Abstract
Aim
miRNAs are showing utility as biomarkers in urologic disease, however, a rigorous evaluation of their stability in urine is lacking. Here, we evaluate the stability of miRNAs in urine under clinically relevant storage procedures.
Materials & methods
Eight healthy individuals provided clean catch urine samples that were stored at room temperature or at 4°C for 5 days, or subjected to ten freeze–thaw cycles at -80°C. For each condition, two miRNAs, miR-16 and miR-21, were quantitated by quantitative real-time PCR.
Results
All conditions demonstrated a surprising degree of stability of miRNAs in the urine: by the end of ten freeze–thaw cycles, 23–37% of the initial amount remained; over the 5-day period of storage at room temperature, 35% of the initial amount remained; and at 4°C, 42–56% of the initial amount remained. Both miRNAs also showed degradation at approximately the same rate.
Conclusion
miRNAs are relatively stable in urine under a variety of storage conditions, which supports their utility as urinary biomarkers.
Keywords: biomarkers, cancer, miRNA, stability, urine
There is growing evidence that miRNAs, which are small ncRNAs made up of 19–23 nucleotides, play regulatory roles in gene expression. In addition to other diseases, miRNAs have been found to be dysregulated in urologic diseases, such as malignancies of the prostate, bladder and kidney [1], exerting their effects as oncogenes or tumor suppressors [2], and in nephrological diseases such as glomerulonephritis [3] and fibrosis [4]. Circulating blood miRNAs have been investigated as novel approaches for biomarker discovery, and more recently, miRNAs have been detected and quantitated in other body fluids including urine, saliva and cerebrospinal fluid [5]. However, the few published reports of urinary miRNAs as potential biomarkers [5,6] have no supporting studies evaluating the stability and possible degradation of miRNAs in urine or optimal storage conditions for urine, in contrast to what is reported for blood miRNAs [7,8].
Given that urine is one of the most easily accessible and noninvasive biofluids available in urology, nephrology and primary care clinics, our laboratory has been searching for compounds that can be stably identified in the urine and can be used as specific biomarkers for renal disease. While there is little question that a metabolomic approach has resulted in stable urinary biomarker prospects in the form of small molecule metabolites [9–11], the possibility that miRNAs may be similar candidates is not at all clear due to the potential instability of miRNA in the harsh urinary milieu.
Supporting the likelihood of identifying clinically useful urinary miRNAs, more than 40 such miRNAs have now been identified and linked to specific urological pathologies, including intrinsic kidney disease, renal cell carcinoma and bladder cancer [1,5,12,13]. However, while there has been some attempt to identify urinary miRNA biomarkers for nephrological disease such as lupus [14], IgA nephropathy [15] and chronic allograft nephropathy [16], there has been no assessment of miRNA stability in these studies as a function of storage times and conditions; this would appear to be a missing yet necessary piece of information going forward with urinary miRNA biomarker studies. The purpose of the current study was to attempt to remedy this situation and to assess urinary miRNA stability under various clinically relevant conditions: room temperature and 4°C, as well as serial freeze–thaw conditions. We now show that urinary miRNA is relatively stable under all of the conditions evaluated. While there is some degradation over time, such degradation results in levels of two representative miRNAs well within the upper limit of qPCR detection at the cycle threshold (CT) value of 35 [17,18], suggesting excellent utility as biomarkers. In addition, the rate of degradation of the miRNAs examined was similar and independent of miRNA sequences of the representative miRNAs, suggesting that the relative stability of miRNA is a common property and that multiplexing using several miRNAs is indeed feasible. This work sets the stage for future research on identifying specific urinary miRNA biomarkers for a variety of kidney diseases, and reinforces the robustness and clinical utility of existing as well as forthcoming studies utilizing urinary miRNA for biomarker studies.
Materials & methods
Urine collection
After appropriate Institutional Review Board approval, clean catch midstream urine samples were obtained from eight healthy individuals (four males and four females), ranging from ages 25–57 years. Voids from the same subjects were taken multiple times for each of the parts of the study: to assess stability of miRNA levels, for pellet versus uncentrifuged urine miRNA quantification and for the urinary exosome experiments. For miRNA stability experiments, aliquots of 1 ml from a 30–50-ml clean catch void were prepared and stored at 4°C, room temperature (∼25°C) or subjected to freeze–thaw cycles from -80°C to room temperature. Each urine sample aliquot for the 4°C and room temperature experiments were processed for RNA isolation and quantification via reverse transcriptase quantitative PCR for miR-16 and miR-21 at time points 0, 1, 3, 6 and 12 h then daily (i.e., every 24 h) until day five. Ten freeze–thaw cycles were completed for sample aliquots kept at -80°C.
Isolation of miRNA from urine
For the study of pellet versus uncentrifuged urinary miRNA isolation, two 1-ml aliquots of urine were set aside from three subjects. One aliquot was centrifuged at 25,000 × g at 4°C for 15 min. After separating the supernatant from the pellet into a new tube, the pellet, supernatant and uncentrifuged aliquots were all processed for miRNA isolation per the manufacturer's protocol (miRNeasy Mini Kit, Qiagen, CA, USA). For the stability experiments, each freeze–thaw aliquot was first thawed on ice prior to proceeding with centrifugation. In all subsequent experiments, 1-ml aliquots were first centrifuged at 25,000 × g at 4°C for 15 min, the supernatant was discarded and the manufacturer's protocol from the miRNeasy Mini Kit was adapted for purification and isolation of miRNA.
Quantitative real-time PCR techniques
Quantification and real-time PCR was carried out via quantitative real-time PCR (qRT-PCR) using miRNA-specific TaqMan® MicroRNA Assays (Applied Biosystems, CA, USA) for miR-16 (UAGCAGCACGUAAAUAUUGGCG) and miR-21 (UAGCUUAUCAGACUGAUGUUGA) per the manufacturer's protocol. The reverse transcription reactions were performed in conjunction with the TaqMan miRNA Reverse Transcription Kit (Applied Biosystems) with RNA eluate to attain cDNA on the Peltier thermal cycler (MJ Research, MA, USA) at 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. Next, combining the miRNA-specific primers and probe mix of the miRNeasy Mini Kit, TaqMan® Universal PCR Master Mix, No AmpErase® UNG (Applied Biosystems), nuclease-free water and reverse transcription product per sample, qRT-PCR reactions were set up in triplicate in a 384-well plate. The plate was sealed using MicroAmp™ Optical Adhesive Film (Applied Biosystems) and briefly centrifuged. Reactions were carried out on the ViiA 7 Real-Time PCR System (Applied Biosystems) at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. No template and no reverse transcription controls were performed for each qPCR reaction plate.
Urinary exosome isolation
Urinary exosomes were isolated from 1-ml aliquots of fresh urine from five of the eight randomly selected subjects, as has been described using differential centrifugation, which involves multiple centrifugations of increasing speeds allowing for the isolation and purification of nanosized urinary exosomes [19]. In the case of urine treated with trypsin to elute possible exosome-bound structures or macromolecules [20], 0.25% trypsin was added to 1 ml of fresh urine and allowed to incubate for 15 min at 37°C prior to isolation of exosomes.
Statistical analysis
For urinary miRNA studies, changes in CT values over time or over multiple freeze–thaw cycles were analyzed using linear models including effects for time (or number of freeze–thaw cycles), subject and their interaction. For the pellet versus uncentrifuged urine study, CT values were compared between miRNA isolated from urine pellets versus uncentrifuged urine using a linear mixed effects model including fixed effects for treatment and sex, and a random intercept term for subject. Post-hoc pairwise comparisons between treatments were made using the Tukey honestly significant difference approach. All analyses were conducted in R, Version 2.13.0 (R Development Core Team, 2011). Linear fits were used to estimate the average rate of degradation because, although the rate varies over time, it does not do so in a systematic way that could be modeled from a statistical stand point. The exponentiated slope from regression was used to derive percent changes in CT values; for example, the regression slope calculated from CT data was transformed to 2-slope in order to reflect changes in quantity on the 2-CT scale.
Results
Optimal processing conditions of urine samples & preparation of RNA
When compared with mRNA and other cellular RNAs, miRNA has shown a surprising degree of stability for reasons that are as yet unknown. Even though urine contains abundant nucleases [21], others have reported miRNAs to be identifiable in the urine, with their stability attributed to nuclease resistance due to smaller nucleic acid size [6] and/or microvesicular containment [22].
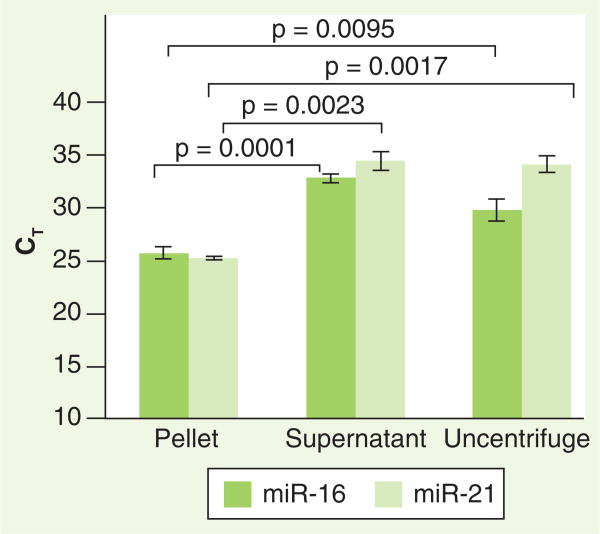
In order to determine the optimal initial preparation conditions for subsequent examination of urinary miRNA (i.e., urinary pellet vs uncentrifuged urine), we first obtained freshly voided, clean-catch urine samples from three healthy volunteers and immediately processed the aliquots for total RNA isolation. Two 1-ml aliquots of urine from each void were obtained. One of these aliquots was subjected to centrifugation, and the pellet and supernatant were separated for further analysis, and the other aliquot was not centrifuged. Total RNA was isolated from each of these fractions and all were analyzed for miR-16 and miR-21 content by qRT-PCR as described in the ‘Materials & methods’ section. For all samples, there were significantly higher levels of both miR-16 and miR-21 (manifested by lower CT values) in the urinary pellet compared with levels in the supernatant and uncentrifuged urine (Figure 1). Whether these differences were due to free-floating urinary miRNA or to that contained within cells or other debris that would appear in the centrifugation pellet was not addressed owing to the lack of relevance to these initial biomarker studies. However, based on these data, in subsequent studies of miRNA stability we analyzed miRNA in the higher concentration urinary pellet, as would most closely mimic an ideal clinical assay conditions.
Figure 1. Higher levels of miRNA are found in the microcentrifuged urinary pellet.
Two 1-ml aliquots of clean-catch urine from a normal subject were processed for RNA isolation. One aliquot was subjected to maximum speed centrifugation (25,000 × g) in a table-top microcentrifuge for 15 min at 4°C. The pellet and supernatant were separated and total RNA was isolated. The second aliquot was processed immediately for total RNA isolation without centrifugation. From these fractions, quantitation of miR-16 and miR-21 was performed as described in the ‘Materials & methods’ section. Error bars indicate standard deviation (p < 0.05).
CT: Cycle threshold.
Relative CT values are proportional to urinary miRNA quantity
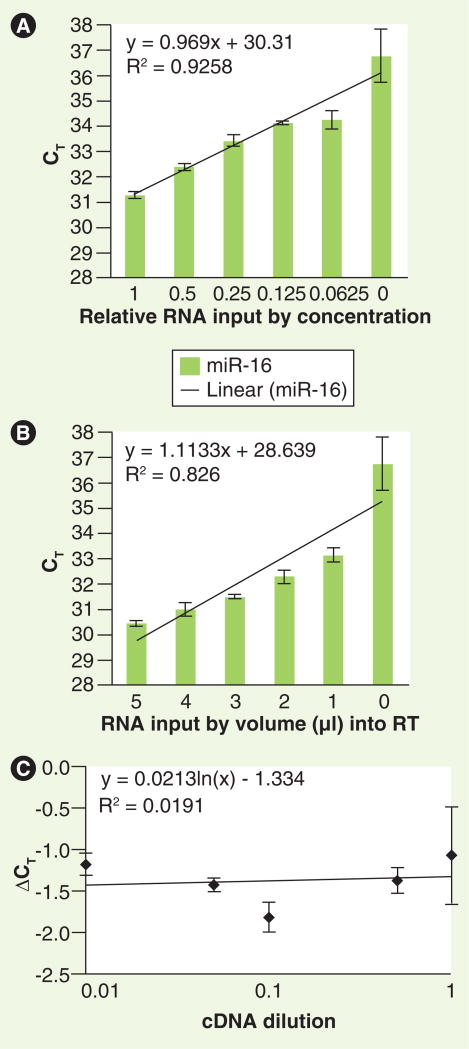
To confirm that the miRNA-specific qRT-PCR assays utilized in this study accurately reflect relative miRNA levels within the urine, we performed serial dilution studies of RNA isolated for the miR-16 analysis. After total RNA was isolated from one volunteer's urine, miR-16 was quantitated by qRT-PCR as a function of either total RNA concentration input into the reverse transcriptase reaction, or as a function of volume of isolated RNA input from the same urine sample into the reverse transcriptase reactions. In the first experiment, isolated total RNA was added to the reverse transcriptase reaction at different relative concentrations via serial dilutions of RNA solution by adding nuclease-free water (Figure 2A); in the second experiment, different volumes of RNA solution (from the same initial RNA solution as the first experiment) were added into the reverse transcriptase reaction (Figure 2B). From the reverse transcription reactions, miR-16 content was quantitated by qPCR as described in the ‘Materials & methods’ section. As is evidenced by the linear relationships between CT values and urinary concentrations (R2 = 0.9258 and 0.826), subsequent experiments measuring relative miRNA levels are quantitatively accurate.
Figure 2. Relative quantitation of urinary miRNA is reflected by cycle threshold values.
Total RNA was isolated from one normal subject and was added to the RT reaction at: (A) various concentrations via serial dilutions of RNA by adding nuclease-free water; or (B) various volumes of the same RNA. Subsequently, miR-16 was quantitated from these reactions as described in the ‘Materials & methods’ section. Error bars indicate standard deviation. (C) Target (miR-16) and reference (U6) amplifications were performed, at the cDNA dilutions indicated, from a single urine sample. The change in CT value between miR-16 and U6 at each cDNA dilution (ΔCT) is shown.
CT: Cycle threshold; RT: Reverse transcription.
To validate and generalize the amplification efficiencies of miRNA in urine, we evaluated U6 levels and assessed the ΔCT relative to template dilution for miR-16 as has been previously reported for validating relative gene expression [23]. U6 was used as a housekeeping miRNA as it has been shown to remain conserved interspecies [24]. Both target and reference amplifications were performed at log cDNA dilutions from a single urine sample. The slope of ΔCT relative to template dilution being close to zero (Figure 2C) confirms that the amplification efficiency of a reference miRNA is similar to that of a reference RNA when detected under the same conditions.
Human urinary miRNA remains relatively stable under various storage conditions
To evaluate urinary miRNA stability under various clinically relevant storage conditions, we isolated total RNA from urine samples and subsequently quantified miRNA by qRT-PCR. The changes in the CT values over time or over multiple freeze–thaw cycles from the qRT-PCR output (in CT values) are expressed in terms of percent change in actual miRNA in this section (see Tables 1 & 2 and statistical analysis section of the ‘Materials & methods’ section) [25]. However, the analysis units were CT values that are shown in Supplementary Figures 1–3 (see online at www.futuremedicine.com/doi/suppl/10.2217/BMM.13.44).
Table 1.
Relative quantitation of miRNA after freeze–thaw storage.
| miRNA | Estimated remaining after each freeze–thaw cycle, % (95% CI) | Estimated remaining after ten freeze–thaw cycles, % (95% CI) |
|---|---|---|
| miR-16 | 86 (82–91) | 23 (14–39) |
| miR-21 | 90 (86–95) | 37 (23–59) |
Table 2.
Relative quantitation of miRNA after room temperature and 4°C storage.
| Condition | miRNA | Estimated remaining after each day, % (95% CI) | Estimated decrease over 5 days, % (95% CI) |
|---|---|---|---|
| Room temperature | miR-16 | 81 (77–86) | 35 (26–46) |
| miR-21 | 81 (75–87) | 35 (24–50) | |
|
| |||
| 4°C | miR-16 | 84 (79–90) | 42 (31–58) |
| miR-21 | 89 (83–95) | 56 (39–79) | |
We utilized two representative miRNAs: miR-21, postulated to be altered within renal cell carcinoma tissue compared with normal tissue [21]; and miR-16, a putative reference miRNA [26]. After subjecting to freeze–thaw (at -80°C to room temperature), storage at room temperature (∼25°C) or at 4°C, total RNA was isolated and miRNA was quantitated. When evaluated under the condition of freeze–thawing (daily cycles from -80°C to room temperature), the quantity of miR-16 showed a decrease of 86% of the previous value after each cycle (95% CI: 82–91; Table 1 & Supplementary Figure 1A), and miR-21 showed a decrease of 90% of the previous value for each cycle (95% CI: 86–95; Table 1 & Supplementary Figure 1B). After ten freeze–thaw cycles, the quantity of miR-16 declined to 23% of the initial value (95% CI: 14–39) and the quantity of miR-21 declined to 37% of the initial value (95% CI: 23–59; Table 1).
Next, the degradation rate of the miRNAs was examined at room temperature (∼25°C). The quantity of miR-16 in urine was shown to decrease each day on average to 81% of the amount the previous day (95% CI: 77–86; Table 2 & Supplementary Figure 2A). Over a 5-day period, the quantity of miR-16 decreased on average to 35% of the initial amount (95% CI: 26–46; Table 2). The quantity of miR-21 in urine was shown to decrease each day on average to 81% of the amount of the previous day (95% CI: 75–87; Table 2 & Supplementary Figure 2B). Over a 5-day period, the quantity of miR-21 decreased on average to 35% of the initial amount (95% CI: 24–50; Table 2).
When stored at 4°C for up to 5 days, the quantity of miR-16 decreased each day on average to 84% of the amount the previous day (95% CI: 79–90; Table 2 & Supplementary Figure 3A). Over a 5 day period, the quantity of miR-16 was shown to decrease on average to 42% of the initial amount (95% CI: 31–58; Table 2). The quantity of miR-21 at 4°C decreased each day on average to 89% of the amount of the previous day (95% CI: 83–95; Table 2 & Supplementary Figure 3B), and over a 5 day period, the amount of miR-21 was shown to decrease on average to 56% of the initial amount (95% CI: 39–79; Table 2).
Thus, under all potential storage conditions evaluated, there was relative stability in the amount of detectable miRNA for up to 5 days at room temperature, 4°C and 10 freeze–thaw cycles. The maximum degradation in miRNA quantity, with ten freeze–thaw cycles, was 28.1% for miR-16. Our finding that the CT values under all storage conditions remained well below the detectable CT value of 35 [17,18] supports the use of urine as a clinically reliable biomarker.
Different miRNAs show similar relative degradation & amplification efficiencies
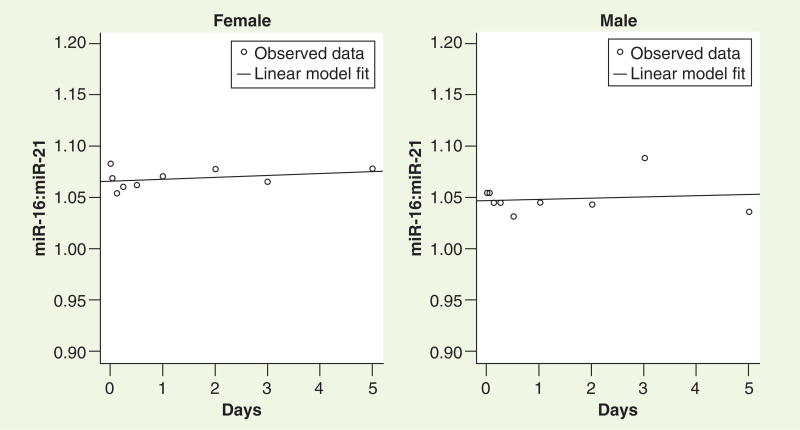
The utility of biomarkers, especially those in the urine, will likely lie in multiplexing rather than in using a single miRNA as a marker of disease. For this reason, it is important to determine whether the rate of degradation of miRNAs of different lengths and/or sequences is similar when measured under the same storage conditions. The rates of degradation of the two miRNAs evaluated in this study at one representative condition (room temperature) were compared, and both of the measured miRNAs appeared to have similar degradation rates as observed by CT ratios (calculated change in ratio of -0.0006/day; 95% CI: -0.005–0.003; Figure 3). To support the generalizability of this phenomenon, these miRNAs were aligned using an online BLAST [101] and no homology was noted, thus these two miRNAs represent completely different sequences.
Figure 3. Nonhomologous miRNAs degrade at similar rates.
From the room temperature data (Supplementary Figure 2), the ratios of the cycle threshold values of both miRNAs were calculated and data from a representative male and female volunteer are shown here. The two miRNAs remained constant over time with the calculated change in ratio of -0.0006 per day with a 95% CI of -0.005–0.003, implying that miRNAs most likely degrade at a constant rate over time.
Urinary miRNA is resistant to trypsin
To test the possibility that urinary miRNAs are associated with urinary proteins or whether they exist as an integral component of proteins on the exterior of exosomes, we assessed the effect of trypsin digestion on the quantity of urinary miRNA [20]. For these experiments, we utilized differential centrifugation because exosomes, released from the cells into the urinary space [19], are difficult to isolate and purify to homogeneity using slow-speed centrifugation alone [27].
Exosomes from five volunteers' urine were isolated using differential centrifugation as described in the ‘Materials & methods’ section; no normalization to concentration of urine was necessary in these experiments because the same void was used for each experiment. qRT-PCR was performed on miRNA isolated from these fractions using primers for miR-16 and miR-21. There was no change in the levels of miR-16 or miR-21 (Table 3) as assessed by CT value when the same fraction of urine was treated with 0.25% trypsin. Thus, the unusual stability of urinary miRNA is not due to its association with exosome-bound structures or macromolecules in urine; these data further support the robustness of urinary miRNA as potential biomarkers.
Table 3.
Comparison of concentrations of miR-16 and miR-21 by treatment of urine with trypsin.
| miRNA | Ratio (95% CI) | p-value |
|---|---|---|
| miR-16: exosome versus exosome with trypsin | 1.19 (0.31–4.54) | 0.988 |
| miR-21: exosome versus exosome with trypsin | 1.12 (0.24–5.20) | 0.998 |
Discussion
While there exist some promising leads from the fields of proteomics [28,29] and metabolomics [10,11], there are currently no specific biomarkers for the early detection of many renal diseases, and there is a paucity of urinary biomarker tests for any urologic disease [6]. The use of a urine RNA-based test may at first appear to be fraught with problems given the known liability of RNA; however, there is increasing evidence that miRNA shows surprising stability in situations in which total RNA or mRNA have been shown to be degraded [22]. In the present study, we have evaluated the relative stability of miRNAs in urine from healthy subjects with an eye towards developing an miRNA-based bioassay for genitourinary and renal pathologies; we found a surprising degree of stability under clinically relevant storage conditions.
miRNAs are 18–25 nucleotides long, small noncoding RNAs that are found to have regulatory effects on messenger RNA expression and activity [30]. As regulators of post-transcriptional expression of a variety of genes, miRNAs impact many renal diseases including glomerulonephritis and fibrosis. There are reports of detection of miRNAs in various and disparate nonurine biofluids including plasma, serum, cerebrospinal fluid, breast milk, and tears [5,7,8], yet none of these, to date, has resulted in the validation of a successful biomarker. The data in our study supports broad applicability of urinary miRNA for a variety of diseases of the genitourinary tract. While the bar is high in terms of specificity for any clinically applicable screening test, limiting the sampling to a population at high risk for the specific disease can make such considerations achievable.
In pursuit of potential applications for miRNAs as biomarkers in genitourinary disease, several miRNAs have already been shown to be associated with urologic cancers. For example, miR-126 and miR-182 were recently shown as dysregulated in bladder cancer and successfully detected in urine of patients with this disease [6], and another report identifies 65 miRNAs that were found to be altered in metastatic and primary renal cell carcinomas in formalin-fixed, paraffin-embedded tissues [31]. miR-210, observed to increase under hypoxic conditions, has been shown to play a role in pancreatic, breast and renal cancers [19,32–34]. For these reasons, and in light of the stability data shown in this study, miRNAs would now appear to have great potential as markers in genitourologic malignancies; examination of specific potential urinary miRNA biomarkers of kidney cancer is currently being pursued in our laboratory.
As far as intrinsic renal disease is concerned, there have been a few studies looking at miRNAs in the urine. In IgA nephropathy, it has been shown that urinary levels of miR-146a and miR-155 were significantly elevated, and that the degree of upregulation correlated with clinical and histological severity of the disease [15,35]. This same group evaluated urinary miRNAs in lupus, and found that some miRNAs were lower in urine of lupus patients compared with controls [14]. In the field of renal transplantation, there are several studies correlating urinary miRNA levels with acute rejection [36] and with chronic allograft nephropathy [16]. However, our study presented here was one of the very few in which both stability of miRNA and reproducibility of its quantitation were precisely evaluated. The study which evaluated stability, in contrast to our data, showed no ‘discernible’ loss of miR-21 expression in the urine of three healthy volunteers after four and 24 h of room temperature storage or after four freeze–thaw cycles [36]. It is not clear from that study whether the cellular and protein debris was removed prior to analysis, which may account for the differences between this study and ours. Furthermore, since there are currently many available kits to isolate miRNA, it is possible that the use of alternative kits may have an effect on quantity and/or quality of miRNA isolated from urine samples. However, since most kits currently available use similar protocols for serum, urine, plasma and other biofluids, we did not utilize such alternative methods. The use of these kits would not affect our principal findings of urine stability under various conditions; in fact, the use of the same kit for all experiments as we did avoids introducing external analytical variability.
While we observed consistent and detectable miRNA over time and freeze–thaw cycles, we found degradation over time of the two miRNAs studied, both of which degraded relatively consistently under all conditions. Our data show that urinary miRNA is best measured within the first 24 h for the most accurate representation of miRNAs in the urine milieu; of the conditions evaluated, storage at 4°C was found to have the least degradation (42–56% reduction in miRNA after 5 days). We did not evaluate miRNA stored over time at -80°C, but this would likely be more stable than 4°C. In all storage conditions evaluated, our finding that the CT values were well below the detectable CT value of 35 [17,18] supports the use of urine as a clinically reliable biomarker. It should be noted that we used linear fits to estimate average rate of degradation of miRNA in our samples, which is a standard procedure. Essentially, though the rates varied over time, they did not do so in a systemic way that we could model. Rather, this was representative of differences between individuals. We accounted for these differences between individuals in the analysis, and these differences would not be removed by increasing the sample size. Rather, increasing the sample size could further narrow the confidence intervals by decreasing effects of standard deviations, however, they are already sufficiently narrow to support our hypothesis.
It should also be noted that there are many factors that may play a role in the observed person-to-person variability of the pattern of CT changes seen in our experiments; for example, unforeseen slight variations in room temperature; or varying 4°C temperature with use (opening and closing of the refrigerator). This study assessing two miRNAs does not allow us to draw firm conclusions on whether or not all miRNAs degrade in a similar manner. However, supporting our data, different miRNAs have shown similar stability when evaluated in plasma and serum [8]. In addition, our data suggest similar degradation patterns and amplification efficiencies over time as a factor of temperature and freeze–thaw cycles, although it should be kept in mind that other miRNAs could have somewhat disparate stability parameters than what we have observed.
Recent studies have also shed light onto the role of exosomes in carrying miRNAs for intercellular communication. Exosomes have been found to originate from endocytosis of the plasma membrane [19]. Exosomes arise when a microvesicular body fuses with the cell membrane, hence exosomes are essentially lipid bilayers but also contain a large number of surface proteins [20]. The unusual stability of miRNA in several biofluids as well as tissues has been attributed to their location within exosomes [12,20], with these organelles theoretically providing a barrier to nucleases. In some cases, miRNAs have been found to bind to exosome-associated surface adaptor proteins such as HDL [37] or Ago2 [38], a situation that could also contribute to their stability. Because exosomes have been previously shown to contain cellular miRNA upon its exit from the cell [12,39], our focus was not whether urinary miRNA was found within exosomes but rather whether miRNA is contained on exosome-associated or other proteins, as this could contribute to its stability. Using trypsin digestion, we showed that degradation of proteins that have been previously associated with miRNA did not alter miRNA stability.
Conclusion & future perspective
In this study we have shown that miRNA is relatively stable in the harsh urinary environment. After storage for 5 days at varied temperatures and after ten freeze–thaw cycles, despite modest degradation, there remained sufficient miRNA for quantitative analysis. Furthermore, the rates of degradation were similar in two different nonhomologous miRNAs, supporting the concept of miRNA multiplexing. Given the growing interest and data concerning the utility of miRNA as potential biomarkers, these molecules are now eminently suitable for closer examination as specific biomarkers of renal disease. Over the next few years, it is likely that specific urinary miRNA biomarkers, used either singly or multiplexed, will become a standard of clinical practice in kidney disease.
Executive summary.
Relative cycle threshold values are proportional to urinary miRNA quantity.
Human urinary miRNA remains relatively stable under various storage conditions.
Different miRNAs show similar relative degradation and amplification efficiencies.
Urinary miRNA is resistant to trypsin.
Urinary miRNAs can be considered useful biomarkers for a variety of nephrological and urological diseases.
Footnotes
Financial & competing interests disclosure: This work was supported by NIH grants 1R01CA135401-01A1 and 1R01DK082690-01A1, and the Medical Service of the US Department of Veterans' Affairs (all to RH Weiss). The authors have no other relevant affliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research: The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1▪.Catto JW, Alcaraz A, Bjartell AS, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59:671–681. doi: 10.1016/j.eururo.2011.01.044. Important as a proof of identification of urological miRNAs. [DOI] [PubMed] [Google Scholar]
- 2▪▪.Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27(Suppl 2):S52–S57. doi: 10.1038/onc.2009.353. Discusses the probable applicability of urinary miRNAs as biomarkers of cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3▪.Kato M, Park JT, Natarajan R. MicroRNAs and the glomerulus. Exp Cell Res. 2012;318:993–1000. doi: 10.1016/j.yexcr.2012.02.034. Important as a proof of concept that miRNAs from the kidney can be found in urine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macconi D, Tomasoni S, Romagnani P, et al. MicroRNA-324–3p promotes renal fibrosis and is a target of ACE inhibition. J Am Soc Nephrol. 2012;23:1496–1505. doi: 10.1681/ASN.2011121144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5▪.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. Demonstrates that miRNAs are detectable in urine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6▪▪.Hanke M, Hoefig K, Merz H, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. Discusses miRNAs found in urine as biomarkers of disease (bladder cancer) [DOI] [PubMed] [Google Scholar]
- 7.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8▪.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. Presents the possibility of miRNAs in exosomes to be the reason for their stability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor SL, Ganti S, Bukanov NO, et al. A metabolomics approach using juvenile cystic mice to identify urinary biomarkers and altered pathways in polycystic kidney disease. Am J Physiol Renal Physiol. 2010;298:F909–F922. doi: 10.1152/ajprenal.00722.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganti S, Taylor S, Kim K, et al. Urinary acylcarnitines are altered in kidney cancer. Int J Cancer. 2012;130(12):2791–2800. doi: 10.1002/ijc.26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K, Taylor SL, Ganti S, Guo L, Osier MV, Weiss RH. Urine metabolomic analysis identifies potential biomarkers and pathogenic pathways in kidney cancer. OMICS. 2011;15:293–303. doi: 10.1089/omi.2010.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12▪▪.Miranda KC, Bond DT, McKee M, et al. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010;78:191–199. doi: 10.1038/ki.2010.106. Presents the possibility that urinary miRNAs are contained in exosomes for stability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada Y, Enokida H, Kojima S, et al. miR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011;102:522–529. doi: 10.1111/j.1349-7006.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang G, Tam LS, Kwan BC, et al. Expression of miR-146a and miR-155 in the urinary sediment of systemic lupus erythematosus. Clin Rheumatol. 2012;31:435–440. doi: 10.1007/s10067-011-1857-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang G, Kwan BC, Lai FM, Chow KM, Li PK, Szeto CC. Elevated levels of miR-146a and miR-155 in kidney biopsy and urine from patients with IgA nephropathy. Dis Markers. 2011;30:171–179. doi: 10.3233/DMA-2011-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scian MJ, Maluf DG, David KG, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am J Transplant. 2011;11:2110–2122. doi: 10.1111/j.1600-6143.2011.03666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55:1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 18.Kwok PY, Gu Z. Single nucleotide polymorphism libraries: why and how are we building them? Mol Med Today. 1999;5:538–543. doi: 10.1016/s1357-4310(99)01601-9. [DOI] [PubMed] [Google Scholar]
- 19▪▪.Gonzales PA, Zhou H, Pisitkun T, et al. Isolation and purification of exosomes in urine. Methods Mol Biol. 2010;641:89–99. doi: 10.1007/978-1-60761-711-2_6. Demonstrates methods of the urinary exosomal miRNA isolation protocol. [DOI] [PubMed] [Google Scholar]
- 20.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 21.Juan D, Alexe G, Antes T, et al. Identification of a microRNA panel for clear-cell kidney cancer. Urology. 2010;75:835–841. doi: 10.1016/j.urology.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 22.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- 25.Gusev Y, editor. MicroRna Profiling in Cancer: a Bioinformatics Perspective. Pan Stanford Publishing; Singapore: 2010. [Google Scholar]
- 26.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids – the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Minamida S, Iwamura M, Kodera Y, et al. 14-3-3 protein beta/alpha as a urinary biomarker for renal cell carcinoma: proteomic analysis of cyst fluid. Anal Bioanal Chem. 2011;401:245–252. doi: 10.1007/s00216-011-5057-5. [DOI] [PubMed] [Google Scholar]
- 29.Yokomizo A, Takakura M, Kanai Y, Sa, et al. Use of quantitative shotgun proteomics to identify fibronectin 1 as a potential plasma biomarker for clear cell carcinoma of the kidney. Cancer Biomark. 2011;10:175–183. doi: 10.3233/CBM-2012-0243. [DOI] [PubMed] [Google Scholar]
- 30.Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol. 2010;10:543–550. doi: 10.1016/j.coph.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 31.White NM, Khella HW, Grigull J, et al. miRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br J Cancer. 2011;105:1741–1749. doi: 10.1038/bjc.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho AS, Huang X, Cao H, et al. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol. 2010;3:109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camps C, Buffa FM, Colella S, et al. Hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 34.Fasanaro P, D'Alessandra Y, Di SV, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G, Kwan BC, Lai FM, Chow KM, Kam-Tao LP, Szeto CC. Expression of microRNAs in the urinary sediment of patients with IgA nephropathy. Dis Markers. 2010;28:79–86. doi: 10.3233/DMA-2010-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenzen JM, Volkmann I, Fiedler J, et al. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant. 2011;11(10):2221–2227. doi: 10.1111/j.1600-6143.2011.03679.x. [DOI] [PubMed] [Google Scholar]
- 37.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.BLAST. http://blast.ncbi.nlm.nih.gov/Blast.cgi.