Abstract
DNA of bacteriophage λ recombines in a cell-free extract prepared from an induced Escherichia coli lysogen of bacteriophage λ. The assay for recombination in vitro takes advantage of the ability of such an extract to package λ DNA and to assemble complete phage particles. For example, when λ DNA that has been extracted from phage with the immunity of 434 is added to an extract, infectious λ imm 434 particles are produced. The precursor DNA molecule in this packaging reaction is a multichromosomal polymer; circular monomers, for example, are not packaged.
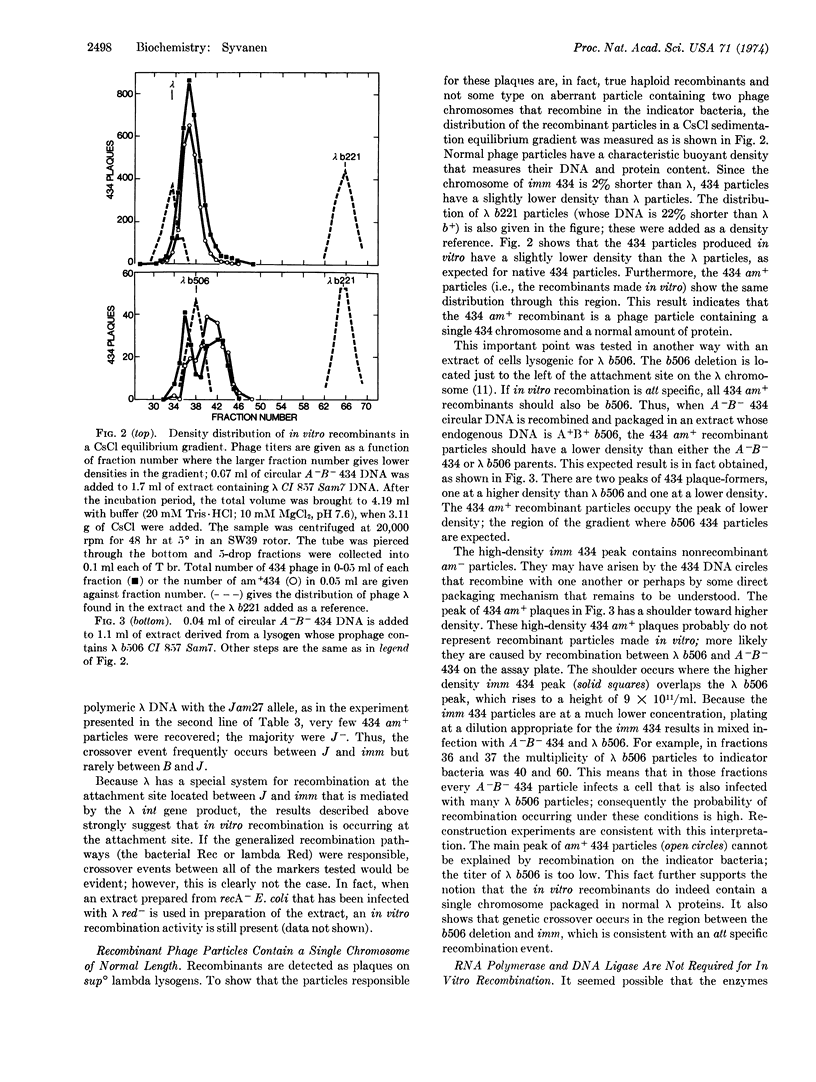
Nevertheless, when 434 circular DNA monomers are added to an extract, some phage that contain the imm 434 marker are produced. In this case, the circular DNA had recombined with λ DNA in the extract and thereby had become part of a polymeric structure, which by the normal packaging process could give rise to infectious particles with the imm 434 marker. Genetic recombination is demonstrated when imm 434 circular monomer DNA carries amber mutations in genes A and B; then most of the 434 plaque formers produced in vitro are A+B+, the genotype of the endogenous λ DNA. Genetic crossing-over occurs through a region that contains the prophage attachment site, suggesting that recombination is carried out by the λ Int functions. The 434 recombinant plaque formers are particles physically identical to wild-type 434 particles, as judged by their buoyant density in a CsCl equilibrium gradient.
Keywords: phage λ DNA, in vitro head assembly, site specific recombination
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbour S. D., Clark A. J. Biochemical and genetic studies of recombination proficiency in Escherichia coli. I. Enzymatic activity associated with recB+ and recC+ genes. Proc Natl Acad Sci U S A. 1970 Apr;65(4):955–961. doi: 10.1073/pnas.65.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttin G., Wright M. Enzymatic DNA degradation in E. coli: its relationship to synthetic processes at the chromosome level. Cold Spring Harb Symp Quant Biol. 1968;33:259–269. doi: 10.1101/sqb.1968.033.01.030. [DOI] [PubMed] [Google Scholar]
- Echolas H., Gingery R. Mutants of bacteriophage lambda defective in vegetative genetic recombination. J Mol Biol. 1968 Jul 14;34(2):239–249. doi: 10.1016/0022-2836(68)90249-0. [DOI] [PubMed] [Google Scholar]
- Enquist L. W., Skalka A. Replication of bacteriophage lambda DNA dependent on the function of host and viral genes. I. Interaction of red, gam and rec. J Mol Biol. 1973 Apr 5;75(2):185–212. doi: 10.1016/0022-2836(73)90016-8. [DOI] [PubMed] [Google Scholar]
- Freifelder D., Levine E. E. Requirement for transcription in the neighborhood of the phage attachment region for lysogenization of Escherichia coli by bacteriophage lambda. J Mol Biol. 1973 Mar 15;74(4):729–733. doi: 10.1016/0022-2836(73)90060-0. [DOI] [PubMed] [Google Scholar]
- Kaiser D., Masuda T. In vitro assembly of bacteriophage Lambda heads. Proc Natl Acad Sci U S A. 1973 Jan;70(1):260–264. doi: 10.1073/pnas.70.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modorich P., Lehman I. R. Deoxyribonucleic acid ligase. A steady state kinetic analysis of the reaction catalyzed by the enzyme from Escherichia coli. J Biol Chem. 1973 Nov 10;248(21):7502–7511. [PubMed] [Google Scholar]
- Parkinson J. S. Deletion mutants of bacteriophage lambda. II. Genetic properties of att-defective mutants. J Mol Biol. 1971 Mar 14;56(2):385–401. doi: 10.1016/0022-2836(71)90472-4. [DOI] [PubMed] [Google Scholar]
- Radding C. M. The role of exonuclease and beta protein of bacteriophage lambda in genetic recombination. I. Effects of red mutants on protein structure. J Mol Biol. 1970 Sep 28;52(3):491–499. doi: 10.1016/0022-2836(70)90415-8. [DOI] [PubMed] [Google Scholar]
- Shulman M. J., Hallick L. M., Echols H., Signer E. R. Properties of recombination-deficient mutants of bacteriophage lambda. J Mol Biol. 1970 Sep 28;52(3):501–520. doi: 10.1016/0022-2836(70)90416-x. [DOI] [PubMed] [Google Scholar]
- Signer E. R., Weil J. Recombination in bacteriophage lambda. I. Mutants deficient in general recombination. J Mol Biol. 1968 Jul 14;34(2):261–271. doi: 10.1016/0022-2836(68)90251-9. [DOI] [PubMed] [Google Scholar]
- Sippel A., Hartmann G. Mode of action of rafamycin on the RNA polymerase reaction. Biochim Biophys Acta. 1968 Mar 18;157(1):218–219. doi: 10.1016/0005-2787(68)90286-4. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., McMilin K. D., Stahl M. M., Malone R. E., Nozu Y., Russo V. E. A role for recombination in the production of "free-loader" lambda bacteriophage particles. J Mol Biol. 1972 Jul 14;68(1):57–67. doi: 10.1016/0022-2836(72)90262-8. [DOI] [PubMed] [Google Scholar]
- Szpirer J., Brachet P. Relations physiologiques entre les phages tempérés lambda et phi80. Mol Gen Genet. 1970;108(1):78–92. doi: 10.1007/BF00343187. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Davidson N. Thermodynamic and kinetic studies on the interconversion between the linear and circular forms of phage lambda DNA. J Mol Biol. 1966 Jan;15(1):111–123. doi: 10.1016/s0022-2836(66)80213-9. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Degree of superhelicity of covalently closed cyclic DNA's from Escherichia coli. J Mol Biol. 1969 Jul 28;43(2):263–272. doi: 10.1016/0022-2836(69)90266-6. [DOI] [PubMed] [Google Scholar]
- Young E. T., 2nd, Sinsheimer R. L. Vegetative lambda DNA. 3. Pulse-labeled components. J Mol Biol. 1968 Apr 14;33(1):49–59. doi: 10.1016/0022-2836(68)90280-5. [DOI] [PubMed] [Google Scholar]



