SUMMARY
Wnt5a directs the assembly of “Wnt-receptor-actin-myosin-polarity (WRAMP)” structure, which integrates cell adhesion receptors with F-actin and myosin to form a microfilament array associated with multivesicular bodies. The WRAMP structure is polarized to the cell posterior, where it directs tail-end membrane retraction, driving forward translocation of the cell body. Here, we define constituents of the WRAMP proteome, including regulators of microfilament and microtubule dynamics, protein interactions, and enzymatic activity. IQGAP1, a scaffold for F-actin nucleation and crosslinking, is necessary for WRAMP structure formation, potentially bridging microfilaments and MVBs. Vesicle coat proteins, including coatomer-I subunits, localize to and are required for the WRAMP structure. Electron microscopy and live imaging demonstrate movement of ER to the WRAMP structure and plasma membrane, followed by elevation of intracellular Ca2+. Thus, Wnt5a controls directional movement by recruiting cortical ER to mobilize a rear-directed, localized Ca2+ signal, activating actomyosin contraction and adhesion disassembly for membrane retraction.
INTRODUCTION
Directional movement in migrating cells requires integrating processes which control motility and cell polarization, allowing cells to distinguish front from back. At the leading edge are events controlling F-actin assembly, mediated by the activation of Rac and Cdc42, Arp2/3 and formins, PIP kinases, and actin severing proteins. These leading edge components drive actin nucleation and polymerization, forming membrane protrusions and sheet extensions which in turn facilitate attachments between integrin receptors and extracellular matrix proteins (Parsons et al., 2010). Also important are processes controlling microtubule assembly and orientation as well as localized vesicle exocytosis to replenish cell surface integrins, modulated through transient Ca2+ fluctuations at the front end of cells (Wei et al., 2009). Cell polarity effectors, e.g. Scribble and PARtition protein complexes, direct trafficking of proteins to the front of migrating cells, enabling localized activation of Rac and Cdc42 (Petrie et al., 2009).
At the cell trailing edge are RhoA and Rho kinase dependent processes leading to contractility driven by F-actin and myosin II, which reduce cell volume and create transient mechanical force to drive forward translocation (Cramer, 2010). Rear-end dynamics are driven by phosphorylation of myosin light chain via Ca2+-calmodulin-dependent myosin light chain kinase (MLCK), as well as recruitment of the Ca2+ protease, calpain, which degrades focal adhesion proteins and allows focal adhesion disassembly (Petrie et al., 2009). Thus, directional movement involves coordination between localized membrane protrusions and formation of cell adhesions at the leading edge, and membrane retraction and disassembly of adhesions at the rear. At the tail end of moving cells, free Ca2+ is needed to activate multiple enzymes, and in some polarized migrating cell types, a gradient of Ca2+ elevated at the rear and decreasing to the front has been demonstrated (Hahn et al., 1992; Brundage et al., 1991). However, little is known about signaling mechanisms which recruit proteins and allow Ca2+ release, polarized to the rear.
We reported that Wnt5a promotes the assembly of the “Wnt-receptor-actin-myosin-polarity (WRAMP)” structure, which includes melanoma cell adhesion molecule (MCAM/CD146/MUC18), as well as Frizzled-3, F-actin, and myosin II (Witze et al., 2008). The WRAMP structure is associated with a dense microfilamentous array surrounding a vesicle pool consisting of multivesicular bodies (MVBs). Its formation requires dynamin and Rab4, revealing a role for vesicle trafficking in Wnt5a-regulated cell polarization. The WRAMP structure forms dynamically and is directed towards the cell posterior, followed within minutes by tail end membrane retraction, allowing nucleokinesis in the direction of cell movement (Fig. 1A, Movie S1). Thus, Wnt5a controls forward cell movement in part by polarized assembly of the WRAMP structure, which integrates receptors, cytoskeletal proteins and MVBs through a mechanism involving receptor endocytosis and endosome trafficking, and directs membrane retraction at the trailing edge.
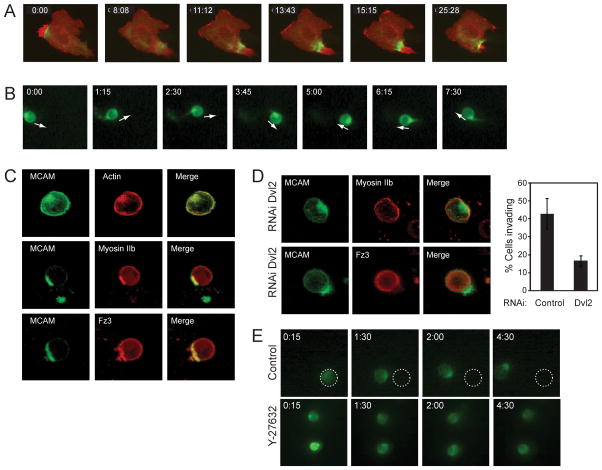
Fig. 1. The WRAMP structure polarizes to the cell posterior during invasion.
(A) WM239A cells transfected with MCAM-RFP and GFP-myosin IIb heavy chain on 2D plates show the WRAMP structure, characterized by polarization of MCAM and myosin IIb, followed by membrane retraction (see Movie S1). Time indicates min:sec. See also Figure S1. (B) In cells transfected with MCAM-GFP invading in 3D hydrogels, MCAM stably localizes at the cell rear relative to the direction of movement, indicated by arrows (see Movie S1). (C) Immunocytochemistry shows that endogeneous myosin IIb, Frizzled-3, and F-actin localize with MCAM to the WRAMP structure in hydrogels. (D) Dvl2 knockdown blocks myosin IIb and Frizzled-3 recruitment to the WRAMP structure in hydrogels. The bar graph quantifies percentages of cells which form WRAMP structures in this experiment (± s.d., n=200–400 cells). (E) ROCK inhibitor Y27632 interferes with MCAM-GFP polarization and inhibits cell invasion (see Movie S1). Time in panels B-E indicate hr:min.
Little is known about the composition or function of the WRAMP structure or how it promotes membrane retraction. Here we present a proteomics strategy to define constituents of the WRAMP structure proteome by mass spectrometry, and report proteins involved in microfilament and microtubule dynamics, as well as membrane organellar function. In particular, subunits of the COP-I coatomer predict a potential involvement of Golgi-endoplasmic reticulum (ER). The WRAMP structure modulates rear-directed recruitment of cortical ER followed by the elevation of free Ca2+, explaining how Ca2+ signaling can occur in a polarized manner in order to promote substrate detachment and actomyosin contractility, events needed for membrane retraction.
RESULTS
In response to Wnt5a, cells form the WRAMP structure, which assembles dynamically and coordinates the colocalization of MCAM, F-actin, and myosin IIb, followed by membrane retraction (Fig. 1A, Fig. S1A, Movie S1). Although the experiments in this study are performed in WM239A melanoma cells, we observe the WRAMP structure in other cell types, such as HUVEC and C2C12 (Fig. S1B, C). In 2D cultured cells, WRAMP structures polarize distal to Golgi, consistent with its rear-directed localization at the trailing edge (Witze et al., 2008).
Because little is known about the WRAMP structure in 3D environments, we examined cells using an engineered extracellular matrix in order to limit other signaling influences (Fairbanks et al., 2009). WM239A cells expressing MCAM-GFP were encapsulated in photopolymerizable hydrogels crosslinked to matrix metalloproteinase (MMP)-degradable peptides and pendant peptides containing the Arg-Gly-Asp (RGD) integrin recognition motif. The resulting 3D hydrogel promotes cell spreading and migration through protease and adhesion-dependent processes.
Live imaging showed cells invading through the 3D matrix with clear evidence for stable rear polarization of MCAM-GFP (Fig. 1B, Movie S1). Immunocytochemistry revealed colocalization of MCAM with F-actin, myosin IIb, and the Wnt5a receptor, Frizzled-3 (Fig. 1C). Recruitment of myosin IIb and Frizzled-3 to the WRAMP structure was inhibited when Wnt signaling was disrupted by RNAi knockdown of Dishevelled (Fig. 1D). Polarization of MCAM was also disrupted when cells were treated with the Rho kinase inhibitor, Y27632 (Fig. 1E, Movie S1), or when RGD ligand was replaced with a scrambled peptide to prevent integrin binding (data not shown). These results demonstrated that the WRAMP structure formed in 3D is analogous to that previously observed in 2D, with polarization being dependent on Wnt signaling via Dishevelled, cell-matrix interactions, and Rho kinase-dependent mechanisms. Surprisingly, the WRAMP structure forms stably at the rear of WM239A cells for many hours in 3D, in contrast to 2D culture where it forms transiently. This provides an important illustration of how local environment profoundly influences the polarity of invasive cancer cells.
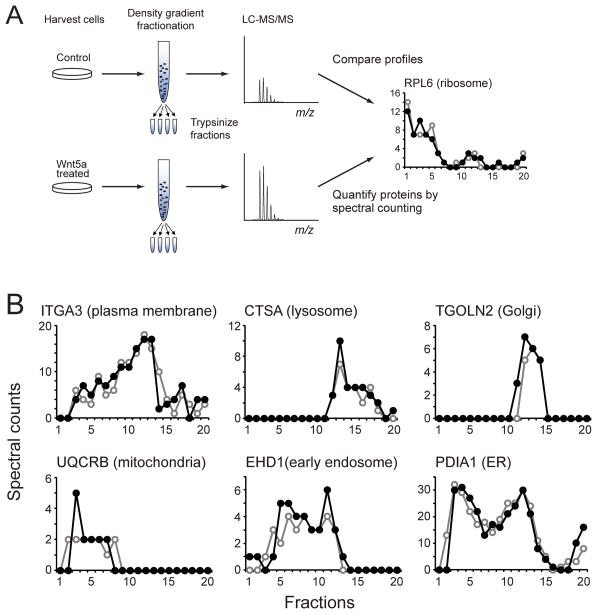
We next looked for other components within the WRAMP structure. Cells were harvested after 30 min of Wnt5a treatment, and post-nuclear membrane preparations were separated by density gradient centrifugation. Proteins in each fraction were trypsinized and peptides identified by LC-MS/MS (Fig. 2A). Label-free quantification by spectral counting (Old et al., 2005) was used to identify proteins altered in abundance or elution in response to Wnt5a. In total, 3,015 proteins were identified (Table S1), which included protein markers for all major organelle classes, including endoplasmic reticulum (ER), Golgi, mitochondria, plasma membrane microsomes, early and late endosomes, lysosomes, and peroxisomes (Fig. 2B). Most proteins were unaffected by Wnt5a, but in 36 proteins, the spectral counts increased by 2-fold or more within a single fraction at high density (fr#2, Fig. 3A–G, Fig. S2). Myosin IIb was also present in the dataset, although with low spectral counts (Fig. 3H). The results suggested that Wnt5a elevates the abundance of certain proteins within a specific organelle pool.
Fig. 2. Proteomics profiling of organelle fractions.
(A) WM239A cells were lysed and postnuclear membranes fractionated by density gradient centrifugation, followed by protein digestion and LC-MS/MS. Protein abundances were quantified by spectral counting. (B) Spectral count profiles of protein organelle markers show few differences between cells treated with (●) or without (○) Wnt5a for 30 min. See also Table S1.
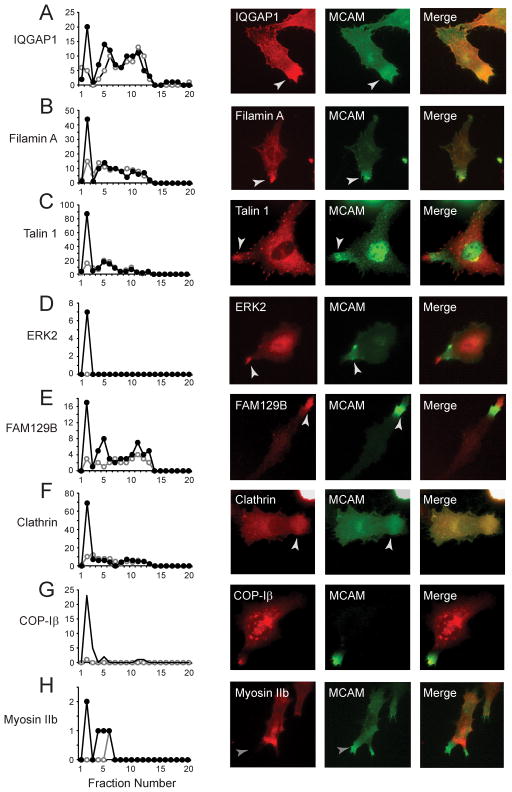
Fig. 3. Components of the WRAMP proteome.
(A–H) Spectral counts of identified proteins vs fraction number (highest density in fr #1), in cells treated with (●) or without (○) Wnt5a. Several proteins showed increased abundance in fr #2 in response to Wnt5a. In each case, immunocytochemistry showed colocalization with endogeneous MCAM, indicating recruitment to the WRAMP structure. See also Figure S2 and Table S2.
We asked whether the proteins within this pool were associated with the WRAMP structure, by monitoring their colocalization with MCAM by indirect immunocytochemistry (Fig. 3A–H). Endogenous forms of every protein tested localized with or in proximity to the WRAMP structure. The results showed that proteins within the WRAMP structure were stable to density gradient purification and could be identified by their increased abundance within a high density fraction in response to Wnt5a. We conclude that proteins identified in this manner represent initial components of the WRAMP proteome.
Constituents of the WRAMP proteome included known regulators of microfilament and microtubule dynamics, cell adhesion and movement, and membrane trafficking, as well as proteins with enzymatic and protein recognition functions (Table S2). Examples included IQGAP1 and filamin-A, microfilament binding proteins which crosslink actin filaments, and gelsolin, an actin severing protein which promotes microfilament turnover and branching (Nakamura et al., 2011; Yin & Stossel 1979). Also present was the microfilament motor, myosin IIa, whose structure and function is related to myosin IIb. Myosin 1b, containing binding domains for calmodulin and phosphatidylinositol phospholipids, is a class I myosin motor which bridges actin-membrane interactions, and also associates with and facilitates trafficking of proteins into MVBs (Salas-Cortes et al., 2005). Other components included focal adhesion proteins, talin-1 and kindlin-3/UNC112, both which mediate protein interactions needed for integrin receptor activation, enabling linkages between F-actin and the extracellular matrix (Burridge & Connell 1983; Moser et al., 2008). Microtubule regulators included tubulin-α1b and dynein, as well as nuclear migration protein nudC, a chaperone which mediates interactions between kinesin, dynein and dynactin (Yamada et al., 2010). Non-cytoskeletal proteins included calpain-2 and the ERK2 MAP kinase, which have been shown to promote focal adhesion disassembly via ERK phosphorylation of calpain and proteolysis of focal adhesion proteins (Glading et al., 2001). FAM129B/Minerva was previously identified as a target of ERK phosphorylation, which mediates oncogenic B-Raf dependent melanoma cell invasion (Old et al., 2009). Finally, the WRAMP proteome contained regulators of vesicle internalization and trafficking, including the vesicle coat proteins, clathrin and α, β, γ and δ subunits of the coatomer protein I complex (COP-I).
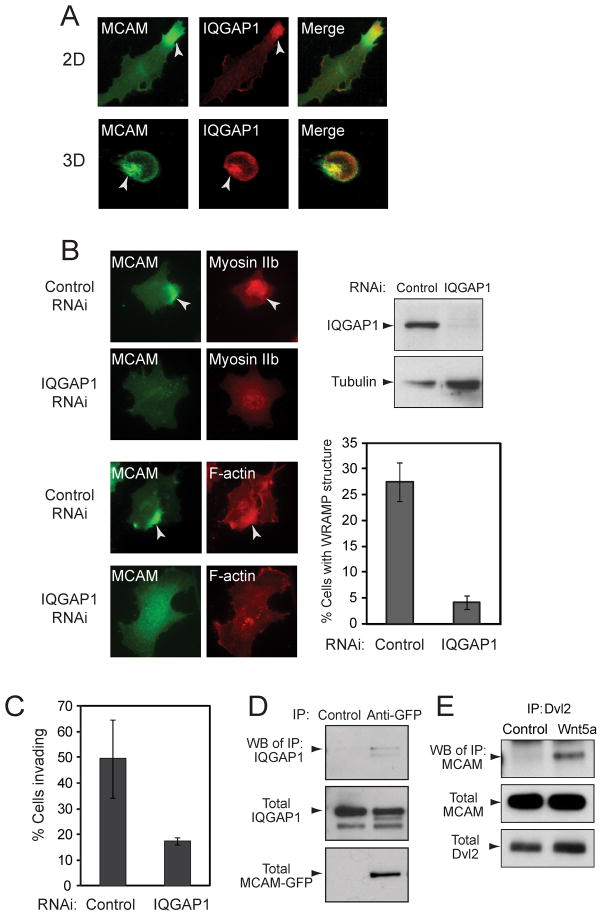
Experiments were performed to test the functional importance of constituent proteins in WRAMP structure formation. IQGAP1 is an adaptor protein which forms a scaffold for actin nucleation by N-WASP/Arp2/3 (Le Clainche et al., 2007). Immunocytochemistry showed that after treatment with Wnt5a, endogeneous IQGAP1 localized to the WRAMP structure marked by MCAM-GFP, in 2D and 3D cell environments (Fig. 4A). RNAi depletion of IQGAP1 interfered with WRAMP structure assembly, blocking polarization of MCAM, myosin IIb and F-actin (Fig. 4B, C). Furthermore, immunoprecipitation of MCAM revealed its interactions with endogeneous IQGAP1 (Fig. 4D). Thus, IQGAP1 is functionally important for WRAMP structure assembly and binds MCAM. Immunoprecipitation of the endogeneous Wnt adaptor protein, Dishevelled, also revealed MCAM binding (Fig. 4E). Thus, our working model is that MCAM is directly linked to Wnt5a/Dvl/Fz signaling, and that assembly of the WRAMP structure is driven in part by adaptor proteins such as IQGAP1, which bridge interactions between MCAM and cytoskeletal proteins.
Fig. 4. IQGAP1 colocalizes with the WRAMP structure and is required for its formation.
(A) MCAM-GFP and endogeneous IQGAP1 were monitored by immunocytochemistry in cells on plates (top) or within hydrogels (bottom). IQGAP1 colocalizes with MCAM at the WRAMP structure in both 2D and 3D culture. (B) Cells in 2D were treated with non-targeting or IQGAP1-RNAi oligonucleotides prior to Wnt5a. Monitoring MCAM-GFP, endogeneous myosin IIb and F-actin shows that knockdown of IQGAP1 inhibits formation of the WRAMP structure. Cells were quantified for the WRAMP structure, monitored with MCAM-GFP (± s.d., n=3, each counting 200–350 cells). (C) Cells in hydrogels were quantified for invasion, which was inhibited by RNAi knockdown of IQGAP1. Invasion was scored by the movement of a cell within the hydrogel by one cell length or more (± s.d., n=3, each counting 330–380 cells). (D) Immunoprecipitation of MCAM-GFP revealed binding to endogeneous IQGAP1. (E) Immunoprecipitation of endogenous Dvl2 revealed association with MCAM-GFP, in cells treated for 30 min with Wnt5a.
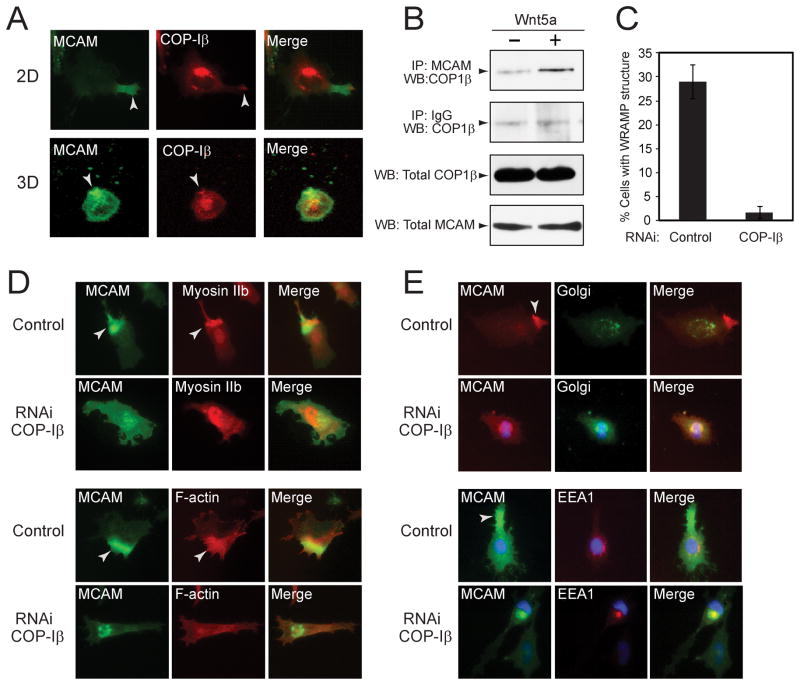
The presence of COP-I subunits within the WRAMP proteome was unexpected, because these proteins have to our knowledge never been linked to cell polarity. Instead, COP-I facilitates retrograde vesicle transport from Golgi to ER within ERGIC organelles, by binding cargo proteins containing C-terminal dilysine motifs (e.g., KKXXC-term, KXKXXC-term) which are canonical signals for ER retrieval. Nevertheless, in response to Wnt5a, COP-I α, β, γ and δ subunits were elevated within the high density fraction (Fig. 3, Fig. S2). Immunocytochemistry of endogeneous COP-Iβ revealed its juxtanuclear localization, consistent with primary recruitment to ERGIC. However, a subfraction of COP-Iβ colocalized with the WRAMP structure at the membrane periphery, in cells cultured on plates or in hydrogels (Fig. 3G, 5A). Coimmunoprecipitation experiments showed that COP-Iβ binds MCAM in a Wnt5a-dependent manner (Fig. 5B). RNAi knockdown of COP-Iβ inhibited formation of the WRAMP structure in response to Wnt5a (Fig. 5C, D). Following COP-Iβ depletion, the levels of MCAM remained constant, however its localization changed to a perinuclear pattern overlapping with markers for Golgi (58 kDa protein) and early endosomes (EEA1) (Fig. 5E). Together, the results suggested a potential role for Golgi or ER in the formation and/or function of the WRAMP structure.
Fig. 5. COP-Iβ colocalizes to the WRAMP structure and is required for its formation.
(A) MCAM-GFP and endogeneous COP-Iβ were monitored by immunocytochemistry in cells on plates (top) or within hydrogels (bottom). COP-Iβ colocalizes with MCAM at the WRAMP structure in 2D and 3D culture. (B) Endogeneous MCAM was immunoprecipitated with αMCAM antibody, and Western blots of pulldowns were probed for MCAM and COP-Iβ. (C) Cells on plates were quantified for the WRAMP structure, monitored by endogenous MCAM polarization (± s.d., n=3, each counting 200–350 cells). (D) Monitoring MCAM-GFP, endogeneous myosin IIb and F-actin shows that RNAi knockdown of COP-Iβ inhibits the WRAMP structure. (E) After COP-Iβ knockdown, endogeneous MCAM appears perinuclear, overlapping the 58 kDa Golgi protein marker and the early endosome marker, EEA1.
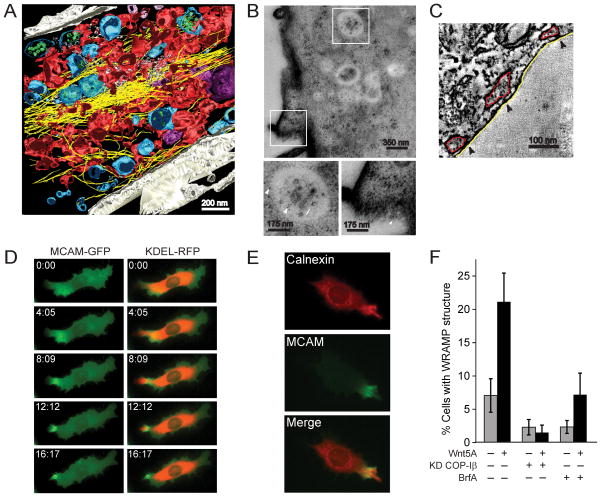
These findings prompted closer examination of the WRAMP ultrastructure. MCAM-GFP expressing cells were treated with Wnt5a for 30 min, and then monitored for the appearance and placement of WRAMP structures. Cells were then fixed and stained and analyzed by correlative electron microscopy (EM) and tomographic reconstruction of 250 nm sections. Fig. 6A shows the 3D reconstruction of a WRAMP structure region. Here, dense filamentous structures consistent with F-actin (yellow) formed extended cables aligned in parallel. MVBs (blue) were identifiable by the presence of intralumenal vesicles of 100–120 nm diameter (green). MCAM-GFP was also monitored after Wnt5a treatment by transmission immunoEM using anti-GFP-coupled gold particles. MCAM was clearly observed in the plasma membrane as well as the limiting membranes and intralumenal vesicles of MVBs appearing within the WRAMP structure (Fig. 6B).
Fig. 6. Cortical ER is recruited to the WRAMP structure.
(A) EM tomography showing the WRAMP structure in a cell treated with Wnt5a. Colors indicate MVBs (blue) and microfilaments (yellow) interspersed among ER tubules and sheets (red) which extend toward the edge of the cell. (B) Immuno-EM using anti-GFP antibodies labeled with gold particles reveals the presence of MCAM-GFP (arrows) within intralumenal vesicles and limiting membranes of MVBs, and at the plasma membrane. Bottom panels show magnified boxes in white. (C) EM tomographs indicate cortical ER (red) juxtaposed within 10 nm of the plasma membrane (white). (D) Live imaging of cells expressing MCAM-GFP and KDEL-RFP reveals localization of KDEL-RFP to the WRAMP structure, preceding membrane retraction (see Movie S2). (E) Indirect immunofluorescense reveals localization of endogeneous calnexin to the WRAMP structure. (F) Pretreatment of cells with Brefeldin A or RNAi knockdown of COP-Iβ blocks WRAMP structure formation. Cells on plates were quantified for the WRAMP structure, monitored by endogenous MCAM polarization (± s.d., n=3, each counting 200–350 cells)
Strikingly, the WRAMP structure was filled with a network of contiguous membrane sheets and tubules (Fig. 6A). These were in some places juxtaposed closely to the plasma membrane (Fig. 6C), suggesting a network of cortical ER. At the cell periphery, the ER localized to the end of the WRAMP structure and bundles of actin extended from the edge of the cell to form larger actin cables. This appearance contrasted that outside of the WRAMP structure, where the ER appeared more sheet like within regions intermediate between the nucleus and trailing edge. Here, endosomal vesicles were present while MVBs were mostly absent, and F-actin appeared less dense with short crossed filament bundles.
Ultrastructural studies revealed that the WRAMP structure is associated with an ER pool in close proximity to the plasma membrane, which forms a network with actin filaments. In order to examine the relationship between ER and the WRAMP structure, cells were co-transfected with MCAM-GFP and a protein fusion between the KDEL ER protein retention motif and monomeric red fluorescent protein (KDEL-RFP). Live imaging showed that KDEL-RFP was recruited dynamically to the WRAMP structure and the membrane edge, prior to membrane retraction (Fig. 6D, Movie S2). We also found that calnexin, an integral membrane protein in ER which functions in protein folding, co-localized with MCAM in the WRAMP structure (Fig. 6E). Treatment of cells with Brefeldin A, which blocks recruitment of COP-I to membrane vesicles, also blocked WRAMP structure formation (Fig. 6F). Whereas KDEL-RFP and calnexin always co-localized with the WRAMP structure in Wnt5a-treated cells, the association of these ER markers was inhibited in cells treated with Brefeldin A or RNAi-COP-Iβ (data not shown). Thus, the WRAMP structure is associated with polarized movement of ER to the plasma membrane, in a manner that depends on COP-I. Recruitment of this ER pool is coordinated with WRAMP structure assembly, where it moves past MCAM to the membrane periphery, and precedes membrane retraction at the trailing edge.
The movement of ER to the WRAMP structure led us to hypothesize its involvement in polarized Ca2+ mobilization. Wnt5a is known to signal via Ca2+ and calmodulin, and Ca2+ release coupled to cortical ER has been implicated during the recruitment of proteins to focal adhesions in response to proinflammatory cytokines (Wang et al., 2006). In addition, ER is juxtaposed against the plasma membrane in response to Ca2+ depletion, enabling interactions between the ER Ca2+ sensor, STIM1, and plasma membrane CRAC-M Ca2+ ion channels (Wu et al., 2006; Park et al., 2009; Orci et al., 2009). Recruitment of cortical ER has been shown to involve Ist2p, an ER protein with a C-terminal dilysine motif which binds COP-I in vitro (Lavieu et al., 2010; Ercan & Seedorf, 2009). Thus, movement of ER to the cell periphery might involve COP-I interactions with proteins found within an ER subdomain.
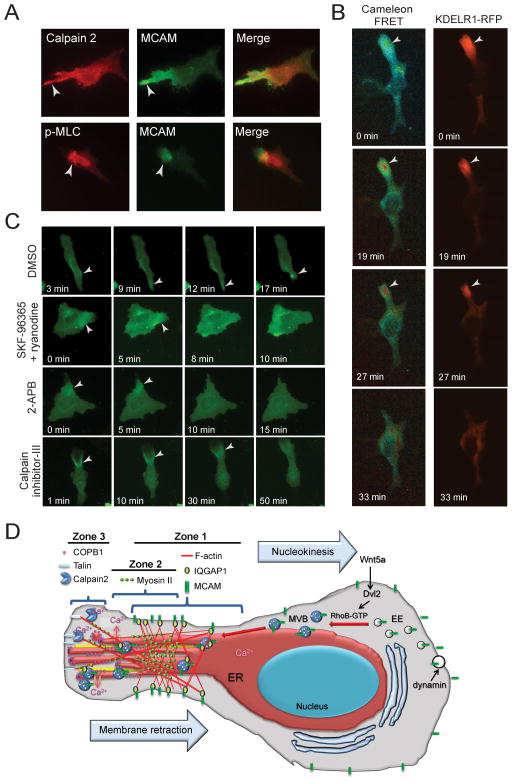
Although our cells were not Ca2+ depleted, the precedent that cortical ER, recruited through COP-I, mediates Ca2+ influx prompted us to hypothesize that the WRAMP structure might similarly function to mobilize Ca2+ at the cell posterior. At the trailing edge, Ca2+ would be needed to promote membrane detachment and retraction catalyzed by Ca2+-dependent proteases and MLCK. This possibility was supported by the appearance of calpain-2 and myosin II within the WRAMP proteome (Fig. 7A, Fig. S2C, M). Immunocytochemistry showed that both calpain-2 and phosphorylated myosin light chain colocalized with MCAM, suggesting that Ca2+-dependent enzymes are recruited to the WRAMP structure and may participate in membrane retraction.
Fig. 7. Coordination between the WRAMP structure and Ca2+ release.
(A) Immunocytochemistry of Wnt5a-treated cells shows recruitment of calpain-2 and phosphorylated myosin light chain to MCAM-GFP in the WRAMP structure. (B) Cells expressing Cameleon and KDEL-RFP show elevation of free Ca2+ accompanying ER recruitment, followed by membrane retraction (see Movie S3). See also Figure S3. (C) In cells expressing MCAM-GFP, Ca2+ channel inhibitors interfere with WRAMP-associated membrane retraction. DMSO control shows normal membrane retraction following WRAMP structure formation. Retraction is blocked by treatment with SKF-96365 + ryanodine prior to Wnt5a (see Movie S3). 2-APB causes MCAM-GFP disassembly when added at the moment of WRAMP structure formation (7 min, see Movie S3). Pretreating cells with calpain inhibitor-III prior to Wnt5a blocks membrane retraction, causing membranes to instead protrude from the WRAMP structure (see Movie S3). (D) A working model for the composition and function of the WRAMP structure, summarizing results from this and previous studies. In response to cell stimulation, MCAM and other receptors are internalized through dynamin-dependent endocytosis. The localization of MCAM within the MVBs and intralumenal vesicles associated with the WRAMP structure suggests that the assembly of the WRAMP structure occurs in part through trafficking of late endosomes which bypass the canonical pathway for lysosome degradation. This may involve RhoB, which is activated by Wnt5a and might inhibit lysosomal fusion. The appearance of tubulin, dynein, and nudC in the WRAMP proteome suggests an involvement of microtubules in polarized vesicle recruitment. The WRAMP structure consists of at least three overlapping subregions, or zones. MCAM, F-actin and IQGAP1 begin assembling in zone 1, located farthest from the trailing edge. Live cell imaging shows that these proteins dynamically translocate to the periphery, moving through zone 2 and finally arriving at zone 3, at the very tip of the trailing edge. We speculate that distinct events may occur within each subregion. For example, zone 1 might organize structures which initiate MVB recruitment and microfilament assembly. Zone 2 might serve to link MVBs to actomyosin, which is in turn linked to focal adhesion proteins in zone 3 which attach to transmembrane proteins and must be disassembled to allow membrane release. In this way, events that are spatially separated may combine to control the dynamics of the WRAMP structure and link organelle trafficking to membrane contractility.
We tested this hypothesis by transfecting cells with the Cameleon FRET biosensor and KDEL-RFP to simultaneously monitor free Ca2+ and ER dynamics. Live cell imaging showed increased Cameleon-FRET signal following the recruitment of KDEL-RFP to the WRAMP structure, and preceding membrane retraction (Fig. 7B, Movie S3). Similar responses were observed in cells co-expressing Cameleon and IQGAP1-RFP (data not shown). The results show that cytosolic Ca2+ increases transiently at the cell trailing edge, concomitant with WRAMP structure assembly and ER recruitment. Within the WRAMP structure, free Ca2+ transiently spiked to 0.8 μM from its basal concentration of 0.1 μM, whereas outside the WRAMP structure, Ca2+ remained unchanged (Fig. S3).
The importance of localized cytosolic Ca2+ for membrane retraction was examined using inhibitors which block Ca2+ mobilization. SKF-96365, 2-aminoethoxydiphenylborane (2-APB), and ryanodine respectively inhibit Ca2+ influx through plasma membrane voltage-gated Ca2+ channels, and ER-associated IP3 and ryanodine receptors. Cells were pre-treated with combination of SKF-96365 + ryanodine for 1 h then treated with Wnt5a and monitored live for MCAM-GFP. In response to SKF-96365 + ryanodine, membrane retraction was consistently repressed, and replaced by membrane ruffles extending from the WRAMP structure (Fig. 7C, Movie S3). Such effects were not observed when the inhibitors were applied separately (not shown), implying that Ca2+ mobilization depends on activation of ion channels both within the ER and plasma membrane. Likewise, 2-APB, an antagonist of IP3 receptors and store-operated Ca2+ entry Trp channels, led to WRAMP structure disassembly and repressed membrane retraction, when drug was added at the moment of WRAMP structure formation (7 min, Fig. 7C, Movie S3). Both events were derepressed when the inhibitor was subsequently removed (24 min, Movie S3). Together, these results demonstrate a role for the WRAMP structure in directing membrane retraction events by elevating free Ca2+ at the trailing edge.
We also tested the importance of calpain, which was recruited to the WRAMP structure and trailing edge membrane (Fig. 7A). The cell permeable calpain inhibitor, MDL-28170 (calpain inhibitor-III), did not affect the integrity of the WRAMP structure, but blocked membrane retraction and in fact led to membrane protrusions outward from the WRAMP structure (Fig. 7C, Movie S3). Calpain inhibitor-III decreased the number of cells exhibiting membrane retraction from 95±5% (n=13) to 36±10% (n=24), after forming a WRAMP structure. Thus, Ca2+ polarization by the WRAMP structure appears important for modulating proteolytic events involved in membrane detachment and retraction.
DISCUSSION
Our study demonstrates that rear-end cell polarization by Wnt5a involves active recruitment of cortical ER in coordination with the WRAMP structure. This finding was prompted by analyses of the WRAMP structure proteome, which revealed the presence of COP-I, suggesting an involvement of ER in WRAMP structure formation. The temporal coordination between movement of KDEL-RFP and calnexin ER markers to the WRAMP structure, enhanced Cameleon signal, and membrane retraction, suggests that the order of events triggered by WRAMP structure assembly involves ER recruitment followed by elevation of free Ca2+ at the cell posterior. This would in turn activate localized Ca2+-dependent enzymes releasing matrix attachments and allowing membrane retraction. The model is supported by the disruption of WRAMP structure assembly by Brefeldin A and COP-Iβ knockdown, and by disruption of WRAMP-associated membrane retraction by inhibitors of ER- and plasma membrane-associated Ca2+ channels. Likely targets include calpain-2, which catalyzes disassembly of integrin-substrate attachments by proteolyzing focal adhesion proteins, and MLCK, which catalyzes myosin light chain phosphorylation and actomyosin contraction. The coupling of these events to the WRAMP structure provides a mechanism that explains how the Wnt5a cell polarity pathway directs intracellular Ca2+ gradients and directional movement via the polarized assembly of receptor-cytoskeletal complexes.
Combining these findings with previous results, we propose a working model for Wnt5a-mediated cell polarity, summarized in Fig. 7D. In response to cell stimulation, MCAM and other receptors are internalized through dynamin-mediated endocytosis (Witze et al., 2008). The appearance of MCAM within MVBs and intralumenal vesicles associated with the WRAMP structure suggests that the polarization of MCAM and the WRAMP structure occurs through trafficking of a late endosomal pool which bypasses the canonical pathway for lysosome-mediated degradation. This may involve RhoB, which we found to be activated by Wnt5a and might serve to prevent lysosomal fusion (Witze et al., 2008, Gampel et al., 1999). The appearance of tubulin and dynein in the WRAMP proteome, as well as nuclear migration protein, nudC, which mediates interactions between kinesin, dynein and dynactin (Yamada et al., 2010), suggests the involvement of microtubules in polarized vesicle recruitment and microtubule motors for vesicle targeting to the WRAMP structure. This is consistent with observations that nocodazole interferes with WRAMP structure formation (E. Witze and N. Ahn, unpublished).
The WRAMP-associated MVBs might in turn recruit actomyosin through adaptor proteins that bind MCAM. For example, RhoB binds early and late endosomes and assembles actin cages around these vesicles by recruiting mDia1/2 formins (Wallar et al., 2006; Fernandez-Borja et al., 2005). IQGAP1, which is associated with and required for WRAMP structure formation, sustains nucleation of actin following formin activation by RhoA (Brandt et al., 2007), and could conceivably mediate vesicle-microfilament interactions by bridging MCAM and F-actin. MCAM binding to COP-I might in turn recruit ER to the WRAMP structure, through interactions between COP-I and dilysine motif-containing proteins such as KDELR.
Recruitment of ER may serve to localize Ca2+ release where it is needed at the site of membrane retraction. It was intriguing that ERK2 is present at the WRAMP structure in its active, phosphorylated form, along with the Ca2+-dependent protease, calpain-2 (Figs. 3D,7A). Localization of phospho-ERK at focal adhesions promotes calpain recruitment and protease activation, leading to proteolysis of focal adhesion proteins as a required step in adhesion disassembly (Carragher & Frame, 2004). ERKs also phosphorylate WRAMP proteins, including FAM129B/Minerva, which promotes melanoma cell invasion (Old et al., 2009).
Other proteins suggest the importance of microfilament and membrane protein interactions, in the WRAMP structure. Filamin-A and gelsolin are actin crosslinking and severing proteins which could stabilize the dense F-actin bundles found within the WRAMP structure (Nakamura et al., 2011; Yin & Stossel 1979). Talin-1 in the WRAMP structure might provide a means to anchor microfilaments to focal adhesions at the cortical plasma membrane. Kindlin3/FERMT3 binds talin-1 and the integrin β receptor, and modulates integrin activation and cell adhesion (Burridge & Connell, 1983; Moser et al., 2008). Filamin-A has a scaffold function, anchoring microfilaments to cell adhesion receptors ICAM1, CEACAM1, and integrin β, and mediating the activation of signaling effectors, such as Rho, ROCK, the RhoGEF, Trio, and the RhoGAP, FilGAP (Nakamura et al., 2011). Calpain cleaves interdomain linker regions in filamin-A, separating domains linking actin binding, homodimerization, and interactions with other binding partners. In melanoma cells, Wnt5a enhanced filamin-A proteolysis by calpain (O’Connell et al., 2009). This would provide a means by which localized Ca2+ release might disassemble the WRAMP structure, by disrupting MCAM-actin-plasma membrane interactions.
The localization of various components identified in this study suggest that the WRAMP structure is not a single homogeneous entity, but contains at least three overlapping subregions, or zones (Fig. 7D). MCAM, actomyosin and IQGAP1 begin assembling in zone 1, located farthest from the trailing edge (Witze et al., 2008; Figs. 1A, 3A). Live imaging shows that these proteins dynamically translocate to the periphery, moving through zone 2 and finally arriving at zone 3, the tip of the trailing edge. Our previous studies have shown that actin accumulation in zone 1 temporally follows MCAM (Witze et al., 2008). Clathrin also overlaps MCAM in zone 1, suggesting its potential association with MVBs. Zone 2 contains myosin IIb, partially overlapping MCAM but typically removed from zone 3 and the membrane edge prior to membrane retraction (Fig. 1A, 3H, Movie S1). Zone 3 contains talin-1, filamin-A, calpain-2, and ERK2, which function in microfilament-membrane attachments and focal adhesion formation/turnover. We speculate that distinct events may occur within each subregion. For example, zone 1 might organize structures which initiate MVB recruitment and microfilament assembly. Zone 2 might link MVBs to actomyosin, which in turn links to focal adhesion proteins in zone 3 that attach to transmembrane proteins and must be disassembled to allow membrane release. In this way, events that are spatially separated may combine to control the dynamics of the WRAMP structure and link organelle trafficking to membrane contractility.
Our findings integrate different lines of evidence explaining how mechanisms controlling cell polarity allow cell movement, suggesting that analogous processes may occur in other systems. Indeed, polarized MCAM-GFP associates with tail end retraction in HUVEC and C2C12 myoblast cells (Fig. S1), and in HT1080 fibrosarcoma and adult hippocampal progenitor cells (data not shown). Polarized actomyosin structures also occur during neuronal migration and axonal pathfinding in cortical neurons, which mediate tail end contraction and nucleokinesis, and are correlated with spatial Ca2+ transients (Martini & Valdeomillos, 2010). In leukocytes and Dictyostelium, rear protrusions called uropods modulate trailing edge movement by recruiting ezrin-radixin-moesin (ERM) proteins which assemble F-actin, myosin, and transmembrane receptors to the cell posterior (Sánchez-Madrid & Serrador). Uropods colocalize talin and F-actin, and myosin II-dependent force generation reorients microtubules to the cell posterior, where retraction is Ca2+-dependent (Eddy et al., 2002). In migrating lymphocytes, Ca2+ promotes release of uropods from the substrate through lysosomal fusion, which is mediated by synaptotagmin vesicle fusion proteins (Colvin et al., 2010). Finally in Dictyostelium, adenylate cyclase is released at uropods during chemotaxis through a mechanism involving vesicle trafficking, secretion of MVBs, and exosome shedding (Kriebel et al., 2008). Such observations share similarities with the presence of actomyosin, MVBs, and Ca2+ in the WRAMP structure and its role in directing tail end retraction during migration. Taken together, it seems likely that the WRAMP structure and its control of tail end polarity is not restricted to melanoma cells, but represents a general mechanism for controlling cell movement. Previous studies showed that Wnt5a, Fz and Dvl modulate Ca2+ release (Kohn & Moon, 2005). This and the noted role of Wnt5a in polarized cell movement might explain why Wnt5a, of all Wnt ligands, can direct the assembly of the WRAMP structure.
A significant finding in our study is the evidence that rear-directed polarization involves dynamic movement of internal membrane pools, including MVBs which recruit MCAM, and cortical ER which promotes localized Ca2+ release. COP-I subunits play a central role in this mechanism, suggesting that Wnt5a-dependent cell polarity may involve recruitment of ER via interactions between COP-I and cortical membrane proteins. COP-I is classically associated with Golgi/ERGIC membranes, although there are a few older controversial reports of its presence on ER or endosomal membranes (Bednarek at al., 1995; Gu et al., 2000). In addition, studies in malignant melanoma cells have shown that melanoma inhibitory activity (MIA) an autocrine protein which promotes cancer progression and metastasis, binds integrins α4β1 and α5β1, mediating membrane detachment (Schmidt et al., 2010). MIA is secreted at the trailing edge of cells through processes involving COP-I, as well as localized KCa3.1 K+ channels which are activated in response to intracellular Ca2+.
It is very likely that the WRAMP structure underlies processes of cancer cell invasion and metastasis. MCAM is upregulated in metastatic melanoma, and promotes cell invasion from epidermis to dermis in skin reconstruct models (Satyamoorthy et al., 2001). Wnt5a strongly enhances invasion of cultured melanoma cells, and its expression is correlated with advanced stage in melanoma specimens (Weeraratna et al., 2002). How Wnt5a promotes cancer cell invasion by controlling cell polarity is currently unknown. In neuronal cell systems, Wnt5a intersects with atypical PKC and PARtition complex proteins which directly bind Dishevelled, and control axon outgrowth through the organization of microtubule and actin dynamics (Zhang et al., 2007; Sun et al., 2001). Our findings show that mechanisms that polarize proteins to the tail end of cells are equally important for Wnt5a-dependent cell movement.
Studies of cancer cell invasion in 3D matrices reveal an important role of rear-end dynamics. Invasion is often classified into separate mechanisms. Mesenchymal mechanisms involve elongated cell morphologies, Rac-dependent leading edge actin polymerization, integrin-β1 polarization and MMP secretion. Amoeboid mechanisms involve rounded cell morphologies which excludes integrin and MMPs, and Rho and ROCK-dependent tail-end retraction (Sanz-Moreno et al., 2008). In A375 melanoma cells Lorentzen et al. (2011) described a uropod-like structure, named the “ezrin-radixin-uropod-like structure (ERULS)”. ERULS mediates amoeboid cell invasion, by tail-end enrichment of ezrin, phosphorylated ezrin, F-actin, myosin light chain, phosphatidylinositol 4,5-bisphosphate and integrin-β1. ERULS differs from other uropod structures in its regulation by protein kinase C and membrane attachment at the trailing edge, which in uropods is normally lifted away from the substrate. Like the WRAMP structure, the ERULS is dependent on Rho kinase (Sahai & Marshall, 2003). In WM239A cells, invasion through 3D collagen is clearly mesenchymal (Old et al., 2009), and invasion through 3D hydrogels requires proteolytic degradation and integrin signaling (Fig. 1), which differs from the standard view of amoeboid movement. Nevertheless, MCAM binds and recruits ezrin/moesin into cell protrusions in melanoma cells, facilitating cell movement by MCAM binding to RhoGDI1, RhoA activation, and ROCK-dependent ERM phosphorylation (Luo et al., 2011). Thus, it is reasonable to hypothesize that ERULS represents one form of the WRAMP structure in cells undergoing amoeboid cell movement, although the WRAMP structure appears to be a more general entity observed across different cell types and different invasion mechanisms.
In summary, polarized formation of the WRAMP structure through MCAM-associated MVBs, cytoskeletal-membrane attachments, and recruitment of cortical ER, provides a working model to explain how Wnt5a promotes invasion, by directing the Ca2+ mobilization needed for actomyosin contraction and tail end membrane retraction. Understanding how the composition and regulation of this machinery varies under different cellular contexts will provide new insight into how Wnt5a controls invasion and metastasis through the control of cell polarity.
EXPERIMENTAL PROCEDURES
Materials
Antibodies used were mouse αMCAM, rabbit αEEA1, rabbit αcalnexin, mouse αDvl2 (Santa Cruz Biotechnology), mouse αIQGAP1 (Invitrogen), mouse αclathrin (BD Transduction), αCOP-Iβ and αGFP (Abcam), rabbit αmyosin IIb and α58kDa Golgi protein (Sigma). Plasmids expressing MCAM-GFP, IQGAP1-RFP, KDEL-RFP (from Eric Snapp, Albert Einstein Univ.), myosin II heavy chain-RFP (from Robert Adelstein, NIH), and Cameleon FRET biosensor (from Roger Tsien, UC San Diego) were described (Witze et al., 2008; Jeong et al., 2006; Snapp et al., 2006; Wei & Adelstein, 2000; Miyawaki et al., 1999). Brefeldin A (Enzo LifeSciences) was added to cells at 10 μg/mL in serum-free RPMI for 2 h prior to treatment ±Wnt5a.
Cell culture
WM239A cells were maintained in RPMI+10% FBS. COP-Iβ, IQGAP1 and Dvl2 knockdowns were performed using siGenome SMARTpool or siRNA duplex oligonucleotides (Dharmacon). Cells were preincubated with inhibitors (2.5 μM) or DMSO for 1 h at 37°C, and media was replaced with RPMI+250 ng/mL Wnt5a. With 2-APB, cells were treated with Wnt5a and monitored live, adding inhibitor at the moment of WRAMP structure formation. For the calpain inhibitor III (EMD Millipore), cells were treated with the inhibitor (12 μM in DMSO) + Wnt5a or control buffer simultaneously and imaged live for one hour. Immunoprecipitations were performed with αGFP, αDvl2, or αCOP-Iβ antibodies, monitoring pulldowns by Western blotting.
Cells were cultured in hydrogels photopolymerized using 4-arm 20,000 MW PEG-norbornene (Fairbanks et al., 2009; Schwartz et al., 2010) crosslinked with peptides KKCGGPQG↓IWGQGCKK, with MMP-degradable sequence (Nagase & Fields, 1996) and CRGDS, with integrin recognition motif (Ruoslahti & Pierschbacher, 1987). A non-bioactive scrambled peptide, CRDGS, was used for control experiments (“0 RGD”). Live images were collected every 0.5 h for up to 36 h using a Nikon TE2000E microscope. Immunostaining was performed after 48 h, collected with a Zeiss LSM5 Pascal scanning confocal microscope. For ROCK inhibition, 10 μM Y27632 (Sigma) was added 2 h before imaging.
Organelle proteomics
WM239A (5.3 × 106) cells were treated ± Wnt5a, and harvested into lysis buffer (10 mM Tris pH 8, 1.5 mM MgCl2, 10 mM KCl). Post-nuclear supernatants were mixed with iodixanol to a final concentration of 17.5% and overlaid onto 700 μL cushions of 20% iodixanol, then overlaid with 700 μL 0.25 M sucrose in lysis buffer and centrifuged without braking (350,000xg, 2 h). 150 μL fractions were collected and added to 150 μL lysis buffer followed by centrifugation (60,000 rpm, 20 min). Pellets were washed, resuspended with 0.1% Rapigest, 50 mM NH4HCO3, and boiled for 10 min. Proteins were reduced, alkylated, and digested with trypsin (Promega) for 3 h at 37°C. Rapigest was cleaved with trifluoroacetic acid followed by centrifugation, and the top layer containing peptides was stored at −80°C.
LC-MS/MS was performed on a Thermo LTQ-Orbitrap mass spectrometer with a Waters nanoAcquity UPLC (BEH-C18 column, 25 cm × 75 μm i.d, 1.7 μm, 100Å, Waters). Peptide digests (2 μg) were separated by a linear gradient from 95% buffer A (0.1% formic acid) to 40% buffer B (0.1% formic acid, 80% CH3CN) over 120 min at 300 nL/min. For each MS scan, the 10 most intense ions were targeted for MS/MS with dynamic exclusion 30 s, 1 Da exclusion width, excluding ions with charge state +1 or unassigned. The maximum injection time was 500 ms for Orbitrap scans (AGC=1×106) and 500 ms for LTQ MS/MS (AGC=1×104).
Microscopy
For immunocytochemistry, 5×105 cells were seeded on glass coverslips for 24 h, washed, and incubated overnight in RPMI. The media was removed, RPMI+150 ng/mL Wnt5a was added, and cells incubated at 37°C for 30 min. Cells were fixed in 4% formalin at room temperature, permeabilized with 0.1% Triton X-100, blocked with 5% BSA, 0.1% Tween/Tris-buffered saline, and incubated with 1° antibody (1:200 dilution) followed by AlexaFluor-488 or -594 donkey αmouse or αrabbit 2° antibody (Invitrogen). Images were collected on an Olympus IX81 inverted microscope and analyzed with Slidebook software.
Live cell imaging was performed in an environmental chamber on an Olympus microscope. Cells were serum-starved, washed with Hanks’ Balanced Salt Solution (HBSS) and incubated in HBSS, 25 mM Hepes pH 7.4. Images were collected every 30–60 s on GFP and RFP channels with stage temperature 35°C. FRET imaging of the Cameleon sensor was performed on a Nikon Eclipse TE2000-S inverted microscope and images acquired with a CoolSNAP ES digital camera (Photometrics) and analyzed with MetaMorph software. Calibrations used Ca2+-free conditions (Rmin) by incubating cells with 5 μM ionomycin, 3 mM EGTA and Ca2+-saturating conditions of 5 μM ionomycin,10 mM CaCl2 (Palmer & Tsien, 2006).
For electron microscopy, cells were prepared as described (McDonald, 1984) with sample fixation in 50 mM sodium cacodylate, 2% glutaraldehyde, and post-fixation with 0.5% osmium tetroxide, 0.8% K3Fe(CN)6. Cells were stained with 0.15% tannic acid, dehydrated in acetone, and embedded in Epon Araldite resin. Images were collected with a Tecnai F-30 electron microscope (FEI Co.) and Gatan CCD camera, recording serial tilts of ± 60° in increments of 1° (Mastronarde, 2005). Each section was imaged in 2 tilt series around orthogonal axes and then assembled into a single reconstruction using IMOD software (Mastronarde, 1997). Tomographic reconstructions were modeled by manual contour tracing using IMOD (Kremer et al., 1996).
Supplementary Material
HIGHLIGHTS.
Wnt5a assembles a WRAMP structure, which directs cell movement via tail retraction.
A proteomics strategy is used to identify constituents of the WRAMP proteome.
WRAMP structure assembly is coordinated with recruitment of cortical ER.
WRAMP structure assembly leads to mobilization of a rear-directed Ca2+ signal.
Acknowledgments
We are indebted to Amy Palmer and William Old for many helpful discussions, to Robert Rogers for assistance with 3D cell culture, to Kevin Dean for assistance with light microscopy, and to Andreas Hoenger for access to The Boulder Laboratory for 3D Electron Microscopy, a National Research Resource. This work was supported by NIH grants F32-CA112847 (ESW), R01-CA118972 (NGA) and P41-RR000592 (AH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bednarek SY, Ravazzola M, Hosobuchi M, Amherdt M, Perrelet A, Schekman R, Orci L. COPI- and COPII-coated vesicles bud directly from the endoplasmic reticulum in yeast. Cell. 1995;83:1183–1196. doi: 10.1016/0092-8674(95)90144-2. [DOI] [PubMed] [Google Scholar]
- Brandt D, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage R, Fogarty K, Tuft R, Fay F. Calcium gradients underlying polarization and chemotaxis of eosinophils. Science. 1991;254:703–706. doi: 10.1126/science.1948048. [DOI] [PubMed] [Google Scholar]
- Burridge K, Connell L. Talin: a cytoskeletal component concentrated in adhesion plaques and other sites of actin-membrane interaction. Cell Motil. 1983;3:405–417. doi: 10.1002/cm.970030509. [DOI] [PubMed] [Google Scholar]
- Carragher N, Frame M. Focal adhesion and actin dynamics: a place where kinases and proteases meet to promote invasion. Trends Cell Biol. 2004;14:241–249. doi: 10.1016/j.tcb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Cramer L. Forming the cell rear first: breaking cell symmetry to trigger directed cell migration. Nat Cell Biol. 2010;12:628–632. doi: 10.1038/ncb0710-628. [DOI] [PubMed] [Google Scholar]
- Colvin R, Means T, Diefenbach T, Moita L, Friday R, Sever S, Campanella G, Abrazinski T, Manice L, Moita C, et al. Synaptotagmin-mediated vesicle fusion regulates cell migration. Nat Immunol. 2010;11:495–502. doi: 10.1038/ni.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy R, Pierini L, Maxfield F. Microtubule asymmetry during neutrophil polarization and migration. Mol Biol Cell. 2002;13:4470–4483. doi: 10.1091/mbc.E02-04-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan E, Momburg F, Engel U, Temmerman K, Nickel W, Seedorf M. A conserved, lipid-mediated sorting mechanism of yeast Ist2 and mammalian STIM proteins to the peripheral ER. Traffic. 2009;10:1802–1818. doi: 10.1111/j.1600-0854.2009.00995.x. [DOI] [PubMed] [Google Scholar]
- Fairbanks B, Schwartz M, Halevi A, Nuttelman C, Bowman C, Anseth K. A versatile synthetic extracellular matrix mimic via thiol-norbornene photopolymerization. Adv Mater. 2009;21:5005–5010. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Borja M, Janssen L, Verwoerd D, Hordijk P, Neefjes J. RhoB regulates endosome transport by promoting actin assembly on endosomal membranes through Dia1. J Cell Sci. 2005;118:2661–2670. doi: 10.1242/jcs.02384. [DOI] [PubMed] [Google Scholar]
- Gampel A, Parker P, Mellor H. Regulation of epidermal growth factor receptor traffic by the small GTPase rhoB. Curr Biol. 1999;9:955–958. doi: 10.1016/s0960-9822(99)80422-9. [DOI] [PubMed] [Google Scholar]
- Glading A, Uberall F, Keyse S, Lauffenburger D, Wells A. Membrane proximal ERK signaling is required for M-calpain activation downstream of epidermal growth factor receptor signaling. J Biol Chem. 2001;276:23341–23348. doi: 10.1074/jbc.M008847200. [DOI] [PubMed] [Google Scholar]
- Gu F, Gruenberg J. ARF1 regulates pH-dependent COP functions in the early endocytic pathway. J Biol Chem. 2000;275:8154–8160. doi: 10.1074/jbc.275.11.8154. [DOI] [PubMed] [Google Scholar]
- Hahn K, DeBiasio R, Taylor D. Patterns of elevated free calcium and calmodulin activation in living cells. Nature. 1992;359:736–738. doi: 10.1038/359736a0. [DOI] [PubMed] [Google Scholar]
- Jeong H, Li Z, Brown M, Sacks D. IQGAP1 binds Rap1 and modulates its activity. J Biol Chem. 2007;282:20752–20762. doi: 10.1074/jbc.M700487200. [DOI] [PubMed] [Google Scholar]
- Kohn AD, Moon RT. Wnt and calcium signaling: beta-catenin-independent pathways. Cell Calcium. 2005;38:439–446. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Kremer J, Mastronarde D, McIntosh J. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kriebel P, Barr V, Rericha E, Zhang G, Parent C. Collective cell migration requires vesicular trafficking for chemoattractant delivery at the trailing edge. J Cell Biol. 2008;183:949–961. doi: 10.1083/jcb.200808105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavieu G, Orci L, Shi L, Geiling M, Ravazzola M, Wieland F, Cosson P, Rothman J. Induction of cortical endoplasmic reticulum by dimerization of a coatomer-binding peptide anchored to endoplasmic reticulum membranes. Proc Natl Acad Sci USA. 2010;107:6876–6881. doi: 10.1073/pnas.1002536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, Didry D, Le D, Egile C, Carlier M, Kroschewski R. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem. 2007;282:426–435. doi: 10.1074/jbc.M607711200. [DOI] [PubMed] [Google Scholar]
- Lorentzen A, Bamber J, Sadok A, Elson-Schwab I, Marshall C. An ezrin-rich, rigid uropod-like structure directs movement of amoeboid blebbing cells. J Cell Sci. 2011;124:1256–1267. doi: 10.1242/jcs.074849. [DOI] [PubMed] [Google Scholar]
- Luo Y, Zheng C, Zhang J, Lu D, Zhuang J, Xing S, Feng J, Yang D, Yan X. Recognition of CD146 as an ERM-binding protein offers novel mechanisms for melanoma cell migration. Oncogene. 2011 doi: 10.1038/onc.2011.244. [DOI] [PubMed] [Google Scholar]
- Martini F, Valdeolmillos M. Actomyosin contraction at the cell rear drives nuclear translocation in migrating cortical interneurons. J Neurosci. 2010;30:8660–8670. doi: 10.1523/JNEUROSCI.1962-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Mastronarde D. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McDonald K. Osmium ferricyanide fixation improves microfilament preservation and membrane visualization in a variety of animal cell types. J Ultra Res. 1984;86:107–118. doi: 10.1016/s0022-5320(84)80051-9. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Griesbeck O, Heim R, Tsien R. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc Natl Acad Sci USA. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M, Nieswandt B, Ussar S, Pozgajova M, Fässler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med. 2008;14:325–330. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- Nagase H, Fields G. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Stossel T, Hartwig J. The filamins: organizers of cell structure and function. Cell Adh Migr. 2011;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M, Fiori J, Baugher K, Indig F, French A, Camilli T, Frank B, Earley R, Hoek K, Hasskamp J, et al. Wnt5A activates the calpain-mediated cleavage of filamin A. J Invest Dermatol. 2009;129:1782–1789. doi: 10.1038/jid.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old W, Meyer-Arendt K, Aveline-Wolf L, Pierce K, Mendoza A, Sevinsky J, Resing K, Ahn N. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- Old W, Shabb J, Houel S, Wang H, Couts K, Yen C, Litman E, Croy C, Meyer-Arendt K, Miranda J, Brown R, Witze E, Schweppe R, Resing K, Ahn N. Functional proteomics identifies targets of phosphorylation by B-Raf signaling in melanoma. Mol Cell. 2009;34:115–131. doi: 10.1016/j.molcel.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Le Coadic M, Shen W, Demaurex N, Cosson P. STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2009;106:19358–19362. doi: 10.1073/pnas.0911280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AE, Tsien RY. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc. 2006;1(3):1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- Park C, Hoover P, Mullins F, Bachhawat P, Covington E, Raunser S, Walz T, Garcia K, Dolmetsch R, Lewis R. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J, Horwitz A, Schwartz M. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R, Doyle A, Yamada K. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoslahti E, Pierschbacher M. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- Sahai E, Marshall C. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- Salas-Cortes L, Ye F, Tenza D, Wilhelm C, Theos A, Louvard D, Raposo G, Coudrier E. Myosin Ib modulates the morphology and the protein transport within multi-vesicular sorting endosomes. Cell Sci. 2005;118:4823–32. doi: 10.1242/jcs.02607. [DOI] [PubMed] [Google Scholar]
- Sánchez-Madrid F, Serrador J. Bringing up the rear: defining the roles of the uropod. Nat Rev Mol Cell Biol. 2009;10:353–359. doi: 10.1038/nrm2680. [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall C. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Satyamoorthy K, Muyrers J, Meier F, Patel D, Herlyn M. Mel-CAM-specific genetic suppressor elements inhibit melanoma growth and invasion through loss of gap junctional communication. Oncogene. 2001;20:4676–4684. doi: 10.1038/sj.onc.1204616. [DOI] [PubMed] [Google Scholar]
- Schmidt J, Friebel K, Schönherr R, Coppolino M, Bosserhoff A. Migration-associated secretion of melanoma inhibitory activity at the cell rear is supported by KCa3.1 potassium channels. Cell Res. 2010;20:1224–1238. doi: 10.1038/cr.2010.121. [DOI] [PubMed] [Google Scholar]
- Schwartz MP, Fairbanks BD, Rogers RE, Rangarajan R, Zaman MH, Anseth KS. A synthetic strategy for mimicking the extracellular matrix provides new insight about tumor cell migration. Integr Biol. 2010;2:32–40. doi: 10.1039/b912438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp EL, Sharma A, Lippincott-Schwartz J, Hegde RS. Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc Natl Acad Sci U S A. 2006;103:6536–6541. doi: 10.1073/pnas.0510657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Lu B, Feng J, Reinhard C, Jan Y, Fantl W, Williams L. PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat Cell Biol. 2001;3:628–636. doi: 10.1038/35083016. [DOI] [PubMed] [Google Scholar]
- Wallar B, Deward A, Resau J, Alberts A. RhoB and the mammalian Diaphanous-related formin mDia2 in endosome trafficking. Exp Cell Res. 2007;313:560–571. doi: 10.1016/j.yexcr.2006.10.033. [DOI] [PubMed] [Google Scholar]
- Wang Q, Herrera-Abreu M, Siminovitch K, Downey G, McCulloch C. Phosphorylation of SHP-2 regulates interactions between the endoplasmic reticulum and focal adhesions to restrict interleukin-1-induced Ca2+ signaling. J Biol Chem. 2006;281:31093–31105. doi: 10.1074/jbc.M606392200. [DOI] [PubMed] [Google Scholar]
- Weeraratna A, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, Trent J. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–288. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- Wei C, Wang X, Chen M, Ouyang K, Song L, Cheng H. Calcium flickers steer cell migration. Nature. 2009;457:901–905. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Adelstein R. Conditional expression of a truncated fragment of nonmuscle myosin II-A alters cell shape but not cytokinesis in HeLa cells. Mol Biol Cell. 2000;11:3617–3627. doi: 10.1091/mbc.11.10.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witze E, Litman E, Argast G, Moon R, Ahn N. Wnt5a control of cell polarity and directional movement by polarized redistribution of adhesion receptors. Science. 2008;320:365–369. doi: 10.1126/science.1151250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Buchanan J, Luik R, Lewis R. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Toba S, Takitoh T, Yoshida Y, Mori D, Nakamura T, Iwane A, Yanagida T, Imai H, Yu-Lee L, et al. mNUDC is required for plus-end-directed transport of cytoplasmic dynein and dynactins by kinesin-1. EMBO J. 2010;29:517–531. doi: 10.1038/emboj.2009.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Stossel T. Control of cytoplasmic actin gel-sol transformation by gelsolin, a calcium-dependent regulatory protein. Nature. 1979;281:583–586. doi: 10.1038/281583a0. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhu J, Yang G, Wang Q, Qian L, Chen Y, Chen F, Tao Y, Hu H, Wang T, Luo Z. Dishevelled promotes axon differentiation by regulating atypical protein kinase C. Nat Cell Biol. 2007;9:743–754. doi: 10.1038/ncb1603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.