Abstract
Spinal muscular atrophy (SMA) is a neurodegenerative disease caused by a deficiency in the survival motor neuron (SMN) protein. SMN mediates the assembly of spliceosomal small nuclear ribonucleoproteins (snRNPs) and possibly other RNPs. Here we investigated SMN requirement for the biogenesis and function of U7—an snRNP specialized in the 3′-end formation of replication-dependent histone mRNAs that normally are not polyadenylated. We show that SMN deficiency impairs U7 snRNP assembly and decreases U7 levels in mammalian cells. The SMN-dependent U7 reduction affects endonucleolytic cleavage of histone mRNAs leading to abnormal accumulation of 3′-extended and polyadenylated transcripts, followed by downstream changes in histone gene expression. Importantly, SMN deficiency induces defects of histone mRNA 3′-end formation in both SMA mice and human patients. These findings demonstrate that SMN is essential for U7 biogenesis and histone mRNA processing in vivo, and identify a novel RNA pathway disrupted in SMA.
Introduction
Spinal muscular atrophy (SMA) is an inherited neurodegenerative disease caused by reduced expression of the survival motor neuron (SMN) protein due to homozygous mutation of the SMN1 gene (Burghes and Beattie, 2009). SMN is part of a macromolecular complex that functions in the biogenesis of small nuclear ribonucleoproteins (snRNPs) critical for pre-mRNA splicing (Neuenkirchen et al., 2008; Pellizzoni, 2007). The SMN complex mediates the assembly of a heptameric ring of Sm proteins (B, D1, D2, D3, E, F and G) onto spliceosomal snRNAs to build functional snRNPs (Meister et al., 2001; Pellizzoni et al., 2002). Importantly, SMN deficiency decreases snRNP levels (Gabanella et al., 2007; Zhang et al., 2008) and induces select splicing defects that contribute to motor system dysfunction in animal models of SMA (Lotti et al., 2012). The SMN complex is thought to have other roles in RNA regulation that could also be relevant to SMA, but are poorly defined (Burghes and Beattie, 2009). Identification of novel RNA pathways controlled by SMN, and characterization of associated post-transcriptional gene regulatory events altered in SMA, are critical to understanding mechanisms of motor neuron disease.
Sm proteins, together with the structurally related LSm (Sm-like) proteins, form a family of ~20 ubiquitously expressed and evolutionarily conserved RNA binding proteins, paralogs of which are found in all three branches of life (Tharun, 2009). Evolutionary diversification allowed formation of Sm/LSm protein complexes of different composition that associate with distinct RNAs and function in diverse RNA pathways such as pre-mRNA splicing, histone mRNA 3′-end processing, and cytoplasmic mRNA decay. Given both SMN’s essential role in Sm core assembly and the structural similarities between Sm and LSm cores, it is plausible that SMN may participate in the assembly of other Sm/LSm protein complexes onto their target RNAs. To date, however, the only established target of SMN function in vivo is spliceosomal snRNPs.
Here, we focused on U7 snRNP biogenesis and function to establish a more general role for SMN in the RNP assembly of the Sm/LSm protein family in vivo and to determine if this RNA pathway is disrupted in SMA. U7 and splicesomal snRNPs follow an analogous biogenesis pathway but differ in both protein composition and function (Schumperli and Pillai, 2004). U7 contains a unique mixed Sm/LSm core comprising Sm proteins B, D3, E, F, G and two LSm proteins (LSm10 and LSm11) instead of the D1 and D2 of spliceosomal snRNPs (Pillai et al., 2003; Pillai et al., 2001). An SMN complex containing LSm10 and Lsm11 proteins has been implicated in U7 snRNP assembly in vitro (Pillai et al., 2003). However, it is unknown if SMN is required for U7 biogenesis and function in vivo. Unlike spliceosomal snRNPs, U7 functions not in pre-mRNA splicing, but in the 3′-end processing of metazoan replication-dependent histone mRNAs (Dominski and Marzluff, 2007). Histone transcripts of this class contain no introns and are the only known eukaryotic mRNAs that lack a poly(A) tail. Instead, these mRNAs end with a conserved 3′-end stem-loop (SL) structure generated by a single U7-dependent endonucleolytic cleavage (Marzluff et al., 2008). Proper histone mRNA 3′-end processing is critical for regulation of histone synthesis, which in turn is essential for genome replication and function.
Using cellular and animal models, we demonstrate the essential role of SMN for the biogenesis of U7 snRNP and 3′-end processing of histone mRNAs in vivo. We also show that these SMN-dependent RNA processing events are disrupted in tissues from a mouse model of SMA as well as in human patients. Our results expand the repertoire of RNA pathways regulated by SMN and provide a molecular framework for exploring alterations of U7 function and histone gene regulation in the etiology of SMA.
Results
SMN is required for U7 snRNP biogenesis
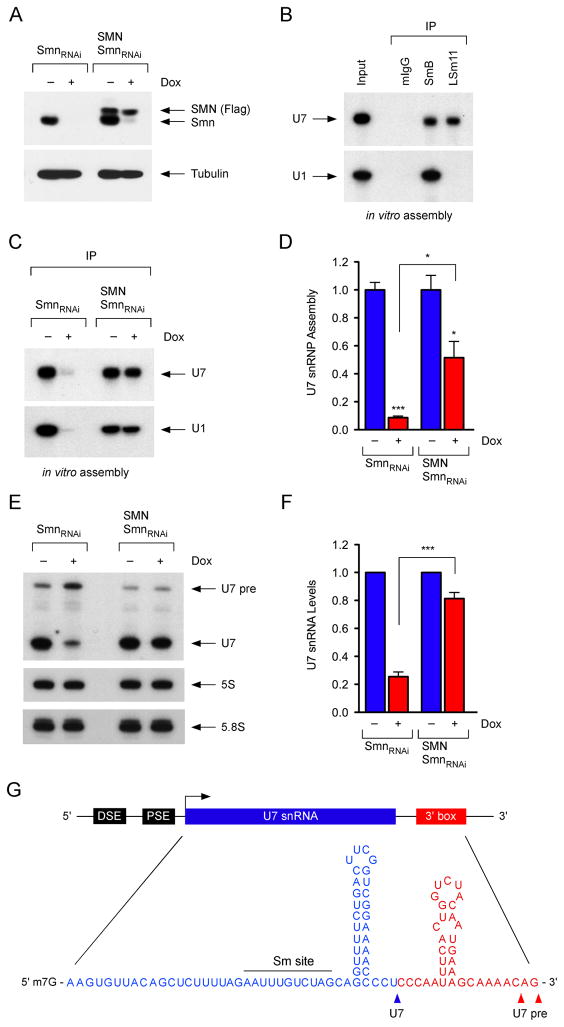
To investigate SMN requirement for U7 snRNP assembly, we utilized NIH3T3-SmnRNAi cells in which addition of doxycycline (Dox) causes depletion of endogenous mouse Smn (Figure 1A) (Lotti et al., 2012; Ruggiu et al., 2012). NIH3T3-SMN/SmnRNAi cells expressing RNAi-resistant human SMN were used to control for potential off-target effects of shRNA expression (Figure 1A). We also developed a novel monoclonal antibody against LSm11 and confirmed its specificity by Western blot as well as selective immunoprecipitation of endogenous U7 snRNA, but not spliceosomal U1 snRNA from NIH3T3 cell extracts (Figure S1A–C). To determine the specificity of U7 snRNP assembly in our experimental conditions, in vitro transcribed radioactive U1 and U7 snRNAs were incubated with NIH3T3 cell extracts followed by immunoprecipitation with SmB and LSm11 antibodies as well as mouse immunoglobulins. SmB associated with both U1 and U7 while LSm11 associated with U7 only (Figure 1B), demonstrating efficient and specific formation of Sm cores in vitro. We then analyzed the effects of SMN deficiency on U7 snRNP assembly using extracts from NIH3T3 cells with either normal or reduced SMN levels. These experiments revealed a strong reduction of U7 snRNP assembly in extracts from SMN-deficient NIH3T3-SmnRNAi cells (Figure 1C–D), which was corrected proportionally to human SMN expression in extracts from NIH3T3-SMN/SmnRNAi cells and similar to that of U1 snRNP (Figure 1C–D) (Lotti et al., 2012). Consistent with a previous study (Pillai et al., 2003), these experiments demonstrate SMN requirement for U7 snRNP assembly in vitro.
Figure 1. SMN is required for U7 snRNP biogenesis.
(A) Western blot analysis of NIH3T3-SmnRNAi and NIH3T3-SMN/SmnRNAi cells cultured with or without Dox for 5 days. (B) In vitro U1 and U7 snRNP assembly with wild-type NIH3T3 cell extracts. (C) In vitro U1 and U7 snRNP assembly with extracts from NIH3T3-SmnRNAi and NIH3T3-SMN/SmnRNAi cells cultured with or without Dox for 5 days. (D) Quantification of U7 snRNP assembly from three independent experiments as in (C). (E) Northern blot analysis of NIH3T3-SmnRNAi and NIH3T3-SMN/SmnRNAi cells cultured with or without Dox for 5 days. (F) Quantification of U7 levels normalized to 5S rRNA from three independent experiments as in (E). (G) Schematic of the U7 gene and sequence of the U7 snRNA precursor.
Data in all graphs are represented as mean and SEM. See also Figure S1.
Next, we studied the effects of SMN deficiency on the accumulation of U7 snRNP in NIH3T3 cells. Northern blot showed that SMN deficiency caused a strong reduction in U7 snRNA to ~25% the amount in NIH3T3-SmnRNAi cells with normal SMN expression and that this reduction was SMN-dependent as U7 levels were restored in NIH3T3-SMN/SmnRNAi cells (Figure 1E–F). Furthermore, immunoprecipitation experiments demonstrated a corresponding SMN-dependent decrease in the levels of U7 snRNPs containing the Sm core and 5′ trimethylated guanosine (TMG) cap (Figure S1D). Thus, SMN is required for the accumulation of U7 snRNP in mammalian cells. These experiments also revealed the association of SMN with U7 snRNA in NIH3T3 cells (Figure S1D), albeit to a lesser extent compared to SmB or TMG antibodies, consistent with U7 snRNA transient association with SMN while undergoing Sm core assembly.
In addition to mature U7 snRNA, Northern blot analysis revealed a larger RNA species that increases in abundance upon Smn deficiency (Figure 1E) and neither associates with Sm proteins nor contains a TMG cap (Figure S1D). We isolated, cloned and sequenced this RNA from wild-type NIH3T3 cells and identified it as a U7 snRNA precursor with a ~35 nucleotide 3′-end extension predicted to form a hairpin structure (Figure 1G), similar to that of spliceosomal pre-snRNAs (Yong et al., 2010). This U7 pre-snRNA can function as a substrate for U7 snRNP assembly in vitro (Figure S1E) and accumulates in SMN-deficient NIH3T3 cells (Figure 1E), consistent with the impairment of U7 snRNP biogenesis at a step preceding Sm core formation. Furthermore, the presence of detectable levels of U7 pre-snRNA under normal conditions suggests that SMN-mediated snRNP assembly is a limiting step of U7 synthesis.
SMN is required for 3′-end formation of histone mRNAs
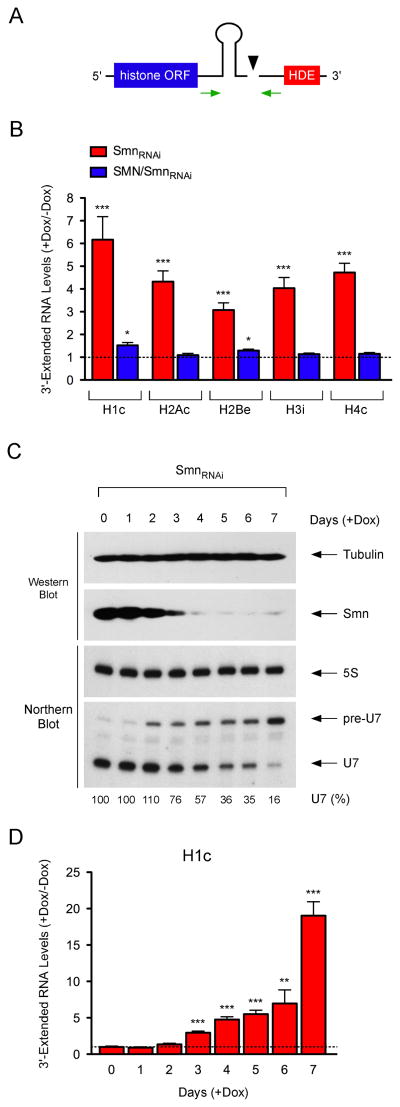
U7 snRNP functions in the 3′-end processing of replication-dependent histone mRNAs (Schumperli and Pillai, 2004). U7 binds the histone downstream element (HDE) and facilitates recruitment of processing factors for endonucleolytic cleavage of histone mRNAs between the 3′ SL and the HDE (Dominski and Marzluff, 2007). To investigate the effects of SMN-dependent disruption of U7 snRNP biogenesis on histone mRNA processing, we measured the levels of 3′-extended histone transcripts that may accumulate due to impaired cleavage (Figure 2A). RT-qPCR revealed that SMN deficiency caused a strong increase in unprocessed histone mRNAs in NIH3T3-SmnRNAi cells, which is corrected by human SMN expression in NIH3T3-SMN/SmnRNAi cells (Figure 2B). To investigate the correlation among SMN protein depletion, decreased U7 snRNA levels, and onset of histone mRNA processing defects, we carried out a temporal analysis following RNAi induction in NIH3T3-SmnRNAi cells (Figure 2C–D). The earliest detectable defect of SMN deficiency was the accumulation of U7 precursors beginning at day 2, which was then followed at day 3 by a reduction in mature U7 levels and the appearance of histone mRNA 3′-end formation defects. Each of these SMN-dependent events displayed a time-dependent accumulation consistent with the progressive SMN depletion and began prior to the onset of cell proliferation defects (Lotti et al., 2012). These experiments indicate that SMN deficiency impairs the biogenesis of functional U7 snRNP leading to disruption of histone mRNA 3′-end formation.
Figure 2. SMN is required for 3′-end formation of histone mRNAs.
(A) Schematic of the 3′-end structure of histone mRNAs and position of RT-qPCR primers. (B) RT-qPCR analysis of 3′-extended histone mRNAs in NIH3T3-SmnRNAi and NIH3T3-SMN/SmnRNAi cells cultured with or without Dox for 5 days. RNA levels in Dox-treated cells were expressed relative to untreated cells (dashed line). (C) Temporal analysis of SMN and U7 levels in NIH3T3-SmnRNAi cells cultured with Dox for the indicated number of days. (D) RT-qPCR analysis of the time-dependent accumulation of 3′-extended histone mRNAs in NIH3T3-SmnRNAi cells cultured as in (C). RNA levels in Dox-treated cells were expressed relative to untreated cells (dashed line).
Data in all graphs are represented as mean and SEM.
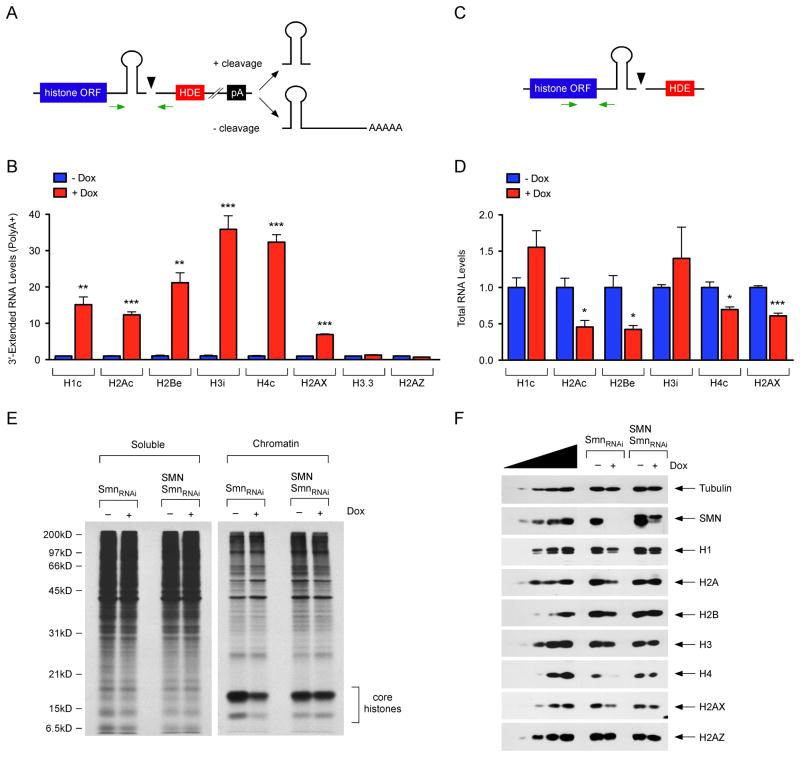
Replication-dependent histone transcripts that do not undergo the proper 3′-end cleavage are aberrantly extended and subject to a downstream polyadenylation signal (Figure 3A) (Lanzotti et al., 2002; Narita et al., 2007). We investigated if polyadenylated histone mRNAs accumulate in SMN-deficient cells by RT-qPCR analysis of poly(A)+ mRNA isolated from NIH3T3-SmnRNAi cells with normal and reduced SMN. SMN deficiency caused a remarkable accumulation of polyadenylated replication-dependent histone mRNAs (Figure 3B), but had no effects on replication-independent H3.3 and H2A.Z histone mRNAs (Figure 3B), which are not processed by U7 and are normally polyadenylated (Marzluff et al., 2008). However, SMN deficiency increased the levels of polyadenylated transcripts from the H2AX gene (Figure 3B). H2AX is an atypical histone variant that contains a 3′-end SL characteristic of replication-dependent histone mRNAs and undergoes 3′-end cleavage in addition to being polyadenylated (Marzluff et al., 2008). Thus, SMN-dependent defects in the 3′-end formation of histone mRNAs lead to the accumulation of their polyadenylated mRNAs.
Figure 3. SMN deficiency alters histone gene expression.
(A) Schematic of histone mRNA 3′-end processing events and position of RT-qPCR primers. (B) RT-qPCR analysis of 3′-extended histone mRNAs in poly(A)+ RNA from NIH3T3-SmnRNAi cells cultured with or without Dox for 5 days. RNA levels in Dox-treated cells were expressed relative to untreated cells. (C) Schematic of the position of RT-qPCR primers for quantification of total histone mRNAs. (D) RT-qPCR analysis of total histone mRNA levels in NIH3T3-SmnRNAi cells cultured with or without Dox for 5 days. RNA levels in Dox-treated cells were expressed relative to untreated cells. (E) Analysis of protein synthesis by [35S]-methionine/cysteine pulse labeling in NIH3T3-SmnRNAi and NIH3T3-SMN/SmnRNAi cells cultured with or without Dox for 5 days. (F) Western blot analysis of histones in NIH3T3-SmnRNAi and NIH3T3-SMN/SmnRNAi cells cultured with or without Dox for 5 days. Total levels of core histones and the linker histone H1 are measured as the antibodies do not distinguish the individual members of each multigene family.
Data in all graphs are represented as mean and SEM. See also Figure S2.
SMN deficiency disrupts histone gene expression
Defective 3′-end processing can lead to a reduction in the mRNA and protein levels of replication-dependent histones (Gruber et al., 2012; Hsin et al., 2011; Sullivan et al., 2009). We therefore investigated the consequences of SMN deficiency on histone gene expression. RT-qPCR showed that the steady-state levels of several histone mRNAs were altered in SMN-deficient NIH3T3-SmnRNAi cells (Figure 3C–D). Analysis of pulse labeled proteins from both soluble and chromatin-bound fractions indicated that SMN deficiency had no effects on global protein synthesis, yet caused a ~60% reduction in the synthesis of core histones in NIH3T3 cells; this reduction was corrected by human SMN expression (Figure 3E). Further, Western blot of SMN-deficient NIH3T3-SmnRNAi cells revealed a decrease in the levels of the core histones and linker H1 protein (Figure 3F). Consistent with defective 3′-end formation and mRNA expression, the amount of the histone H2AX, but not H2AZ, was also reduced. Collectively, these results indicate that SMN-dependent disruption of histone 3′-end formation can alter histone gene expression.
SMN depletion induces cell proliferation defects in NIH3T3 cells (Li et al., 2013; Lotti et al., 2012). We found that these defects are associated with a decrease in the proportion of cells in S phase and a corresponding increase of those in G1 (Figure S2A), akin to the effects of U7 snRNP inhibition on the cell cycle (Wagner and Marzluff, 2006). Given the essential need for histones in genome replication, the SMN-dependent impairment of histone synthesis may contribute to these phenotypes. To further investigate the effects of histone dysregulation induced by SMN deficiency, we analyzed nucleosome repeat length in SMN-deficient NIH3T3 cells and found no evidence for gross chromatin alterations (Figure S2B). We then focused on the histone variant H2AX that has a well-established role in the cellular response to genotoxic stress (Bonner et al., 2008). H2AX is rapidly phosphorylated at serine 139 upon DNA damage, and this phosphorylated form (γH2AX) acts as an initiating signal to recruit various chromatin remodeling complexes and DNA repair factors to the site of damaged DNA for efficient repair. Consistent with the levels of H2AX expression, immunofluorescence experiments showed that γH2AX induction in NIH3T3 cells treated with various DNA damaging agents was reduced by SMN deficiency (Figure S2C–D) and restored by expression of human SMN (Figure S2E–F). Thus, disruption of histone gene expression induced by SMN deficiency might impair cell division and the cellular ability to respond to DNA damage.
SMN deficiency impairs U7 snRNP biogenesis and histone 3′-end formation in SMA mice
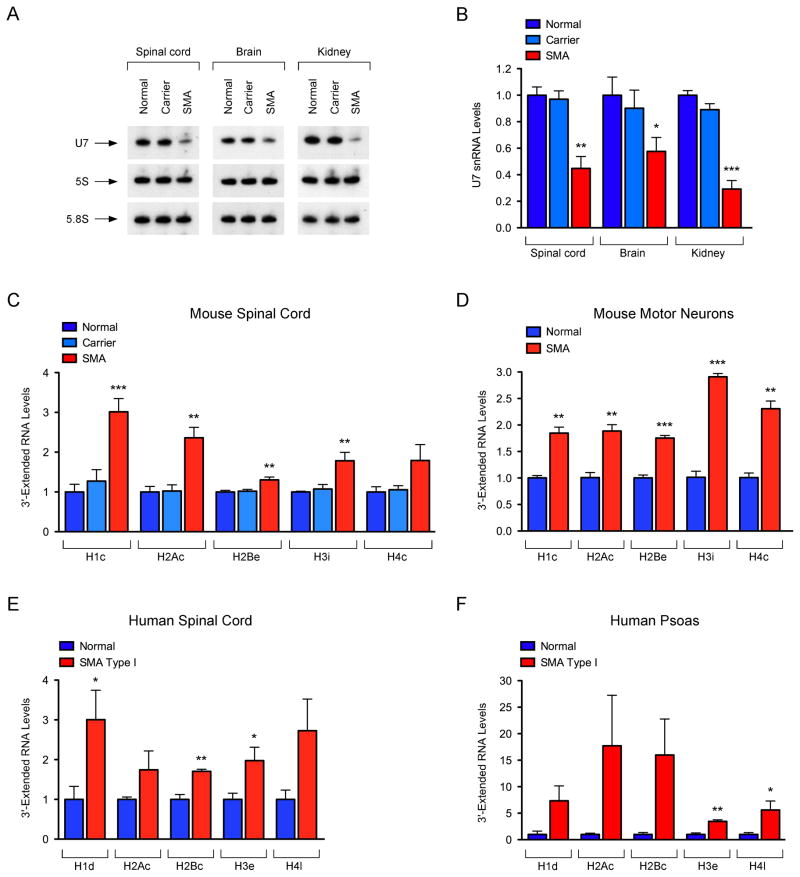
To investigate the effects of SMN deficiency on U7 snRNP biogenesis and histone mRNA 3′-end formation in vivo, we used the SMNΔ7 mouse model that has ubiquitously low levels of SMN expression and manifests features of the human motor neuron disease SMA (Le et al., 2005). First, we analyzed U7 snRNA levels in spinal cord, brain and kidney from control (Smn+/+;SMN2+/+;SMNΔ7+/+), carrier (Smn+/−;SMN2+/+;SMNΔ7+/+), and SMNΔ7 SMA (Smn−/−;SMN2+/+;SMNΔ7+/+) mice at postnatal day 6 (P6), an early symptomatic stage in this animal model. Northern blot showed a strong decrease in U7 snRNA levels in all tissues from SMA mice when compared to both control and carrier mice that are phenotypically normal (Figure 4A–B). These results demonstrate an SMN requirement for U7 expression in vivo and its disruption in a mouse model of SMA.
Figure 4. Disruption of U7 snRNP biogenesis and histone mRNA 3′-end formation in SMA.
(A) Northern blot analysis of U7 expression in tissues from normal, carrier and SMA mice at P6. (B) Quantification of U7 levels normalized to 5S rRNA from three independent experiments as in (A). (C) RT-qPCR analysis of 3′-extended histone mRNAs in spinal cord from normal, carrier, and SMA mice at P6. (D) RT-qPCR analysis of 3′-extended histone mRNAs in LCM motor neurons from normal and SMA mice at P6. (E) RT-qPCR analysis of 3′-extended histone mRNAs in the spinal cord from controls (n=3) and SMA patients (n=4). (F) RT-qPCR analysis of 3′-extended histone mRNAs in the psoas from controls (n=3) and SMA patients (n=4).
Data in all graphs are represented as mean and SEM. See also Figure S3.
Next, we analyzed 3′-end formation of histone mRNAs in SMA mice. RT-qPCR revealed remarkable defects of histone mRNA processing in the spinal cord as well as other tissues from SMA mice compared to normal controls at P6 (Figures 4C and S3A-B). In the spinal cord of SMA mice, histone 3′-end formation defects are already present at a pre-symptomatic stage, further accumulate in a time-dependent manner, and are followed by alterations in the total levels of histone mRNAs similar to those in SMN-deficient NIH3T3 cells (Figure S3C–D). Thus, SMN is required for histone 3′-end formation in vivo.
To study the effect of SMN deficiency on histone mRNA processing in motor neurons, we injected fluorescently labeled CTb into the iliopsoas of both control and SMA mice at P2 and then selectively isolated lumbar motor neurons innervating this axial muscle by laser capture microdissection at P6 (Lotti et al., 2012). Remarkably, RT-qPCR demonstrated that SMN deficiency impairs 3′-end formation of histone mRNAs in SMA motor neurons compared to normal motor neurons (Figure 4D), revealing SMN-dependent disruption of this RNA pathway in disease-relevant neurons of SMA mice.
Disruption of histone 3′-end formation in SMA patients
Given the requirement of SMN for histone mRNA processing in vivo, we sought to investigate if defects in this pathway are apparent in SMA patients. As expected, RT-qPCR showed reduction in the levels of full-length SMN mRNA in both spinal cord and psoas muscle from SMA patients compared to age-matched controls (Figures S3E and S3G). Moreover, upregulation of CDKN1A mRNA—a well-established event induced by SMN deficiency in both SMA mice and human patients (Olaso et al., 2006; Ruggiu et al., 2012)—was readily observed in human SMA tissue while the levels of the ubiquitously expressed ATP6 mRNA, used as additional control, did not change (Figures S3E and S3G). Of key importance, RT-qPCR revealed the accumulation of 3′-extended histone mRNAs in both skeletal muscle and spinal cord from SMA patients relative to controls (Figure 4E–F). Although variable and more pronounced in psoas muscle than in spinal cord, a consistent increase in the levels of misprocessed histone mRNAs was found in tissue from all SMA patients relative to controls (Figures S3F and S3H). Thus, SMN deficiency of the magnitude of human SMA causes defects in histone mRNA 3′-end formation.
Discussion
A key function of the SMN protein, mutation of which is responsible for SMA, is as a molecular chaperone for the assembly of RNP complexes involved in RNA processing. Knowledge of the full repertoire of in vivo SMN functions is thus central for understanding basic mechanisms of post-transcriptional gene regulation and the basis for this motor neuron disease. Here, we demonstrate that SMN is essential for U7 snRNP biogenesis and proper 3′-end processing of histone mRNAs in vivo. SMN deficiency disrupts this fundamental RNA pathway in a mouse model of SMA as well as human patients. These results conclusively establish the in vivo requirement for SMN in the assembly of Sm/LSm class RNPs beyond spliceosomal snRNPs and identify novel SMN-dependent RNA processing events affected in SMA. These findings may have important implications in identifying molecular mechanisms of this neurodegenerative disorder, may open new avenues of therapeutic targeting, and may lead to early disease biomarkers that are essential to efficient clinical trials for SMN-restoring therapies.
Our studies establish SMN requirement for the biosynthesis of functional U7 snRNP in vivo. We show that SMN associates with U7 snRNAs and that formation of the Sm/LSm core on U7 is strongly impaired by SMN deficiency, consistent with depletion of a specialized SMN complex containing LSm10 and LSm11 proteins (Pillai et al., 2003). Importantly, we demonstrate for the first time that the SMN-dependent disruption of U7 snRNP assembly leads to a severe reduction in the levels of mature U7 and concomitant accumulation of a previously uncharacterized U7 pre-snRNA. The functional consequence of the U7 snRNP reduction induced by SMN deficiency is the disruption of replication-dependent histone mRNA processing as evidenced by accumulation of uncleaved, 3′-extended transcripts that are aberrantly polyadenylated. The production of polyadenylated histone mRNAs indicates that the SMN-dependent reduction in functional U7 snRNP levels does not result simply in slower processing of histone transcripts, but in a switch to a different modality of 3′-end processing. While leaving open the possibility that SMN may also influence the expression of other factors involved in histone mRNA processing through its role in splicing with consequent exacerbation of the effects of U7 reduction, loss of the unique 3′-end structure of histone mRNAs through this series of RNA processing defects induced by SMN deficiency ultimately leads to dysregulated histone gene expression.
Complete loss of SMN function is incompatible with life at the cellular and organism levels in mammals (Burghes and Beattie, 2009). Although this has been thought to result from disruption of SMN function in RNA splicing, our findings provide an alternative but not mutually exclusive explanation. Since histones are crucial for DNA replication, defective histone synthesis induced by U7 deficiency in the absence of SMN would likely halt cell division at an early stage and contribute to embryonic lethality. Accordingly, severe cell proliferation defects ensue in response to U7 dysfunction (Ideue et al., 2009; Wagner and Marzluff, 2006) and SMN depletion (Lotti et al., 2012) in mammalian cells.
Our results demonstrate that U7 snRNP biogenesis and function are disrupted in SMA—a childhood neurodegenerative disease characterized by ubiquitous SMN deficiency (Burghes and Beattie, 2009). The observation that histone mRNA 3′-end processing defects accumulate in multiple tissues of both SMA mice and human patients points to this RNA pathway as a candidate source for biomarkers in SMA. Biomarkers are greatly needed to serve as molecular readouts for the efficacy of a variety of therapeutics currently entering SMA clinical trials (Van Meerbeke and Sumner, 2011). Additionally, both U7 reduction and histone 3′-end formation defects can be used in preclinical studies to evaluate restoration of SMN function in SMA mice.
Our discovery that SMN deficiency alters histone mRNA processing in SMA has potential important implications for understanding the molecular mechanisms of the disease. While motor neuron loss is a hallmark of SMA, it is becoming increasingly clear that other networked neurons participate in motor system dysfunction induced by SMN deficiency (Imlach et al., 2012; Mentis et al., 2011) and other cell types inside and outside the nervous system may also contribute to SMA pathology (Hamilton and Gillingwater, 2013). In addition to RNA splicing defects (Lotti et al., 2012; Ruggiu et al., 2012; Zhang et al., 2008), altered histone gene regulation induced by SMN deficiency could potentially have profound cellular consequences and contribute to some of these deficits. Reduced cell proliferation in the hippocampus of SMA mice causing defects in brain development (Wishart et al., 2010) may be the downstream consequence of defects in histone synthesis. The effects of SMN-dependent histone dysregulation need not be limited to impaired proliferation, however. Consistent with increased susceptibility to genotoxic stress of SMA patient-derived fibroblasts (Wang et al., 2005), we show that SMN deficiency can lead to decreased expression of histone H2AX and lower levels of its phosphorylated form upon DNA damage. Importantly, we also show that SMN deficiency causes disruption of histone mRNA 3′-end formation in SMA motor neurons, demonstrating that SMN-dependent histone dysregulation is not restricted to cycling cells. Histones play essential roles in the regulation of gene expression by facilitating dynamic changes in the structural and functional organization of chromatin. Even subtle alterations in the cellular pool of histones could have significant effects on gene regulation and detrimental consequences in SMA neurons.
In summary, our findings identify a novel RNA pathway that is disrupted by SMN deficiency in vivo in experimental models and human disease tissues, and provide the initial foundation for a potential involvement of histone dysregulation in the etiology of SMA. Future studies will determine whether SMN-dependent U7 snRNP dysfunction and histone 3′-end formation defects have a role in SMA pathology. In light of our findings and emerging connections between SMN and genes associated with amyotrophic lateral sclerosis (Yamazaki et al., 2012), investigation of a more general role for histone biology in human motor neuron disease will also deserve consideration.
Experimental Procedures
NIH3T3 cell lines
The NIH3T3 cell lines used in this study have been described previously (Lotti et al., 2012; Ruggiu et al., 2012). Treatments and methods for extract preparation and immunoprecipitation are described in Supplemental Experimental Procedures.
SMA mice and LCM
All experiments with mice were conducted as approved by the Institutional Laboratory Animal Care and Use Committee of Columbia University. FVB.Cg-Tg(SMN2*delta7)4299Ahmb Tg(SMN2)89Ahmb Smn1tm1Msd/J (JAX Stock number 005025) mice were interbred to obtain SMA mutant mice (Le et al., 2005). Alexa-488-conjugated cholera toxin subunit-β (CTb) was injected in the iliopsoas muscle of normal and SMA mice at P2 and motor neurons were isolated by LCM from spinal cords of injected mice at P6 (Lotti et al., 2012).
Human SMA tissue
SMA and control human tissue was collected at autopsy following parental informed consent as approved by the Johns Hopkins Medicine Institutional Review Board. Some control human tissue was also obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD. Further information is provided in Supplemental Experimental Procedures.
RNA analysis
Total RNA was isolated using Trizol reagent (Invitrogen) followed by digestion with RNase-free DNaseI (Ambion). Poly (A)+ RNA was purified from total RNA using oligo(dT)+ beads (New England Biolabs). RNA from LCM neurons was purified using the Absolutely RNA Nanoprep Kit (Agilent) and linear amplification was performed with the MessageAmp II aRNA Amplification Kit (Ambion) according to the manufacturer’s instructions. RT-qPCR and Northern blot experiments were carried out as previously described (Lotti et al., 2012). The sequence of primers and probes used in this study are listed in Table S1. In vitro snRNP assembly experiments were carried out as described previously with minor modifications (Gabanella et al., 2007; Pellizzoni et al., 2002). Additional information is provided in Supplemental Experimental Procedures.
Supplementary Material
Research Highlights.
SMN is required for U7 snRNP biogenesis
SMN is required for 3′-end formation of replication-dependent histone mRNAs
SMN deficiency alters histone gene expression
U7 snRNP biogenesis and histone mRNA processing are disrupted in SMA
Acknowledgments
This work was supported by grants from NIH-NINDS R01NS069601 and R21NS067448 (to L.P.), R01NS078375 (to G.Z.M.), R01NS062869 (to C.J.S), F31NS079002 (to S.T.); DoD W81XWH-11-1-0689 (to G.Z.M.); FSMA A. Lewis Young Investigator Award (to G.Z.M.); and SMA Foundation (to C.J.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber JJ, Olejniczak SH, Yong J, La Rocca G, Dreyfuss G, Thompson CB. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol Cell. 2012;45:87–98. doi: 10.1016/j.molcel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton G, Gillingwater TH. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol Med. 2013;19:40–50. doi: 10.1016/j.molmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334:683–686. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideue T, Hino K, Kitao S, Yokoi T, Hirose T. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009;15:1578–1587. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlach WL, Beck ES, Choi BJ, Lotti F, Pellizzoni L, McCabe BD. SMN is required for sensory-motor circuit function in Drosophila. Cell. 2012;151:427–439. doi: 10.1016/j.cell.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti DJ, Kaygun H, Yang X, Duronio RJ, Marzluff WF. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol Cell Biol. 2002;22:2267–2282. doi: 10.1128/MCB.22.7.2267-2282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- Li DK, Tisdale S, Espinoza-Derout J, Saieva L, Lotti F, Pellizzoni L. A Cell System for Phenotypic Screening of Modifiers of SMN2 Gene Expression and Function. PLoS ONE. 2013;8:e71965. doi: 10.1371/journal.pone.0071965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti F, Imlach WL, Saieva L, Beck ES, Hao le T, Li DK, Jiao W, Mentis GZ, Beattie CE, McCabe BD, et al. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151:440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Mentis GZ, Blivis D, Liu W, Drobac E, Crowder ME, Kong L, Alvarez FJ, Sumner CJ, O’Donovan MJ. Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron. 2011;69:453–467. doi: 10.1016/j.neuron.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Olaso R, Joshi V, Fernandez J, Roblot N, Courageot S, Bonnefont JP, Melki J. Activation of RNA metabolism-related genes in mouse but not human tissues deficient in SMN. Physiol Genomics. 2006;24:97–104. doi: 10.1152/physiolgenomics.00134.2005. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Grimmler M, Meister G, Will CL, Luhrmann R, Fischer U, Schumperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Will CL, Luhrmann R, Schumperli D, Muller B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M, McGovern VL, Lotti F, Saieva L, Li DK, Kariya S, Monani UR, Burghes AH, Pellizzoni L. A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol Cell Biol. 2012;32:126–138. doi: 10.1128/MCB.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumperli D, Pillai RS. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci. 2004;61:2560–2570. doi: 10.1007/s00018-004-4190-0. [DOI] [PubMed] [Google Scholar]
- Sullivan KD, Mullen TE, Marzluff WF, Wagner EJ. Knockdown of SLBP results in nuclear retention of histone mRNA. RNA. 2009;15:459–472. doi: 10.1261/rna.1205409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int Rev Cell Mol Biol. 2009;272:149–189. doi: 10.1016/S1937-6448(08)01604-3. [DOI] [PubMed] [Google Scholar]
- Van Meerbeke JP, Sumner CJ. Progress and promise: the current status of spinal muscular atrophy therapeutics. Discov Med. 2011;12:291–305. [PubMed] [Google Scholar]
- Wagner EJ, Marzluff WF. ZFP100, a component of the active U7 snRNP limiting for histone pre-mRNA processing, is required for entry into S phase. Mol Cell Biol. 2006;26:6702–6712. doi: 10.1128/MCB.00391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Dimatteo D, Funanage VL, Scavina M. Increased susceptibility of spinal muscular atrophy fibroblasts to camptothecin-induced cell death. Mol Genet Metab. 2005;85:38–45. doi: 10.1016/j.ymgme.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Wishart TM, Huang JP, Murray LM, Lamont DJ, Mutsaers CA, Ross J, Geldsetzer P, Ansorge O, Talbot K, Parson SH, et al. SMN deficiency disrupts brain development in a mouse model of severe spinal muscular atrophy. Hum Mol Genet. 2010;19:4216–4228. doi: 10.1093/hmg/ddq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Chen S, Yu Y, Yan B, Haertlein TC, Carrasco MA, Tapia JC, Zhai B, Das R, Lalancette-Hebert M, et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012;2:799–806. doi: 10.1016/j.celrep.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Kasim M, Bachorik JL, Wan L, Dreyfuss G. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol Cell. 2010;38:551–562. doi: 10.1016/j.molcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.