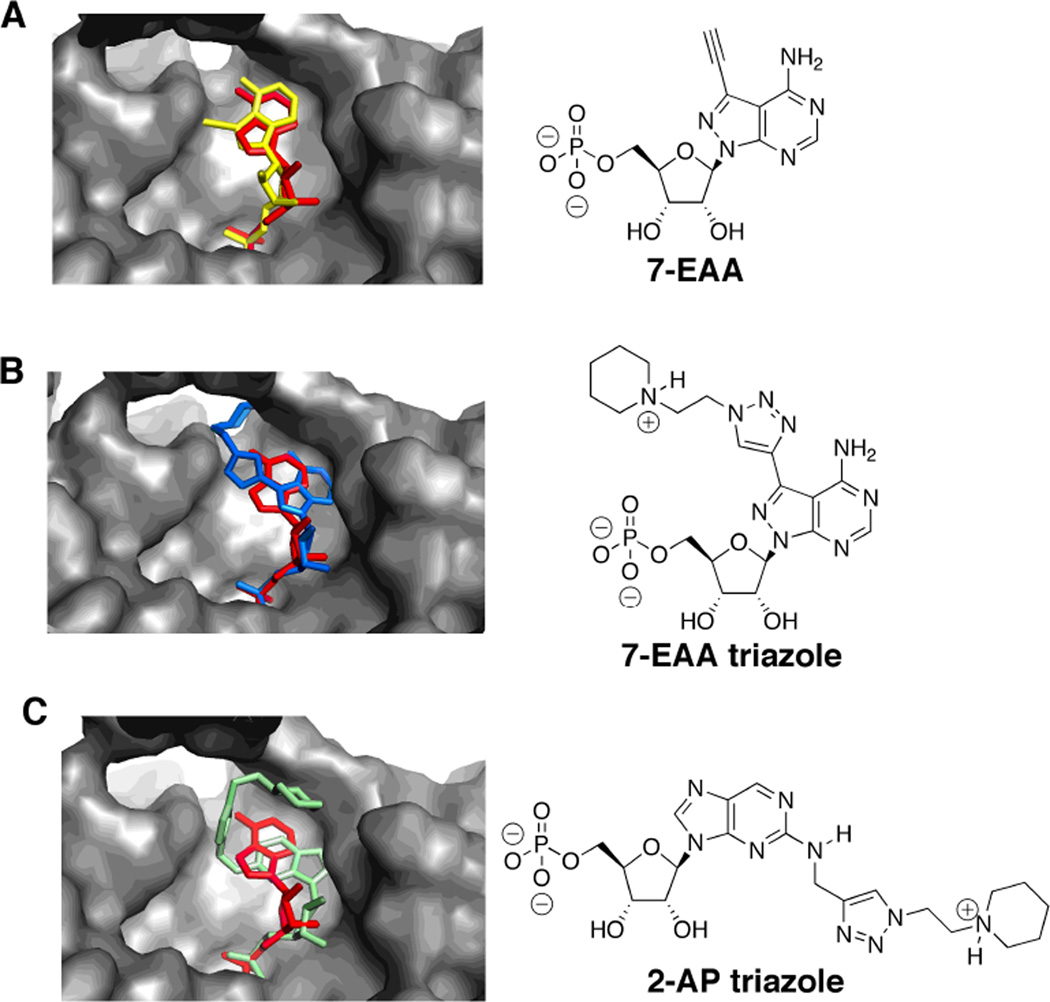
Our lab has prepared adenosine analogs that direct substituents into either the minor8,16,17 or major groove18 when present in duplex RNA. Since adenine (along with uracil) is a preferred natural nucleobase ligand for the Ago2 MID domain, we initially screened a library of adenosine analogs for good hAgo2-binding scores as described above.13 For instance, 7-ethynyl-8-aza-7-deazaadenosine (7-EAA) is readily incorporated into RNA via the phosphoramidite and serves as a precursor to 7-triazole derivatives by CuAAC reactions (Figure 2).or major groove18 However, 7-EAA was predicted to fit poorly into the 5’-nucleotide binding pocket of hAgo2 (Figure 2) and did not score well relative to adenosine in the computational screening (Table 1). Indeed, the lowest-energy binding pose for 7-EAA has the ethyne clashing with the aromatic ring of Tyr 815 of the adjacent PIWI domain (Figure 1, Figure 2A). Triazoles formed from reaction of this analog with different azides were also screened (57 total) (see Supplementary Figure 1 and Supplementary Table 1) and generally had poor scores and inferior fits in the binding site compared to adenosine (see 7-EAA triazole in Figure 2B, Table 1).
Figure 2.
Predicted hAgo2 binding modes for purine analogs positioned at a guide strand 5’ end. Each analog is shown in overlay with adenosine (red) in the guide strand 5’-end binding site of hAgo2 shown as a gray surface.10 (A) 7-EAA (yellow), (B) 7-EAA triazole (blue), (C) 2-AP triazole (green).
Table 1.
| Predicted hAgo2 bindinga | siRNA activityb | |
|---|---|---|
| Adenosine | 1.0 | ++ |
| 7-EAA | 8.8 | + |
| 7-EAA triazole | 9.2 | + |
| 2-AP-triazole | 9.5 | + |
Scores normalized from1–10 from several scoring methods (see Experimental Section). A lower number represents better-predicted binding to guide strand 5’-end.
Experimental siRNA activity determined at 100 pM. +++ = <10% luciferase activity remaining after knockdown; ++ = 10–40%; + = 41–70%; - = >70% (Figure 3).
2-Aminopurine analogs can be used in duplex RNA as adenosine replacements that project substituents into the minor groove.8,16 Our earlier work showed that an N-ethylpiperidine triazole derived from 2-propargylaminopurine (i.e. 2-AP-triazole) is well tolerated at position 14 of the guide strand but not at position 2 (Figure 2C).8 When evaluated computationally for binding into the hAgo2 guide strand 5’-end binding site, this structure also did not fit well and scored poorly relative to adenosine (Figure 2C) (Table 1).
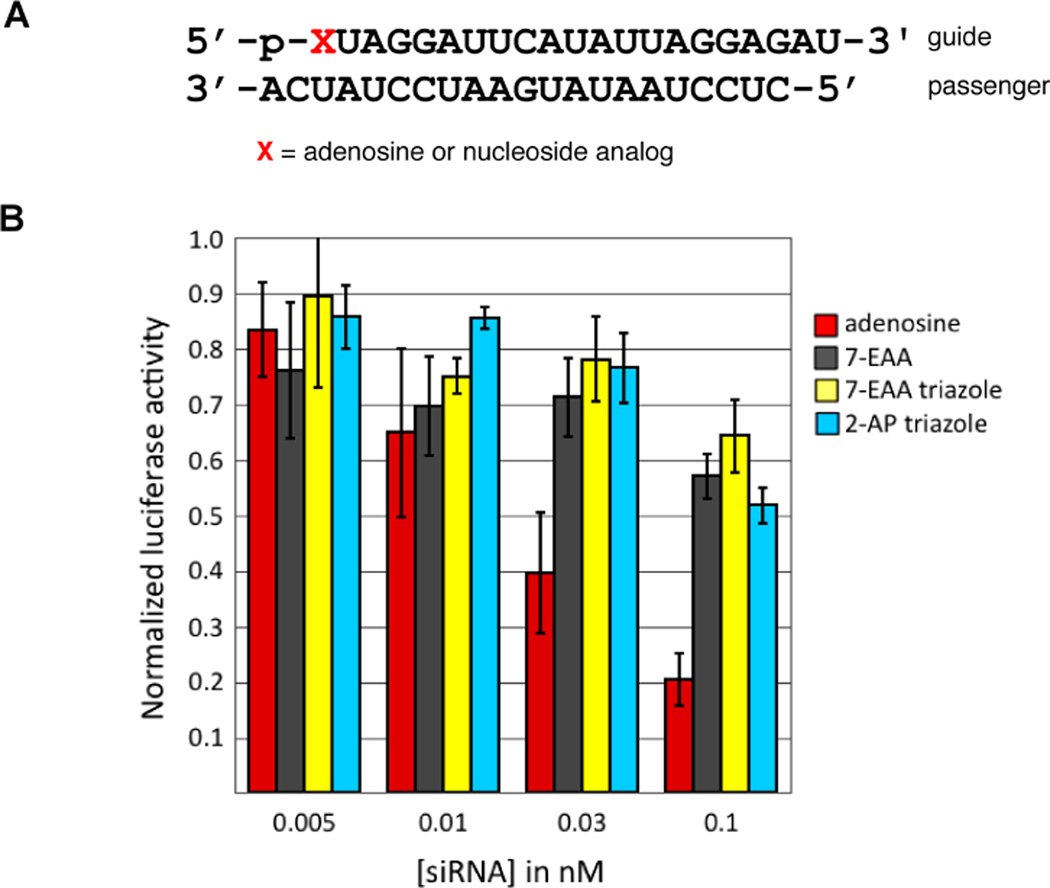
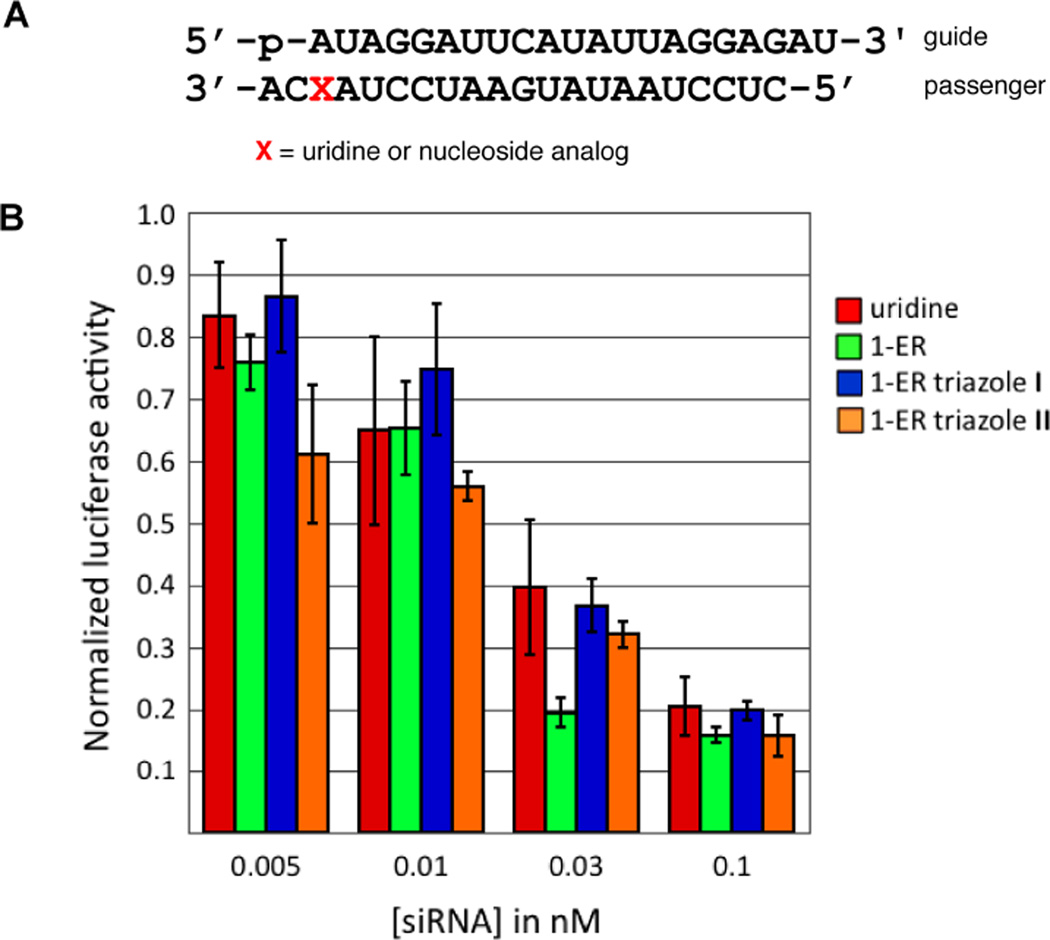
These computational screening results suggested that the adenosine analogs would likely inhibit RNAi activity when positioned at the guide strand 5’ end. To determine the effect of the 2-AP triazole at this position, it was incorporated into an siRNA that targets the PIK3CB sequence present on a luciferase reporter plasmid and activity for the modified siRNA was measured in HeLa cells (Figure 3). Consistent with the computational results, the 2-AP-triazole inhibited RNAi when present at guide position 1 (Figure 3). The 7-EAA and the 7-EAA-triazole have been tested for their impact on RNAi activity when positioned at the PIK3CB guide strand 5’-end and were re-evaluated here for direct comparison to the 2-AP triazole.18 Again consistent with the predictions described above, both of these modifications inhibit activity relative to the parent siRNA with adenosine at position 1 of the guide strand (Figure 3).
Figure 3.
(A) Sequence of siRNA used in this study. X indicates guide strand position 1. All siRNAs were prepared with a 5’-phosphorylated guide strand (p). (B) Knockdown activity of siRNAs with adenosine, 7-EAA, 7-EAA triazole or 2-AP triazole at the guide strand 5’ end.18 Activity is reported as the ratio of Renilla/firefly luciferase signal at various concentrations of transfected siRNA in HeLa cells.
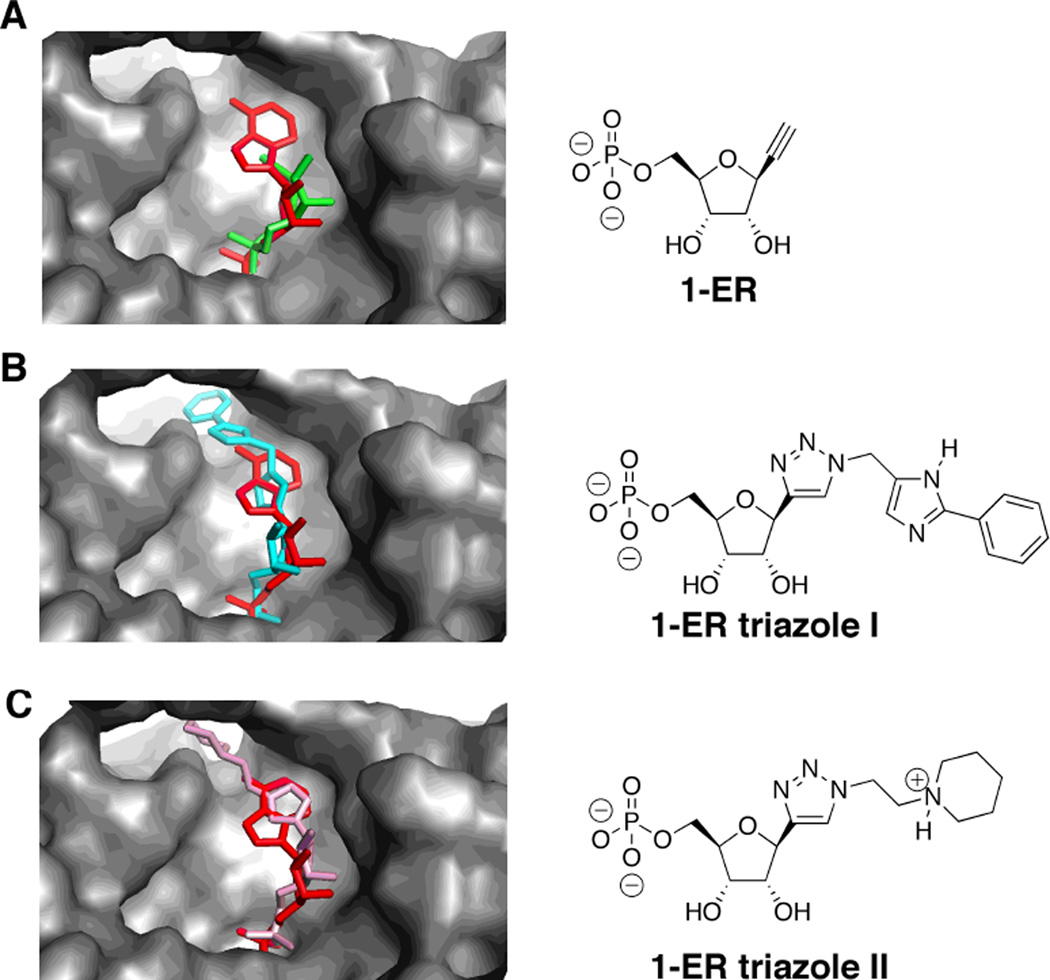
While these results suggest computational screening is useful for predicting the effect of different 5’-end modifications, our previously described adenosine derivatives were shown to be poor choices as nucleobase replacements for the siRNA guide strand 5’ end. Others have shown efficient knockdown with an siRNA bearing a nucleobase analog that is a good mimic of the shape of uridine at this position.19 However, some loss of activity was reported with a size-expanded adenosine analog at the 5’ end of the guide strand.20 Considering these observations and the size and shape of the hAgo2 guide strand 5’-end binding pocket, we next evaluated analogs that replace the natural base entirely. We chose to focus on 1,2,3-triazol-4-yl bases since these could be readily prepared from 1-ethynylribose (1-ER) containing precursor RNA and different azides (Figure 4). Such analogs could stack with Tyr 529 in the Ago2 5’-end binding pocket but cannot participate in W/C H-bonding. However, the lack of W/C H-bonding was not considered to be a significant concern for guide strand position 1 because this nucleotide does not directly contact the target RNA strand. We carried out computational screening on 58 different 1,2,3-triazol-4-yl bases, along with the parent 1-ethynylribose, and found compounds that scored well and significantly better than the adenosine derivatives described above (e.g. 1-ER triazoles I and II) (Figure 4)(Table 2) (Supplementary Figure 1) (Supplementary Table 1). The apparent ability of 1-ER and derivative triazoles to fit the hAgo2 guide 5’-end binding site stimulated our synthesis of the required phosphoramidite.
Figure 4.
Predicted hAgo2-binding modes for 1-ER derivatives positioned at a guide strand 5’ end. Each analog is shown in overlay with adenosine (red) in the guide strand 5’-end binding site of hAgo2 shown as a gray surface.10 (A) 1-ER (green), (B) 1-ER triazole I (blue), (C) 1-ER triazole II (pink).
Table 2.
| Predicted hAgo2 bindinga | siRNA activityb | |
|---|---|---|
| adenosine | 1.0 | ++ |
| 1-ER | 2.1 | ++ |
| 1-ER triazole I | 1.2 | +++ |
| 1-ER triazole II | 1.3 | +++ |
Scores normalized from 1–10 from several scoring methods (see Experimental Section). A lower number represents better-predicted binding to guide strand 5’-end.
Experimental siRNA activity determined at 100 pM. +++ = <10% luciferase activity remaining after knockdown; ++ = 10–40%; + = 41–70%; - = >70% (Figure 5).
Synthesis of 1-ER phosphoramidite and generation of modified guide strands
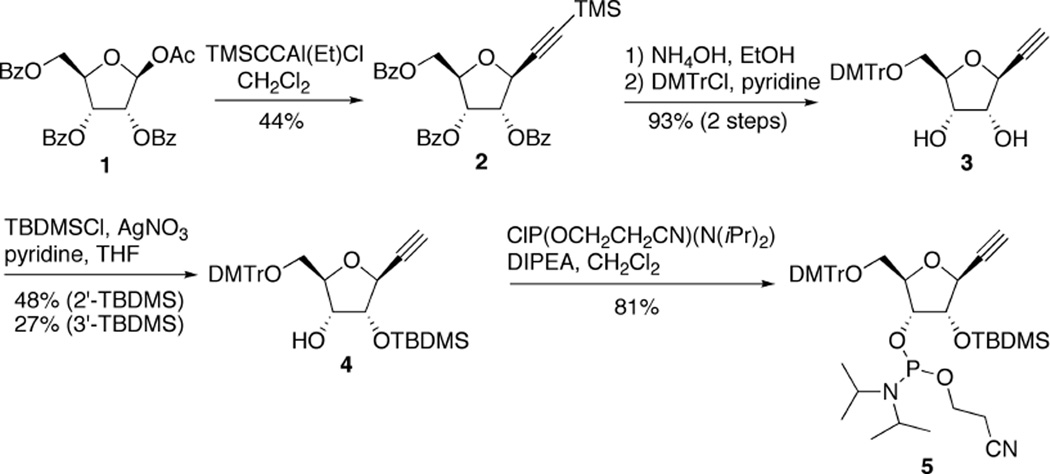
The DNA type 1-ethynyl 2-deoxyribose phosphoramidite has been synthesized to investigate the effects of various triazole derivatives on the stability of DNA duplexes and triplex-formation.21,22 Here we prepared the RNA type 1-ethynyl ribose (1-ER) phosphoramidite to incorporate it into siRNA strands. The skeleton of the β-anomer of 1-ER was synthesized from commercially available sugar 1 using ethyl[trimethylsilyl(ethynyl)]aluminum chloride23,24 according to a literature report25(Scheme 1). Only one isomer was observed in this reaction. The trimethylsilyl and benzoyl groups of 2 were removed with NH4OH/EtOH, and the primary hydroxyl was protected with a dimethoxytrityl group to give compound 3. The configuration was determined from 1H NMR NOESY data of compound 3. Since an NOE between H-1 and H-4 was observed, the isomer was assigned as the β-anomer. Phosphoramidite building block 5 was synthesized by normal tert-butyldimethylsilyl protection at the 2-position to give 4 and subsequent phosphitylation.
Scheme 1.
The 1-ER phosphoramidite building block 1 was used to incorporate the analog into the guide strand for an siRNA with PIK3CB targeting sequence (see Figure 3A). The RNA synthesis and cleavage/deprotection were performed using standard methods. The RNAs were purified by denaturing polyacrylamide gel electrophoresis (PAGE) and confirmed by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) analysis. To prepare high scoring 1,2,3-triazol-4-yl bases, two different azides were allowed to react with 1-ethynyl ribose modified RNA under CuAAC conditions.26–29 Products were generated by incubating in HEPES buffer (pH 7) with the single-stranded RNA, tris-[1-(3-hydroxypropyl)-1H-[1,2,3]triazol-4-yl)methyl]amine (THPTA) ligand,30 CuSO4, sodium ascorbate and azide at ambient temperature. The reaction was completed in 2–3 h, and the products were purified from the reaction mixture by denaturing PAGE and confirmed by MALDI-MS analysis.
RNA interference with siRNAs bearing 1-ER or 1,2,3-triazol-4-yl bases
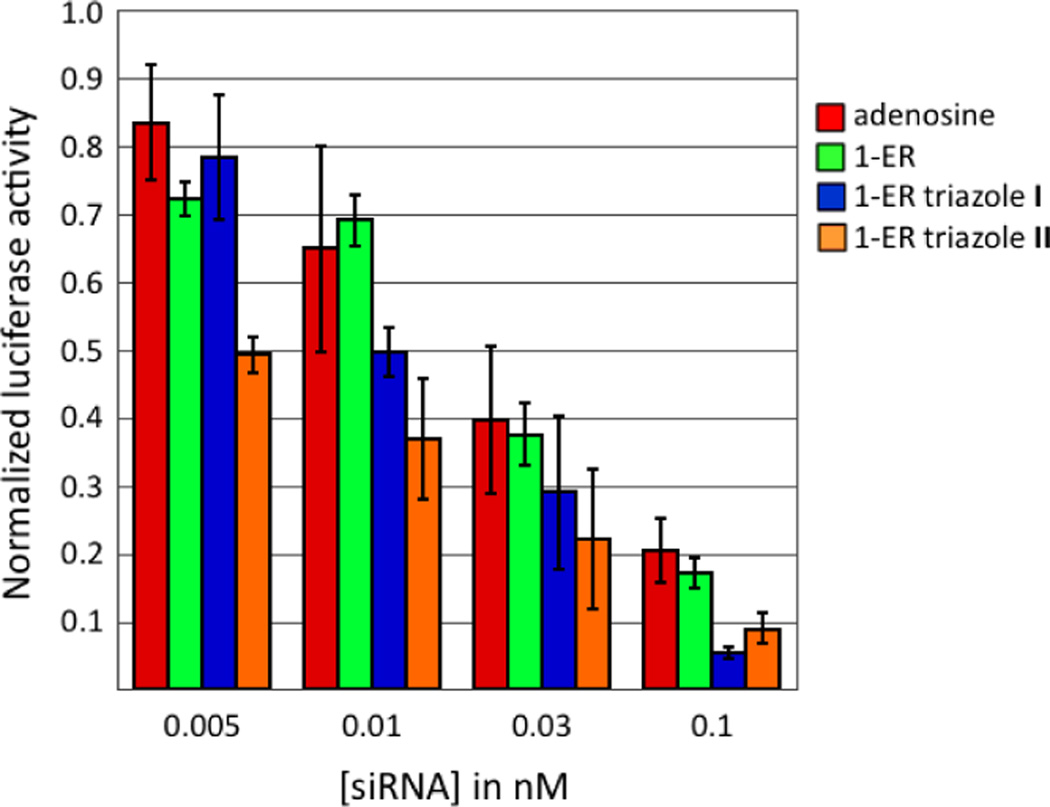
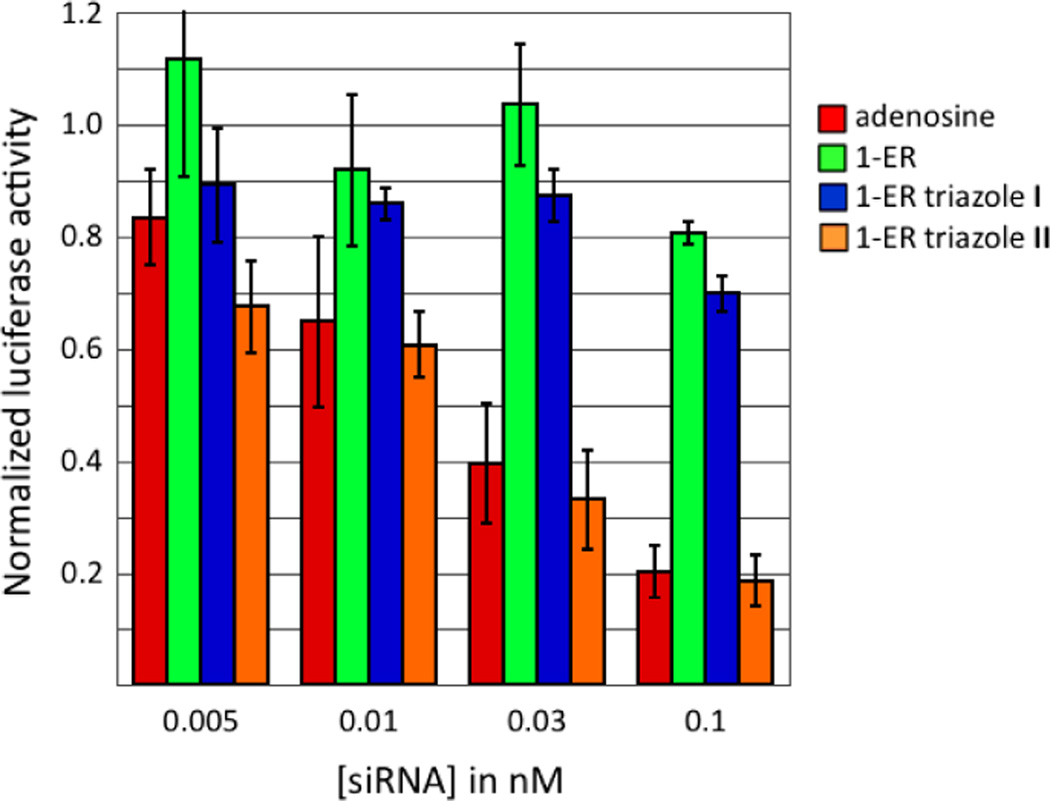
The guide strand of the PIK3CB siRNA was modified with either 1-ER, 1-ER triazole I, or 1-ER triazole II at position 1 (Figure 4). RNAi activity for the modified siRNAs was measured in HeLa cells as described above. As a test of the effect of modification position on activity, we also incorporated these analogs at guide position 12. Less is known about how the base of position 12 interacts with hAgo2 since electron density is lacking for this nucleotide in recently reported hAgo2/guide RNA crystal structures.10,11 However, position 12 of the guide strand lies in the middle of the siRNA where pairing to the passenger strand is important for siRNA stability. Furthermore, pairing of target RNA with guide strand nucleotides 2–8 and 12–17 is believed to define the most efficient sites for RNAi, so base structure at guide position 12 would be expected to be important for activity.31 For each modified siRNA, we carried out a four-point concentration profile in the RNAi assay (Figure 5, Figure 6). Importantly, replacement of the adenine base at position 1 with 1-ER, 1-ER triazole I or 1-ER triazole II was well tolerated and resulted in siRNAs with potency similar to (1-ER) or exceeding that (1-ER triazoles I and II) of the unmodified duplex (Figure 5). This is in sharp contrast to the purine analogs described above and in good agreement with the computational screening results (Table 2). The docking suggests that 1-ER triazoles I and II are capable of occupying a cleft in the protein adjacent to the guide strand 5’-end binding site and between K525 and P526 of the MID domain and Y815 and L817 of the PIWI domain (Figure 4B,C; Supplementary Figure 2). Crystallization of the complex of hAgo2 bound to a 5’-end modified guide strand will be necessary to test this hypothesis. The activity effects observed are clearly guide strand position-dependent since replacement of the adenine base at guide position 12 with either the 1-ER modification or 1-ER triazole I inhibited knockdown (Figure 6).
Figure 5.
Knockdown activity of siRNAs with adenosine, 1-ER, 1-ER triazole I or 1-ER triazole II at the guide strand 5’ end. Activity is reported as the ratio of Renilla/firefly luciferase signal at various concentrations of transfected siRNA in HeLa cells.
Figure 6.
Knockdown activity of siRNAs with adenosine, 1-ER, 1-ER triazole I or 1-ER triazole II at guide strand position 12. Activity is reported as the ratio of Renilla/firefly luciferase signal at various concentrations of transfected siRNA in HeLa cells.
While the novel modifications may be affecting siRNA potency by modulating guide stand binding to hAgo2 as designed, this is not the only effect that could be responsible for the observed results. The nucleoside analogs could also facilitate duplex unwinding and enhance loading of the desired guide strand into hAgo2.32 This is particularly true for analogs placed near the guide strand 5’ end that are duplex destabilizing (as is the case for 1-ER and the triazoles derived from it, see below, Table 3).32 If this were the source of the effects on knockdown efficacy, one would expect to see similar results if the analog were placed at the same end of the duplex but in the opposite strand. We tested this by preparing siRNAs with 1-ER, 1-ER triazole I and 1-ER triazole II at position 19 of the passenger strand for the siRNA targeting the PIK3CB sequence (Figure 7 A). Interestingly, for the siRNAs with 1-ER triazoles I and II at passenger strand position 19, the activity observed is indistinguishable from that of the unmodified duplex (with uridine at this position) (Figure 7B). Therefore the enhanced knockdown observed when these analogs are at the 5’ end of the guide strand appears not to be simply a result of localized duplex destabilization. For the 1-ER-modified passenger strand, more effective knockdown compared to unmodified siRNA is observed at 30 pM so this analog could also be altering strand bias during loading of hAgo2.
Table 3.
Thermal melting temperatures of modified siRNAs
| Guide position 12 modification | ||||
| unmodified | 1-ER | 1-ER triazole I | 1-ER triazole II | |
| siRNA TM, °C | 64.7 ± 0.4 | 56.1 ± 0.1 | 57.0 ± 0.1 | 56.2 ± 0.3 |
Figure 7.
Knockdown activity of siRNAs with uridine, 1-ER, 1-ER triazole I or 1-ER triazole II at passenger strand position 19. A) Sequence of siRNA tested showing site of passenger strand modification B) Activity is reported as the ratio of Renilla/firefly luciferase signal at various concentrations of transfected siRNA in HeLa cells.
We also wished to evaluate the effect these modifications have on the stability of the siRNA to nucleases present in serum since an increase in serum stability could result in increased RNAi activity.33 However, we found that 1-ER, 1-ER triazole I and 1-ER triazole II at the guide strand 5’ end caused a slight decrease in serum stability when compared to unmodified siRNA (Supplementary Figure 4). Thus, enhanced RNAi activity arising from these modifications is unlikely to be the result of increasing resistance to nucleases present in serum.
Surprisingly, efficient knockdown is observed with 1-ER triazole II at position 12 (Figure 6). This could be explained by a stabilizing effect on RNA-RNA interactions of a positive charge on the piperidine present in the 1-ER triazole II structure (Figure 4C). We measured thermal melting temperatures (TM’s) for the modified siRNAs to determine the effect on RNA duplex stability for each of the analogs introduced at guide position 12 (Table 3). All of the modifications made, including 1-ER triazole II, destabilized the siRNA duplex to the same extent. Whereas the unmodified siRNA had a TM = 64.7 ± 0.4 °C, each of the modified siRNAs had TM values of 56–57 °C. Therefore, the ability of 1-ER triazole II to function as an effective base replacement at guide position 12 is not due simply to its positive charge stabilizing RNA-RNA interactions. Earlier work showed that 7-EAA and 7-EAA triazole (Figure 2) are effective adenosine replacements at guide position 12.18 In addition, Li et al. reported that difluorotoluene replacement for uracil at this position resulted in minimal loss in siRNA activity.34 However, Hernandez, Peterson and Kool found that size-expanded nucleobases near the center of an siRNA guide strand (e.g. at positions 9,10,11 and 14) caused a substantial reduction in activity.20 These results, along with our observation that 1-ER or 1-ER triazole I at guide position 12 inhibits RNAi, indicate a clear sensitivity to base structure at this poston wth some nuceobase anaogs but not others functioning we The positive effects of certain nucleobase analogs at this position, such as 1-ER triazole II, appear to be largely independent of their effects on duplex stability and likely arise from beneficial interaction with components of the RNAi pathway, perhaps hAgo2 itself. Further study will be necessary to fully define their origin, however.
Conclusion
In summary, we developed and validated a method using computational screening for identitying functional 5’-end modifiers for siRNA guide strands. 5’-End modifiers need not be capable of W/C H-bonding but should fit the general shape of the 5’-end binding site in MID/PIWI domains of hAgo2. We devised an efficient synthesis of the 1-ER phosphoramidite and generated RNA bearing this modification. 1-ER serves as a precursor to 1,2,3-triazol-4-yl bases via CuAAC reactions with azides and specific 1,2,3-triazol-4-yl bases perform well at position 1 of an siRNA guide strand. This stands in contrast to purine derivatives with modified Hoogsteen faces or N2 substituents that inhibit RNAi when placed at the guide strand 5’ end. We also reported the unexpected discovery of a non-W/C H-bonding analog that performs well at position 12 where H-bonding to target RNA was expected to be important. These studies expand the repertoire of nucleoside analogs useful for siRNAs. In addition, this work sets the stage for future efforts to test our hAgo2 binding hypothesis for 1,2,3-triazol-4-yl bases with high resolution structures, to evaluate the impact of these novel bases on siRNA off-target effects and their duration of action as well as further computational screening to identify additional high performing guide strand 5’-end modifications.
Experimental Section
All reagents were purchased from commercial sources (Sigma-Aldrich or Fisher Scientific) and were used without further purification unless otherwise stated. Reactions were carried out under an atmosphere of dry argon. 1H, 13C, and 31P NMR spectra were recorded with Varian VNMRS600 or Varian Mercury 300 spectrometers. High-resolution ESI mass spectra were obtained at the University of California, Davis mass spectrometry facility, on an Orbitrap FTMS instrument. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectra of oligonucleotides were measured in the positive ion mode, using 6-aza-2-thiothymine as a matrix.
Computational Screening
The overall process of computational screening involved five steps: 1. Library creation, 2. Conformer generation, 3. Receptor creation, 4. Receptor-library docking, and 5. Analysis. All programs mentioned are part of the OpenEye Suite of programs.15 As the approach is aimed at high throughput, molecular dynamics simulations are not part of the docking protocol used.
Library creation: First, a ligand library was created, consisting of molecules thought to be easily accessible from the 7-EAA, 2-AP or 1-ER structures using commercially accessible precursors from Sigma-Aldrich. This was done to narrow our search to molecules that, if thought to be good candidates after screening, could be quickly generated and incorporated into an oligomer for testing.
Conformer generation: Once this library was compiled, the Omega program was used to generate a list of conformers of each molecule, ranging from <20 for smaller, rigid molecules to >5000 for large, flexible molecules. This required expanding the number of maximum conformers from the program default of 200.
Receptor creation: Starting from published crystallographic data (pdb file 4EI1) 10, several similar receptors of various sizes were created, which, along with heavy atom contour surfaces, define how close to the protein surface the heavy atoms in screened ligands can dock. This was accomplished with make_receptor. The receptors were created so as to ensure that even the largest molecules in our computational screen would be accommodated in their most extended conformations. Once the heavy atom contours were created, several constraints were applied, to ensure that docking was forced to place the ribose subunit where it would be if it were at the 5 end of a strand of bound RNA. A constraint was imposed on the ribose 2 oxygen linkage (modeled in our library as a free hydroxyl). An additional constraint was applied to the ribose 4 oxygen, and to the phosphate (see Supplementary Figure 3).
Receptor-library docking: The library of substrate conformers from step 2 was then docked into the receptors from step 3 using FRED, with several scoring functions available in OpenEye. These scoring methods each have strengths and weaknesses, depending on the system of interest. They utilize various proprietary algorithms that model shape, protein-ligand hydrogen-bonding, implicit solvent-ligand hydrogen bonding, and polarity/electrostatics (see the OpenEye website for additional information). Each of these factors are used to give a conformer a score for a specific pose (within the constraints imposed) and once this has been done for all valid poses of all conformers, an overall score and ranking is generated as output, along with atomic coordinates for the corresponding poses.
Analysis: Due to the fact that traditional docking programs such as OpenEye were not specifically designed to model RNA-protein interactions, and the relatively low-level of the calculations involved, we felt further analysis was justified. To account for the fact that we are modeling a flexible protein with a rigid docking method, we analyzed the top ten poses of the top scoring molecules. We reasoned that if a molecule is a good fit, the top poses should all score nearly as well as the highest for each candidate. Additionally, these top poses should all bind in a similar way (i.e., molecules that show large variance in their top poses can be thought as likely to be more fluxional in the real system compared to molecules that show little variance in their top poses). Five sets of scores were created, each differing in the relative importance of the scoring algorithms that contributed to the overall score. For example, the created receptor was scored first with chemgauss4, the latest scoring function available in OpenEye. Another score was generated with the same receptor, but using the slightly older chemgauss3 scoring function. Yet another was derived from the Consensus scoring method, which gives a score based on several available scoring functions, as detailed on the OpenEye web site. Lastly, a method was implemented that utilizes the Consensus method to choose the order of the top poses, which are then actually given a score with chemgauss3, and then the reverse, being chemgauss3 pose selection and Consensus scoring. Due to the fact that each of these scoring methods generates scores within their own numerical scale (each scoring function is different), we normalized these scores and averaged them to obtain an overall score. Finally, an ordering of priority for experimental screening was generated for the top scoring molecules based on the criteria of fluxionality described above, and overall rank across all scoring functions. For example, if a molecule was in the top five of four of the five scoring methods, and showed little fluxionality, it was deemed an excellent candidate. This last step is more subjective than the normalized scores themselves, as chemical intuition, with knowledge of the limitations of our methods constantly in mind, comes into play.
3,6-Anhydro-1,2-dideoxy-1-(trimethylsilyl)-4,5,7-tri-O-benzoyl-D-allo-hept-1-ynitol (2)
To a solution of trimethylsilylacetylene (1.8 mL, 13 mmol) in anhydrous toluene (3 mL) was added a solution of n-BuLi (8.0 mL, 1.6 M solution in hexane, 13 mmol) at 0 °C. After 25 min, to the mixture was added a solution of EtAlCl2 (13 mL, 1.0 M solution in hexane, 13 mmol). After 30 min, a solution of 1-O-acetyl-2,3,5-tri-O-benzoyl-β-D-ribose 1 (806 mg, 1.6 mmol) in anhydrous CH2Cl2 (9 mL) was added, and the mixture was stirred at 0 °C for 2.5 h and at room temperature for 4 h. The reaction mixture was quenched with saturated aqueous NaHCO3 at 0 °C, and filtered through Celite. The filtrate was washed with saturated aqueous NaHCO3, dried over Na2SO4, and the solvent was removed under reduced pressure. The residue was purified by column chromatography on a silica gel with hexane/EtOAc (19:1→9:1) to give 2 (380 mg, 44%) as a pale yellow oil. 1H NMR (CDCl3, 300 MHz, TMS): δ (ppm) 8.08 (d, J = 7.8 Hz, 2H), 8.00 (d, J = 7.8 Hz, 2H), 7.89 (d, J = 7.8 Hz, 2H), 7.60-7.50 (m, 3H), 7.44-7.50 (m, 6H), 5.85-5.78 (m, 2H), 4.96 (d, J = 3.6 Hz, 1H), 4.74-4.69 (m, 1H), 4.63-4.56 (m, 2H), 0.17 (s, 9H). 13C NMR (CDCl3, 150 MHz): δ(ppm) 166.4, 165.4, 165.3, 133.7, 133.6, 133.3, 129.9, 128.6, 100.3, 93.9, 79.4, 76.5, 72.9, 72.2, 64.5, −0.22. ESIHRMS (m/z): calcd for C31H30O7Si [M+H]+ 543.1834, obsd 543.1839.
3,6-Anhydro-1,2-dideoxy-7-O-(4,4’-Dimethoxytrityl)-D-allo-hept-1-ynitol (3)
A solution of 30% NH4OH was added to a solution of 2 (370 mg, 0.68 mmol) in EtOH, and the mixture was stirred at room temperature for 2 days. The reaction mixture was evaporated under reduced pressure, and the residue was coevaporated with anhydrous acetonitrile and anhydrous pyridine. The dried residue was dissolved in anhydrous pyridine (3 mL). To the solution was added DMTrCl (415 mg, 1.2 mmol), and the mixture was stirred at room temperature for 1 h. The reaction mixture was diluted with EtOAc (100 mL), and washed with water (50 mL) and brine (50 mL). The organic phase was dried over Na2SO4, and the solvent was removed under reduced pressure. The residue was purified by column chromatography on a silica gel with hexane/EtOAc (4:1→2:1) containing 0.5% pyridine to give 3 (290 mg, 93%) as a yellow oil. 1H NMR (CD2Cl2, 300 MHz): δ (ppm) 7.47 (d, J = 7.8 Hz, 2H), 7.37-7.19 (m, 7H), 6.83 (d, J = 9.0 Hz, 4H), 4.47 (dd, J = 5.1, 2.1 Hz, 1H), 4.29 (t, J = 5.1 Hz, 1H), 4.19 (t, J = 5.1 Hz, 1H), 3.99-3.94 (m, 1H), 3.77 (s, 6H), 3.34 (brs, 2H), 3.29 (dd, J = 10, 3.9 Hz, 1H), 3.15 (dd, J = 10, 4.5 Hz, 1H), 2.60 (d, J = 2.1 Hz, 1H). 13C NMR (CD2Cl2, 150 MHz): δ (ppm) 158.6, 144.9, 136.1, 136.0, 130.3, 130.2, 128.4, 128.0, 126.9,113.3, 86.5, 83.2, 81.4, 76.8, 75.0, 73.2, 72.8, 64.3, 55.4. ESIHRMS (m/z): calcd for C28H28O6 [M-H]- 459.1813, obsd 459.1786.
3,6-Anhydro-1,2-dideoxy-7-O-(4,4’-Dimethoxytrityl)-4-O-(tert-butyldimethylsilyl)-D-allo-hept-1-ynitol (4)
Compound 3 (203 mg, 0.44 mmol) was coevaporated with anhydrous acetonitrile, and the dried residue was dissolved in anhydrous THF (2 mL). To the solution was added dry pyridine (177 µL, 2.2 mmol) and AgNO3 (112 mg, 0.66 mmol). After 5 min, TBDMSCl (119 mg, 0.79 mmol) was added, and the mixture was stirred at room temperature for 2 h. The reaction mixture was diluted with CH2Cl2, filtered through Celite, and washed with saturated aqueous NaHCO3 (20 mL) and brine (20 mL). The organic phase was dried over Na2SO4, and concentrated under reduced pressure. The residue was purified by column chromatography on a silica gel with hexane/EtOAc (19:1) containing 0.5% pyridine to give 4 (121 mg, 48%) as a pale yellow foam and 3’-TBDMS (68 mg 27%) as a pale yellow foam. 1H NMR (CD2Cl2, 300 MHz): δ (ppm) 7.49 (d, J = 7.8 Hz, 2H), 7.39-7.22 (m, 7H), 6.84 (d, J = 8.7 Hz, 4H), 4.48 (dd, J = 6.6, 4.5 Hz, 1H), 4.42 (dd, J = 6.6, 2.1 Hz, 1H), 4.01-3.98 (m, 2H), 3.78 (s, 6H), 3.33 (dd, J = 10, 3.0 Hz, 1H), 3.04 (dd, J = 10, 3.3 Hz, 1H), 2.64 (d, J = 2.1 Hz, 1H), 2.60 (d, J = 3.3 Hz, 1H), 0.94 (s, 9H), 0.21 (s, 3H), 0.18 (s, 3H). 13C NMR (CD2Cl2, 150 MHz): δ (ppm) 159.0, 145.5,136.5, 136.3, 130.6, 130.5, 128.6, 128.2, 127.1, 113.5, 86.6, 84.5, 82.2, 78.2, 75.0, 73.3, 72.7, 64.7, 55.6, 25.9, 18.4, −4.25, −4.74. ESIHRMS (m/z): calcd for C34H42O6Si [M-H]- 573.2667, obsd 573.2678.
3,6-Anhydro-1,2-dideoxy-7-O-(4,4’-dimethoxytrityl)-5-O-[2-(cyanoethoxy)(N,N-diisopropylamino)phosphino] −4-O- (tert-butyldimethylsilyl)-D-allo-hept-1-ynitol (5)
Compound 4 (80 mg, 0.14 mmol) was coevaporated with anhydrous acetonitrile twice and the dried residue was dissolved in anhydrous CH2Cl2 (1 mL). To the solution was added N,N-diisopropylethylamine (145 µL, 0.83 mmol) and (2-cyanoethyl)-N,N-diisopropylchlorophosphoramidite (80 µL, 0.36 mmol), and the mixture was stirred at room temperature for 1 h. The reaction mixture was quenched with MeOH (50 µL) and saturated aqueous NaHCO3 (15 mL), and extracted with EtOAc (20 mL). The organic phases were washed with brine (15 mL), dried over Na2SO4, and the solvent was removed under reduced pressure. The residue was purified by column chromatography on silica gel with hexane/EtOAc (9:1) containing 0.1% pyridine to give 5 (87 mg 81%) as a white foam. 31P NMR (CD2Cl2, 121 MHz): δ (ppm) 151.2, 149.4. ESIHRMS (m/z): calcd for C43H59N2O7PSi [M+H]+ 775.3902, obsd 775.3916.
2-Phenyl-5-(azidomethyl)-1 H-imidazole (6)
TsCl (164 mg, 0.86 mmol) was added to a solution of (2-phenyl-1H-imidazol-5-yl)methanol (100 mg, 0.57 mmol) and triethylamine (90 µL, 0.65 mmol) in anhydrous CH2Cl2 (4 mL) at room temperature, and the mixture was stirred for 16 h. The reaction mixture was diluted with CH2Cl2 (20 mL), and washed with saturated aqueous NaHCO3 (20 mL) and brine (20 mL). The organic phase was dried over Na2SO4, and concentrated under reduced pressure. The residue was dissolved in anhydrous DMF (1 mL), and sodium azide (112 mg, 1.72 mmol) was added. The mixture was stirred at room temperature for 20 h. The reaction mixture was diluted with ether (20 mL), and washed with water (20 mL) and brine (20 mL). The organic phase was dried over Na2SO4, and the solvent was removed under reduced pressure. The residue was purified by column chromatography on a silica gel with hexane/EtOAc (2:1) to give 6 (16 mg, 14%) as a white solid. 1H NMR (CD3OD, 300 MHz): δ (ppm) 7.87 (d, J = 6.6 Hz, 2H), 7.49-7.40 (m, 3H), 7.22 (s, 1H), 4.36 (s, 1H). 13C NMR (CD3OD, 150 MHz): δ (ppm) 148.9, 131.2, 130.5, 130.1, 130.0, 127.2, 126.5, 49.4. ESIHRMS (m/z): calcd for C10H9N5 (M+H)+ 200.0931, obsd 200.0928.
Synthesis of siRNA guide and passenger strands
Unmodified RNA oligonucleotides were purchased from either Sigma, ChemGenes or at the University of Utah DNA/Peptide Core Facility (Salt Lake City, Utah). RNAs containing modified nucleosides were synthesized at the University of Utah DNA/Peptide Core Facility using the 5’-DMTr protected β-cyanoethyl phosphoramidites on a 200 nmol scale. All RNAs were deprotected as previously described.35 SiRNA guide strands were chemically 5’-phosphorylated during automated synthesis.
Copper-catalyzed azide/alkyne cycloaddition (CuAAC) reactions with RNA
A solution of crude RNA (3.5 nmol) in HEPES buffer (pH 7, 12.77 µL, final concentration 100 mM) was treated with tris-[1-(3-hydroxypropyl)-1H-[1,2,3]triazol-4-yl)methyl]amine ligand (100 mM solution in H2O, 1.75 µL, 175 nmol), either 2-(azidomethyl)-5-phenyl-1 H-imidazole (6)(100 mM solution in DMSO, 0.35 µL, 35 nmol) or 1-(2-azidoethyl)-piperidine(50 mM solution in 0.5 M Tris-HCl, pH 8.0, 0.7 µL, 35 nmol),36 sodium ascorbate (100 mM solution in H2O, 1.75 µL, 175 nmol), and CuSO4 (100 mM solution in H2O, 0.88 µL, 88 nmol). The solution was incubated for 2–3 h at room temperature. The reaction mixture was then diluted with PAGE loading buffer (80% formamide containing 10 mM EDTA) and PAGE-purified (19% denaturing, 20 W, 6–8 h). After electrophoresis, the RNA bands were visualized by UV shadowing (254 nm light, F254 TLC plate as a backing), and extracted from the gel via the crush and soak method at 4 °C overnight in 0.5 M NH4OAc containing 0.1 mM EDTA. Polyacrylamide particles were removed using a Centrex filter (0.2 µm) and the oligonucleotide solution was desalted using C18 Sep-Pak cartridges (Waters), eluting with 1:1 CH3CN/H2O or precipitated with 3:1:0.1 ethanol/crush and soak oligonucleotide solution/3M sodium acetate followed by 70% ethanol wash. 1-ER containing RNAs were purified by gel electrophoresis and isolated as described above for the triazole-containing RNAs.
RNA quantification and mass spectrometry analysis
The oligonucleotide solutions or precipitated pellets were lyophilyzed to dryness, resuspended in nuclease-free water and quantified by absorbance measurements at 260 nm. Click reaction yields for guide position 1 (84 ± 2% for 1-ER triazole I and 81 ± 2% for 1-ER triazole II, values averaged over three trials ± standard deviation) were determined by comparing the side-by-side purified yield of unclicked 1-ER oligonucleotides from the crude mixture to the click reactions on the same crude RNA. Identity of the oligonucleotides containing either 1-ER, triazole I or triazole II modifications was confirmed by MALDI-TOF mass spectroscopy using a saturated solution of 6-aza-2-thiothymine in 0.1 M aqueous dibasic ammonium citrate as a matrix. Mass spectra were recorded in the positive ionization mode and calibrated to an internal DNA standard of 6740.4 Daltons. List of mass values, [M+H]+, (G = guide, P =passenger, number indicates position of modification), for all 1-ER modified siRNAs: G1: Calcd 6718.0, obsd 6719.0; G12: Calcd 6718.0, obsd 6719.2; P19: Calcd 6477.0, obsd 6477.3. For all 1-ER triazole I modified siRNAs: G1: Calcd 6917.2, obsd 6717.7; G12: Calcd 6917.2, obsd 6920.2; P19: Calcd 6676.2, obsd 6676.4. For all 1-ER triazole II modified siRNAs: G1: Calcd 6872.2, obsd 6873.1; G12: Calcd 6872.2, obsd 6872.5; P19: Calcd 6631.2, obsd 6630.1.
Melting temperature analysis
The thermal stability of siRNA duplexes containing either adenosine, 1-ER, 1-ER triazole I or 1-ER triazole II at guide position 12 were analyzed in a similar fashion to our previously described methods.16,18 siRNA duplexes were formed by hybridizing 462.5 pmoles of complementary strands in 925 µL of TE buffer (10 mM Tris-HCl, pH 7.8, 0.1 mM EDTA and 100 mM NaCl). The values reported in Table 3 are an average of three thermal melting experiments (152.5 pmol, 305 µL per experiment), with an experimental temperature range of 30 to 80 °C. The values in Table 3 indicate the average melting temperature ± standard deviation.
Cell culture
HeLa cells (ATCC) were grown at 37 °C in humidified 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) supplemented with 10% fetal bovine serum (FBS, GIBCO) and 1% antibiotic-antimycotic (100x Anti-Anti, GIBCO). The cells were maintained in exponential growth.
Transfection and RNAi activity assay
For the dual-luciferase RNA interference assay, HeLa cells were grown to 80–90% confluence then detatched using Accutase (Innovative Cell Technologies, without calcium or magnesium). Reverse transfection of the HeLa cells was accomplished using siPORT NeoFX (Ambion) in Opti-MEM media (GIBCO) to deliver the psiCHECK-2-PIK3CB plasmid and siRNAs as previously described.18 SiRNA duplex hybridization was accomplished by combining equal amounts of purified passenger and native or modified guide strands to a final concentration of 5 µM in 10 mM Tris-HCl, 50 mM KCl, pH 7.5. The samples were heated at 95 °C for 5 minutes followed by slow cooling to room temperature over a period of approximately 2 h. All siRNAs referenced in Figure 3, Figure 5 and Figure 6 were tested side-by-side in the same 96-well plate assay. Three individual assays were averaged to give the values and standard deviations plotted in those figures.
Supplementary Material
Acknowledgements
P.A.B. and D.J.T. acknowledge the National Institutes of Health for financial support in the form of grant R01-GM080784. We are also grateful for the Research Fellowship from the Japan Society for the Promotion of Science (JSPS) for Young Scientists (K.O.). We also gratefully acknowledge the United States Department of Education; GAANN (Graduate Assistance in Areas of National Need) Fellowship to JGH.
Footnotes
Supporting Information Available: Structures and computational screening scores for analogs evaluated, supplementary figures with docking constraints and putative 1,2,3-triazol-4-yl binding site in hAgo2, NMR spectra for compounds 2–5. This information is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Kurreck J. Angew Chem Int Ed Engl. 2009;48:1378. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shukla S, Sumaria CS, Pradeepkumar PI. ChemMedChem. 2010;5:328. doi: 10.1002/cmdc.200900444. [DOI] [PubMed] [Google Scholar]
- 3.Gaynor JW, Campbell BJ, Cosstick R. Chem Soc Rev. 2010;39:4169. doi: 10.1039/b920362c. [DOI] [PubMed] [Google Scholar]
- 4.Peacock H, Kannan A, Beal PA, Burrows CJ. J Org Chem. 2011;76:7295. doi: 10.1021/jo2012225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manoharan M. Curr Opin Chem Biol. 2004;8:570. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Watts JK, Deleavey GF, Damha MJ. Drug Discov Today. 2008;13:842. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Addepalli H, Meena; Peng CG, Wang G, Fan Y, Charisse K, Jayaprakash KN, Rajeev KG, Pandey RK, Lavine G, Zhang L, Jahn-Hofmann K, Hadwiger P, Manoharan M, Maier MA. Nucleic Acids Res. 2010;38:7320. doi: 10.1093/nar/gkq568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peacock H, Fostvedt E, Beal PA. ACS Chem Biol. 2010;5:1115. doi: 10.1021/cb100245u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lima WF, Wu H, Nichols JG, Sun H, Murray HM, Crooke ST. J. Biol. Chem. 2009;284:26017. doi: 10.1074/jbc.M109.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schirle NT, MacRae IJ. Science. 2012;336:1037. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. Cell. 2012;150:100. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma JB, Ye K, Patel DJ. Nature. 2004;429:318. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank F, Sonenberg N, Nagar B. Nature. 2010;465:818. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 14.Felice KM, Salzman DW, Shubert-Coleman J, Jensen KP, Furneaux HM. Biochem J. 2009;422:329. doi: 10.1042/BJ20090534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.OpenEye Scientific Software, I., version 1.7.4 ed. USA: Santa Fe, NM; [Google Scholar]
- 16.Peacock H, Maydanovych O, Beal PA. Org Lett. 2010;12:1044. doi: 10.1021/ol100019r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peacock H, Fucini RV, Jayalath P, Ibarra-Soza JM, Haringsma HJ, Flanagan WM, Willingham A, Beal PA. J Am Chem Soc. 2011;133:9200. doi: 10.1021/ja202492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibarra-Soza JM, Morris AA, Jayalath P, Peacock H, Conrad WE, Donald MB, Kurth MJ, Beal PA. Org Biomol Chem. 2012;10:6491. doi: 10.1039/c2ob25647a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Frank-Kamenetsky M, Rajeev KG, Egli M, Manoharan M. ACS Chem. Bio. 2006;1:176. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez AR, Peterson LW, Kool ET. ACS Chem. Bio. 2012;7:1454. doi: 10.1021/cb300174c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakahara M, Kuboyama T, Izawa A, Hari Y, Imanishi T, Obika S. Bioorg Med Chem Lett. 2009;19:3316. doi: 10.1016/j.bmcl.2009.04.063. [DOI] [PubMed] [Google Scholar]
- 22.Hari Y, Nakahara M, Pang J, Akabane M, Kuboyama T, Obika S. Bioorg Med Chem. 2011;19:1162. doi: 10.1016/j.bmc.2010.12.049. [DOI] [PubMed] [Google Scholar]
- 23.Itoh Y, Haraguchi K, Tanaka H, Gen E, Miyasaka T. J Org Chem. 1995;60:656. [Google Scholar]
- 24.Haraguchi K, Sumino M, Tanaka H. J Org Chem. 2006;71:4433. doi: 10.1021/jo060194m. [DOI] [PubMed] [Google Scholar]
- 25.Castagnolo D, Botta L, Botta M. Tetrahedron Lett. 2009;50:1526. [Google Scholar]
- 26.El-Sagheer AH, Brown T. Chem Soc Rev. 2010;39:1388. doi: 10.1039/b901971p. [DOI] [PubMed] [Google Scholar]
- 27.Huisgen R, Szeimies G, Mobius L. Chemische Berichte-Recueil. 1967;100:2494. [Google Scholar]
- 28.Kolb HC, Finn MG, Sharpless KB. Angew Chem Int Ed Engl. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 29.Meldal M, Tornoe CW. Chem Rev. 2008;108:2952. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 30.Hong V, Presolski SI, Ma C, Finn MG. Angew Chem Int Ed Engl. 2009;48:9879. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz N, Marenstein DR, De Angelis DA, Wang W-Q, Nelander S, Jacobsen A, Marks DS, Massague J, Sander C. Silence. 2011;2:1. doi: 10.1186/1758-907X-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarz DS, Hutvagner G, Du T, Zu Z, Aronin N, Zamore PD. Cell. 2003;115:199. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 33.Turner JJ, Jones SW, Moschos SA, Lindsay MA, Gait M. J. Mol. Biosyst. 2007;3:43. doi: 10.1039/b611612d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li F, Pallan PS, Maier M, Rajeev KG, Mathieu SL, Kreutz C, Fan Y, Sanghvi J, Micura R, Rozners E, Manoharan M, Egli M. Nucl. Acids Res. 2007;35:6424. doi: 10.1093/nar/gkm664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maydanovych O, Easterwood LM, Cui T, Veliz EA, Pokharel S, Beal PA. Methods Enzymol. 2007;424:369. doi: 10.1016/S0076-6879(07)24017-0. [DOI] [PubMed] [Google Scholar]
- 36.Carboni B, Vaultier M, Carrié R. Tetrahedron Lett. 1988;29:1279. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.