Abstract
The title compound, C15H15NO4S, was obtained by the condensation of 4-aminoacetophenone and 4-methoxybenzenesulfonyl chloride. The dihedral angle between the benzene rings is 86.56 (9)° and the molecule has an approximate V-shaped conformation. The C atom of the methoxy group is roughly coplanar with its attached ring [deviation = 0.177 (3) Å], as is the methyl C atom of the acetyl group with its ring [deviation = 0.065 (2) Å]. An intramolecular C—H⋯O interaction generates an S(6) ring. In the crystal, N—H⋯O and C—H⋯O hydrogen bonds link the molecules into [010] chains. Weak C—H⋯π interactions are also observed.
Related literature
For related structures, see: Li et al. (2006 ▶); Xu et al. (2005 ▶). For background to and applications of sulfonamides, see: Alsughayer et al. (2011 ▶); Dragostin et al. (2013 ▶);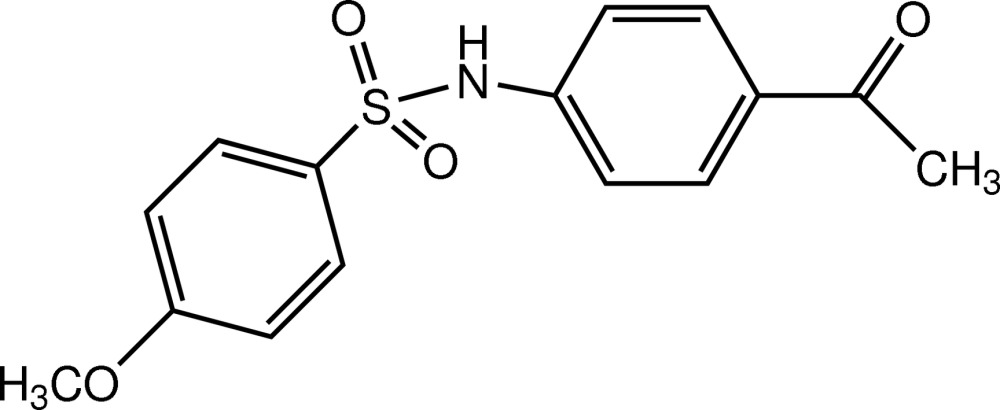
Experimental
Crystal data
C15H15NO4S
M r = 305.35
Monoclinic,
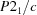
a = 12.8220 (3) Å
b = 8.2709 (2) Å
c = 14.6165 (4) Å
β = 112.841 (1)°
V = 1428.52 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.24 mm−1
T = 298 K
0.48 × 0.44 × 0.33 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.894, T max = 0.924
15571 measured reflections
4120 independent reflections
2593 reflections with I > 2σ(I)
R int = 0.043
Refinement
R[F 2 > 2σ(F 2)] = 0.047
wR(F 2) = 0.135
S = 1.04
4120 reflections
196 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.20 e Å−3
Δρmin = −0.34 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL, PLATON (Spek, 2009 ▶), Mercury (Macrae et al., 2006 ▶) and publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813029875/hb7153sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813029875/hb7153Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813029875/hb7153Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 and Cg2 are the centroids of the C1–C6 and C7–C12 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N1⋯O3i | 0.85 (2) | 2.05 (2) | 2.896 (2) | 172.3 (19) |
| C8—H8A⋯O2 | 0.93 | 2.38 | 3.030 (2) | 127 |
| C9—H9A⋯O1ii | 0.93 | 2.53 | 3.459 (2) | 174 |
| C14—H14A⋯Cg1i | 0.96 | 2.83 | 3.630 (3) | 141 |
| C14—H14C⋯Cg2iii | 0.96 | 2.83 | 3.529 (2) | 130 |
| C15—H15C⋯Cg1iv | 0.96 | 2.99 | 3.804 (3) | 144 |
Symmetry codes: (i) 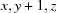 ; (ii)
; (ii)  ; (iii)
; (iii) 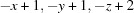 ; (iv)
; (iv) 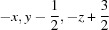 .
.
Acknowledgments
Financial support from the Thailand Research Fund through the Royal Golden Jubilee PhD Program (grant No. PHD/0137/2554) is gratefully acknowledged. CSCK thanks the Universiti Sains Malaysia for a postdoctoral research fellowship. The authors extend their appreciation to Prince of Songkla University and the Universiti Sains Malaysia for the APEX DE2012 grant No. 1002/PFIZIK/910323.
supplementary crystallographic information
1. Comment
Sulfonamides containing an –SO2NH– group can be found in many pharmacologically active compounds: recent reports have described antibacterial (Alsughayer et al., 2011) and antioxidant (Dragostin et al., 2013) behaviour. As part of our ongoing research in this field, the title compound (I), a 4-methoxybenzene-sulfonamide derivative, was synthesized for being used as starting material for various syntheses. Herein the crystal structure of (I) is reported.
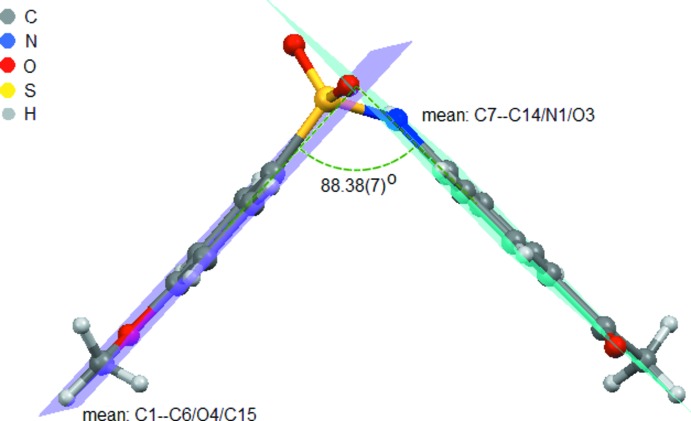
Figure 1 shows the molecular structure of (I), C15H15NO4S, suggesting a V-shaped conformation (Fig. 2). The benzene rings make the dihedral angle of 86.56 (9)°. The methoxy group is almost co-planar with its attached benzene ring with the deviation of 0.0310 (2) Å for the eight non H atoms (C1–C6/O4/C15) and the torsion angle C15–O4–C4–C5 = -2.2 (3)°. The amide group and acetyl substituent also lie in almost the same plane with the bound benzene ring with the deviation of 0.0220 (2) Å for the ten non H atoms (C7–C14/N1/O3) and the torsion angles of C11–C10–C13–O3 = -177.28 (18)° and C11–C10–C13–C14 = 2.6 (3)°. The dihedral angle between these two planes [C1–C6/O4/C15 and C7–C14/N1/O3] is 88.38 (7)° (Fig. 2). An intramolecular C8—H8A···O2 weak interaction generates an S(6) ring (Fig. 1) Bond distances of (I) are comparable with those in related structures (Li et al., 2006 and Xu et al., 2005).
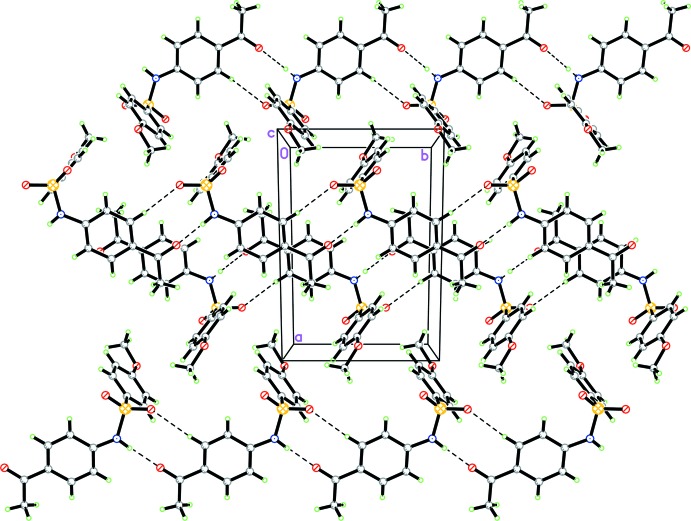
In the crystal (Fig. 3), the molecules are linked by N—H···O hydrogen bonds and C—H···O weak interactions (Table 1) into chains along [010]. Weak C—H···π interactions are also observed (Table 1).
2. Experimental
The title compound was synthesized by condensation of 4-aminoacetophenone (0.40 g, 3 mmol) and 4-methoxybenzenesulfonyl chloride in CH2Cl2 (30 ml) in the presence of pyridine. The reaction mixture was refluxed for 24 hr at 40 °C and monitored with TLC for the completion of the reaction. Water was then added and the concoction was extracted with CH2Cl2. The solvent was evaporated under reduced pressure to yield the resulting solid of the title compound (yield 68%). Yellow blocks of (I) were recrystalized from acetone:CH3OH solution (1:1 v/v) by slow evaporation of the solvent at room temperature after several days, Mp. 448–449 K.
3. Refinement
Amide H atoms was located from the difference maps and refined isotropically. The remaining H atoms were positioned geometrically and allowed to ride on their parent atoms, with d(C—H) = 0.93 Å for aromatic and 0.96 for CH3 atoms. The Uiso values were constrained to be 1.5Ueq of the carrier atom for methyl H atoms and 1.2Ueq for the remaining H atoms. A rotating group model was used for the methyl groups.
Figures
Fig. 1.

The molecular structure of (I), showing 40% probability displacement ellipsoids. The intramolecular C—H···O hydrogen bond is shown as a dashed line.
Fig. 2.
The V-shape conformation of the molecule.
Fig. 3.
The crystal packing of the title compound viewed along the c axis. Hydrogen bonds were shown as dashed lines.
Crystal data
| C15H15NO4S | F(000) = 640 |
| Mr = 305.35 | Dx = 1.420 Mg m−3 |
| Monoclinic, P21/c | Melting point = 448–449 K |
| Hall symbol: -P 2ybc | Mo Kα radiation, λ = 0.71073 Å |
| a = 12.8220 (3) Å | Cell parameters from 4120 reflections |
| b = 8.2709 (2) Å | θ = 1.7–29.9° |
| c = 14.6165 (4) Å | µ = 0.24 mm−1 |
| β = 112.841 (1)° | T = 298 K |
| V = 1428.52 (6) Å3 | Block, yellow |
| Z = 4 | 0.48 × 0.44 × 0.33 mm |
Data collection
| Bruker APEXII CCD diffractometer | 4120 independent reflections |
| Radiation source: sealed tube | 2593 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.043 |
| φ and ω scans | θmax = 29.9°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −17→17 |
| Tmin = 0.894, Tmax = 0.924 | k = −11→11 |
| 15571 measured reflections | l = −20→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.047 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.135 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0625P)2 + 0.0913P] where P = (Fo2 + 2Fc2)/3 |
| 4120 reflections | (Δ/σ)max = 0.001 |
| 196 parameters | Δρmax = 0.20 e Å−3 |
| 0 restraints | Δρmin = −0.34 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.21179 (4) | 1.00630 (5) | 1.08283 (4) | 0.04186 (15) | |
| O1 | 0.21200 (12) | 1.17413 (16) | 1.10664 (11) | 0.0546 (4) | |
| O2 | 0.14687 (11) | 0.89569 (17) | 1.11408 (11) | 0.0529 (4) | |
| O3 | 0.48523 (14) | 0.21499 (16) | 1.11408 (12) | 0.0624 (4) | |
| O4 | 0.05688 (12) | 0.98389 (16) | 0.64850 (11) | 0.0540 (4) | |
| N1 | 0.34483 (13) | 0.95337 (19) | 1.13298 (12) | 0.0408 (4) | |
| C1 | 0.16988 (14) | 0.9867 (2) | 0.95362 (14) | 0.0378 (4) | |
| C2 | 0.22355 (15) | 1.0787 (2) | 0.90451 (14) | 0.0418 (4) | |
| H2A | 0.2849 | 1.1439 | 0.9404 | 0.050* | |
| C3 | 0.18492 (15) | 1.0722 (2) | 0.80272 (15) | 0.0432 (4) | |
| H3A | 0.2209 | 1.1321 | 0.7697 | 0.052* | |
| C4 | 0.09193 (16) | 0.9760 (2) | 0.74878 (14) | 0.0415 (4) | |
| C5 | 0.04067 (17) | 0.8825 (2) | 0.79829 (15) | 0.0487 (5) | |
| H5A | −0.0201 | 0.8161 | 0.7627 | 0.058* | |
| C6 | 0.07979 (16) | 0.8882 (2) | 0.90017 (15) | 0.0466 (5) | |
| H6A | 0.0454 | 0.8254 | 0.9333 | 0.056* | |
| C7 | 0.38891 (14) | 0.7983 (2) | 1.12776 (12) | 0.0353 (4) | |
| C8 | 0.32855 (16) | 0.6554 (2) | 1.12191 (14) | 0.0422 (4) | |
| H8A | 0.2549 | 0.6591 | 1.1191 | 0.051* | |
| C9 | 0.37892 (16) | 0.5091 (2) | 1.12029 (15) | 0.0427 (4) | |
| H9A | 0.3381 | 0.4144 | 1.1157 | 0.051* | |
| C10 | 0.48954 (16) | 0.4993 (2) | 1.12535 (13) | 0.0381 (4) | |
| C11 | 0.54871 (16) | 0.6432 (2) | 1.13233 (14) | 0.0422 (4) | |
| H11A | 0.6230 | 0.6394 | 1.1368 | 0.051* | |
| C12 | 0.49943 (15) | 0.7902 (2) | 1.13275 (14) | 0.0413 (4) | |
| H12A | 0.5400 | 0.8849 | 1.1364 | 0.050* | |
| C13 | 0.54079 (17) | 0.3380 (2) | 1.12393 (14) | 0.0427 (4) | |
| C14 | 0.66124 (18) | 0.3279 (3) | 1.13447 (15) | 0.0542 (5) | |
| H14A | 0.6814 | 0.2168 | 1.1316 | 0.081* | |
| H14B | 0.7094 | 0.3736 | 1.1970 | 0.081* | |
| H14C | 0.6704 | 0.3870 | 1.0815 | 0.081* | |
| C15 | −0.0427 (2) | 0.8943 (3) | 0.59083 (17) | 0.0666 (6) | |
| H15A | −0.0602 | 0.9117 | 0.5215 | 0.100* | |
| H15B | −0.1050 | 0.9300 | 0.6067 | 0.100* | |
| H15C | −0.0297 | 0.7812 | 0.6057 | 0.100* | |
| H1N1 | 0.3889 (18) | 1.031 (2) | 1.1334 (15) | 0.047 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0414 (2) | 0.0396 (3) | 0.0475 (3) | 0.00720 (19) | 0.0204 (2) | −0.00056 (19) |
| O1 | 0.0597 (9) | 0.0422 (8) | 0.0628 (9) | 0.0125 (6) | 0.0248 (7) | −0.0084 (6) |
| O2 | 0.0485 (8) | 0.0588 (9) | 0.0603 (9) | 0.0041 (6) | 0.0309 (7) | 0.0070 (7) |
| O3 | 0.0769 (10) | 0.0329 (7) | 0.0842 (11) | −0.0013 (7) | 0.0386 (9) | 0.0008 (7) |
| O4 | 0.0561 (9) | 0.0552 (9) | 0.0464 (9) | −0.0140 (7) | 0.0152 (7) | −0.0038 (6) |
| N1 | 0.0411 (8) | 0.0332 (8) | 0.0468 (10) | 0.0015 (7) | 0.0156 (7) | −0.0022 (7) |
| C1 | 0.0337 (8) | 0.0337 (9) | 0.0464 (10) | 0.0046 (7) | 0.0158 (8) | 0.0028 (7) |
| C2 | 0.0365 (9) | 0.0369 (10) | 0.0508 (12) | −0.0058 (7) | 0.0156 (8) | −0.0021 (8) |
| C3 | 0.0404 (10) | 0.0378 (10) | 0.0528 (12) | −0.0039 (8) | 0.0196 (9) | 0.0016 (8) |
| C4 | 0.0412 (9) | 0.0374 (10) | 0.0438 (11) | 0.0011 (8) | 0.0143 (8) | −0.0015 (8) |
| C5 | 0.0444 (10) | 0.0437 (11) | 0.0526 (12) | −0.0126 (8) | 0.0130 (9) | −0.0018 (9) |
| C6 | 0.0435 (10) | 0.0425 (10) | 0.0552 (12) | −0.0073 (8) | 0.0209 (9) | 0.0039 (9) |
| C7 | 0.0381 (8) | 0.0344 (9) | 0.0321 (9) | 0.0022 (7) | 0.0121 (7) | −0.0007 (7) |
| C8 | 0.0385 (9) | 0.0405 (10) | 0.0496 (11) | −0.0001 (8) | 0.0193 (8) | 0.0004 (8) |
| C9 | 0.0455 (10) | 0.0337 (9) | 0.0513 (11) | −0.0035 (8) | 0.0213 (9) | 0.0010 (8) |
| C10 | 0.0440 (9) | 0.0346 (9) | 0.0356 (9) | 0.0016 (7) | 0.0153 (8) | 0.0026 (7) |
| C11 | 0.0373 (9) | 0.0411 (10) | 0.0494 (11) | 0.0022 (8) | 0.0179 (8) | 0.0020 (8) |
| C12 | 0.0391 (9) | 0.0346 (9) | 0.0506 (11) | −0.0030 (7) | 0.0180 (8) | 0.0004 (8) |
| C13 | 0.0552 (11) | 0.0377 (10) | 0.0356 (10) | 0.0069 (8) | 0.0180 (9) | 0.0038 (7) |
| C14 | 0.0590 (12) | 0.0497 (12) | 0.0530 (13) | 0.0166 (10) | 0.0208 (10) | −0.0003 (9) |
| C15 | 0.0587 (14) | 0.0790 (16) | 0.0519 (14) | −0.0175 (12) | 0.0103 (11) | −0.0074 (12) |
Geometric parameters (Å, º)
| S1—O2 | 1.4261 (14) | C7—C12 | 1.392 (2) |
| S1—O1 | 1.4308 (13) | C7—C8 | 1.397 (2) |
| S1—N1 | 1.6335 (16) | C8—C9 | 1.376 (2) |
| S1—C1 | 1.7590 (19) | C8—H8A | 0.9300 |
| O3—C13 | 1.218 (2) | C9—C10 | 1.394 (3) |
| O4—C4 | 1.358 (2) | C9—H9A | 0.9300 |
| O4—C15 | 1.433 (3) | C10—C11 | 1.394 (2) |
| N1—C7 | 1.416 (2) | C10—C13 | 1.491 (2) |
| N1—H1N1 | 0.85 (2) | C11—C12 | 1.371 (2) |
| C1—C6 | 1.381 (3) | C11—H11A | 0.9300 |
| C1—C2 | 1.397 (3) | C12—H12A | 0.9300 |
| C2—C3 | 1.375 (3) | C13—C14 | 1.494 (3) |
| C2—H2A | 0.9300 | C14—H14A | 0.9600 |
| C3—C4 | 1.395 (3) | C14—H14B | 0.9600 |
| C3—H3A | 0.9300 | C14—H14C | 0.9600 |
| C4—C5 | 1.387 (3) | C15—H15A | 0.9600 |
| C5—C6 | 1.375 (3) | C15—H15B | 0.9600 |
| C5—H5A | 0.9300 | C15—H15C | 0.9600 |
| C6—H6A | 0.9300 | ||
| O2—S1—O1 | 119.39 (9) | C9—C8—C7 | 119.50 (17) |
| O2—S1—N1 | 108.79 (8) | C9—C8—H8A | 120.3 |
| O1—S1—N1 | 104.35 (9) | C7—C8—H8A | 120.3 |
| O2—S1—C1 | 108.17 (9) | C8—C9—C10 | 121.67 (17) |
| O1—S1—C1 | 108.75 (8) | C8—C9—H9A | 119.2 |
| N1—S1—C1 | 106.72 (8) | C10—C9—H9A | 119.2 |
| C4—O4—C15 | 117.12 (16) | C11—C10—C9 | 117.90 (16) |
| C7—N1—S1 | 126.00 (13) | C11—C10—C13 | 122.34 (17) |
| C7—N1—H1N1 | 113.8 (14) | C9—C10—C13 | 119.76 (16) |
| S1—N1—H1N1 | 112.0 (14) | C12—C11—C10 | 121.26 (17) |
| C6—C1—C2 | 120.09 (18) | C12—C11—H11A | 119.4 |
| C6—C1—S1 | 120.10 (15) | C10—C11—H11A | 119.4 |
| C2—C1—S1 | 119.69 (14) | C11—C12—C7 | 120.25 (16) |
| C3—C2—C1 | 119.49 (17) | C11—C12—H12A | 119.9 |
| C3—C2—H2A | 120.3 | C7—C12—H12A | 119.9 |
| C1—C2—H2A | 120.3 | O3—C13—C10 | 120.55 (18) |
| C2—C3—C4 | 120.26 (18) | O3—C13—C14 | 119.98 (17) |
| C2—C3—H3A | 119.9 | C10—C13—C14 | 119.47 (17) |
| C4—C3—H3A | 119.9 | C13—C14—H14A | 109.5 |
| O4—C4—C5 | 124.41 (17) | C13—C14—H14B | 109.5 |
| O4—C4—C3 | 115.76 (17) | H14A—C14—H14B | 109.5 |
| C5—C4—C3 | 119.82 (18) | C13—C14—H14C | 109.5 |
| C6—C5—C4 | 119.90 (18) | H14A—C14—H14C | 109.5 |
| C6—C5—H5A | 120.0 | H14B—C14—H14C | 109.5 |
| C4—C5—H5A | 120.0 | O4—C15—H15A | 109.5 |
| C5—C6—C1 | 120.40 (18) | O4—C15—H15B | 109.5 |
| C5—C6—H6A | 119.8 | H15A—C15—H15B | 109.5 |
| C1—C6—H6A | 119.8 | O4—C15—H15C | 109.5 |
| C12—C7—C8 | 119.41 (16) | H15A—C15—H15C | 109.5 |
| C12—C7—N1 | 117.40 (16) | H15B—C15—H15C | 109.5 |
| C8—C7—N1 | 123.14 (17) | ||
| O2—S1—N1—C7 | 53.22 (18) | C2—C1—C6—C5 | −1.4 (3) |
| O1—S1—N1—C7 | −178.31 (15) | S1—C1—C6—C5 | 174.65 (15) |
| C1—S1—N1—C7 | −63.27 (17) | S1—N1—C7—C12 | 151.37 (15) |
| O2—S1—C1—C6 | 5.98 (17) | S1—N1—C7—C8 | −31.2 (3) |
| O1—S1—C1—C6 | −125.08 (15) | C12—C7—C8—C9 | −0.6 (3) |
| N1—S1—C1—C6 | 122.88 (15) | N1—C7—C8—C9 | −177.92 (17) |
| O2—S1—C1—C2 | −177.94 (13) | C7—C8—C9—C10 | 0.6 (3) |
| O1—S1—C1—C2 | 51.00 (16) | C8—C9—C10—C11 | 0.1 (3) |
| N1—S1—C1—C2 | −61.03 (15) | C8—C9—C10—C13 | 179.65 (17) |
| C6—C1—C2—C3 | 1.0 (3) | C9—C10—C11—C12 | −1.0 (3) |
| S1—C1—C2—C3 | −175.13 (14) | C13—C10—C11—C12 | 179.50 (17) |
| C1—C2—C3—C4 | 0.8 (3) | C10—C11—C12—C7 | 1.0 (3) |
| C15—O4—C4—C5 | 2.7 (3) | C8—C7—C12—C11 | −0.3 (3) |
| C15—O4—C4—C3 | −176.22 (18) | N1—C7—C12—C11 | 177.24 (17) |
| C2—C3—C4—O4 | 176.85 (16) | C11—C10—C13—O3 | −177.28 (18) |
| C2—C3—C4—C5 | −2.2 (3) | C9—C10—C13—O3 | 3.2 (3) |
| O4—C4—C5—C6 | −177.21 (18) | C11—C10—C13—C14 | 2.6 (3) |
| C3—C4—C5—C6 | 1.7 (3) | C9—C10—C13—C14 | −176.96 (18) |
| C4—C5—C6—C1 | 0.1 (3) |
Hydrogen-bond geometry (Å, º)
Cg1 and Cg2 are the centroids of the C1–C6 and C7–C12 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N1···O3i | 0.85 (2) | 2.05 (2) | 2.896 (2) | 172.3 (19) |
| C8—H8A···O2 | 0.93 | 2.38 | 3.030 (2) | 127 |
| C9—H9A···O1ii | 0.93 | 2.53 | 3.459 (2) | 174 |
| C14—H14A···Cg1i | 0.96 | 2.83 | 3.630 (3) | 141 |
| C14—H14C···Cg2iii | 0.96 | 2.83 | 3.529 (2) | 130 |
| C15—H15C···Cg1iv | 0.96 | 2.99 | 3.804 (3) | 144 |
Symmetry codes: (i) x, y+1, z; (ii) x, y−1, z; (iii) −x+1, −y+1, −z+2; (iv) −x, y−1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB7153).
References
- Alsughayer, A., Elassar, A.-Z. A., Mustafa, S. & Sagheer, F. A. (2011). J. Biomater. Nanobiotechnol 2, 144–149.
- Bruker (2009). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Dragostin, O. M., Lupascu, F., Vasile, C., Mares, M., Nastasa, V., Moraru, R. F., Pieptu, D. & Profire, L. (2013). Molecules, 18, 4140–4157. [DOI] [PMC free article] [PubMed]
- Li, X.-M., Zeng, C.-C., Xu, Y.-S., Yan, H., Zheng, D.-W. & Zhong, R. G. (2006). J. Chem. Crystallogr 36, 357–363.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst D65, 148–155. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Xu, Y.-S., Zeng, C.-C., Li, X.-M. & Zhong, R.-G. (2005). Acta Cryst. E61, o3802–o3804.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813029875/hb7153sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813029875/hb7153Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813029875/hb7153Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report