Abstract
We studied group I introns in sterile cultures of selected groups of lichen photobionts, focusing on Trebouxia species associated with Xanthoria s. lat. (including Xanthomendoza spp.; lichen-forming ascomycetes). Group I introns were found inserted after position 798 (Escherichia coli numbering) in the large subunit (LSU) rRNA in representatives of the green algal genera Trebouxia and Asterochloris. The 798 intron was found in about 25% of Xanthoria photobionts including several reference strains obtained from algal culture collections. An alignment of LSU-encoded rDNA intron sequences revealed high similarity of these sequences allowing their phylogenetic analysis. The 798 group I intron phylogeny was largely congruent with a phylogeny of the Internal Transcribed Spacer Region (ITS), indicating that the insertion of the intron most likely occurred in the common ancestor of the genera Trebouxia and Asterochloris. The intron was vertically inherited in some taxa, but lost in others. The high sequence similarity of this intron to one found in Chlorella angustoellipsoidea suggests that the 798 intron was either present in the common ancestor of Trebouxiophyceae, or that its present distribution results from more recent horizontal transfers, followed by vertical inheritance and loss. Analysis of another group I intron shared by these photobionts at small subunit (SSU) position 1512 supports the hypothesis of repeated lateral transfers of this intron among some taxa, but loss among others. Our data confirm that the history of group I introns is characterized by repeated horizontal transfers, and suggests that some of these introns have ancient origins within Chlorophyta.
Keywords: Lichen, LSU 798 group I intron, Photobiont, rbcL, SSU 1512 group I intron, Trebouxiophyceae, Trebouxia
INTRODUCTION
Group I introns are a distinctive group of RNA sequences that catalyze their own excision from precursor RNA transcripts with concomitant exon ligation. Group I introns have a random and highly biased distribution owing to the two intron transfer mechanisms of reverse splicing and homing (Hoshina & Imamura 2009). Group I introns have been reported in various genes from a variety of organisms (Michel & Westhof, 1990). About 2,939 group I introns (for details, see http://www.rna.icmb.utexas.edu/SAE/2C/Distributions/g12tpt.php) have been identified in eukaryotic genomes (Haugen et al. 2005a, Nielsen & Johansen 2009, Bachar et al. 2013). Of these, 2,311 are found in the nucleus at dozens of sites in small and large subunit (SSU, LSU) ribosomal DNA. About 235 occur in mitochondrial genes and 393 in plastid DNA. Group I introns are characterized by four conserved core sequences (P, Q, R and S) and ten conserved secondary structures (P1 to P10; Burke et al. 1987). Five major subgroups of group I introns, termed IA, IB, IC, ID, and IE are recognized based on secondary structure (Michel & Westhof, 1990) and are always inserted into highly conserved regions of rRNA genes (Johansen et al. 1996). All group I introns catalyze their excision from primary transcripts using guanosine as a cofactor. Many of these ribozymes are autocatalytic, others rely on host-derived factors to facilitate splicing (Cech 1988, Cech et al. 1994). Among two Chlorella photobiont species of the ciliate Paramecium bursaria a monophyletic clade of six introns was found, all of them derived from an ancestral intron (Csw.L2449) that had spread into heterologous sites (Hoshina and Imamura 2009). Some of these introns share common internal guide sequences, but in others these sequence fragments are inserted further upstream (Hoshina & Imamura 2009). These Chlorella photobionts are proposed to be a model system for studies on how group I introns insert at novel sites.
In eukaryotic microorganisms, horizontal transfer of group I introns appears to be the rule, rather than the exception. Therefore, the mapping and phylogenetic characterization of the widespread, but scattered nuclear group I introns can potentially clarify pathways and mechanisms of intron movement among eukaryotes (Einvik et al. 1998). Various studies have focused on the mechanisms underlying group I intron mobility. Many group I introns in the genomes of plastids, mitochondria, phages and eubacteria contain open reading frames (ORFs) that encode endonucleases. These endonucleases mediate sequence-specific ‘homing’ of group I introns into allelic sites (Dujon 1989). Although group I introns do not normally contain more than one ORF, the intron in the cox1 gene of the non-photosynthetic, parasitic chlorophyte Helicosporidium sp. contains two ORFs (Pombert & Keeling 2010). Intron movement can also occur via reverse splicing, as demonstrated for the Tetrahymena thermophila LSU group I intron (Sogin et al. 1986).
Intron loss appears to be common as demonstrated by the ‘optional’ distribution of group I introns within closely related taxa (Bhattacharya et al. 1996a, Bhattacharya et al. 1996b). At the RNA level, intron loss seems to occur by reverse transcription of an intron-less RNA followed by homologous recombination with the intron containing genomic copy of the coding region (Dujon 1989). In some eukaryotic lineages (e.g., plants, including green algae, charophytes, red algae, or ciliates, fungi etc.) mobility conferring - open reading frames (ORFs) have not yet been found in group I introns. This suggests that intron mobility (and loss) is likely to be mediated by reverse-splicing that does not rely on a group I intron-encoded ORF (Bhattacharya et al. 1996b). However in myxomycetes (Amoebozoa) such as Didymium iridis and Physarum polycephalum, nuclear group I introns spread extensively via homing (e.g., Ruoff et al. 1992, Haugen et al. 2005b). Intron sequences encoding homing endonucleases have also been reported from lichenized ascomycetes (Reeb et al. 2007). Putative group I introns were found in nuclear the rSSU gene of the non-lichenized ascomycetes Phialophora americana, P. europaea (both saprotrophs) and P. verrucosa (human pathogen; Dothideomycetes), which were found to be monophyletic along with introns of lichen forming ascomycetes Porpidia crustulata (Lecanoromycetes) and Arthonia lapidicola (Arthoniomycetes; Harris & Rogers 2011). During a large phylogenetic survey of green algae, twenty three distantly related isolates were found to contain at least one group I intron in the chloroplast rbcL gene, all of them with distinct evolutionary origins. These introns were inserted either after position 462 or 699 (McManus et al. 2012).
The lichen symbiosis is particularly interesting with regard to group I intron presence and distribution. In lichen-forming ascomycetes, several group I introns at 13 different insertion positions within the nuclear SSU rDNA have been identified (Bhattacharya et al. 2002). The SSU rDNA intron variation observed by DePriest (1993) in a population of Cladonia chlorophaea was due to optional group I introns, which varied in number, position, restriction pattern and size. Some of these insertion positions are unique, whereas others (516, 943, 1046, and 1506) are also found in other organisms including unicellular green algae (Gargas et al. 1995b). Within the monophyletic Parmeliaceae (Lecanorales; more than 60 species investigated) correlations between intron insertion sites and ecological and geographical parameters were observed (Gutierrez et al. 2007). Because the green algal photobionts of lichen-forming fungi contain numerous nuclear-encoded rDNA group I introns, they are a model group to study the origin and phylogeny of these sequences (Bhattacharya et al. 1998, Bhattacharya et al. 1994, Bhattacharya et al. 1996a, Bhattacharya et al. 1996b). Friedl et al. (2000) concluded that the SSU rDNA 1,512 group I intron was present in the common ancestor of the green algal classes Trebouxiophyceae, Chlorophyceae and Ulvophyceae and that it was laterally transferred at least three times among species of Trebouxia de Puymaly. They also concluded that intron loss was a common event during chlorophyte evolution. The green algal SSU group I introns at insertion sites 1,056, 1,506, and 1,512 (position relative to the E. coli coding region) form distinct phylogenetic lineages based on the insertion site (Bhattacharya et al. 1994, Bhattacharya et al. 1996b). The intron phylogenies were largely congruent with the rDNA (i.e., host cell) phylogeny, suggesting vertical inheritance of the introns rather than lateral transfers during the evolution of green algae, with some exceptions being described by Friedl et al. (2000) and Bhattacharya et al. (2001).
Del Campo et al. (2010) partially sequenced plastid LSU rDNA in fifteen Trebouxia and Asterochloris species and observed a high diversity of group I introns. The resulting phylograms revealed two main clades, one comprising A. erici, A. glomerata, A. irregularis, the other T. arboricola, T. crenulata, T. decolorans, T. shomanii and T. jamesii. The authors suggested combining the phylogeneitic analyses of the chloroplast and nuclear (nrITS) for improving phylogenetic accuracy. (del Campo et al. 2010) described different group I introns (at position cL2449 and cL2504) in chloroplast LSU rDNA in Trebouxia photobiont within the same thallus of Ramalina farinacea (L) Ach. and speculated on presence of different photobiont species within a thallus.
In studies of the genetic diversity among green algal photobionts of Xanthoria s. lat. and Xanthomedoza spp. (lichen-forming ascomycetes), several Trebouxia strains and species were found to contain group I introns (Nyati et al. submitted). Moreover, these lichenized fungi associate with a well-defined, limited number of algal strains. Hence, this group of lichen photobionts represents a model system to study intron gains and losses in Trebouxia. The genus Asterochloris Tscherm.-Woess forms lichen symbioses with Cladonia and other lichenized ascomycetes. We included type strains of Asterochloris to infer whether some introns could be ancient, as evidenced by intron sharing among Trebouxia and Asterochloris lineages. The main aim of this study was to characterize the group I introns and investigate the possibility of intron gains and losses among photobionts associated with lichen-forming ascomycetes of the genera Xanthoria s. lat. and Xanthomendoza. For this purpose, we: 1) characterize a newly found nuclear LSU rDNA group I intron at site 798 (L798); 2) determine the distribution of the L798 and nuclear SSU rDNA 1512 (S1512) group I introns; and 3) investigate congruence between nrITS and intron phylogenies among green algal lichen photobionts of the genera Trebouxia and Asterochloris.
MATERIALS AND METHODS
Lichen collection, storage and photobiont isolation and culture
Photobionts were isolated and cultured as described (Honegger 2003) from several Xanthoria spp. including the broadly sampled X. parietina (Table 1). Type cultures of algal strains were obtained from the Culture Collection of Algae at the University of Innsbruck, Austria (IB/ASIB) (Table 2). All cultured algal strains included in this study including reference strains of sterile algal cultures, are stored in liquid nitrogen at the Institute of Plant Biology, University of Zürich, Switzerland following the method of Honegger (2003).
Table 1.
List of samples analyzed in this study for LSU and SSU introns.
| Trebouxia speciesb | Isolated from Xanthoria sp. |
Country | Voucher Noa | ITS accession no. |
LSU accession no. |
|---|---|---|---|---|---|
| T. arboricola de Puymaly | X. calcicola Oxner | CH | P-105-I-a | AJ969542 | AM261263 |
| T. arboricola | X. calcicola | CH | P-141-II | AJ969552 | |
| T. arboricola | X. capensis Kärnefelt, Arup & L. Lindblom | ZA | P-306-I-a | AJ969591 | AM261277 |
| T. arboricola | X. ectaneoides (Nyl.) Zahlbr. | F | P-158-IV-mc | AJ969560 | |
| T. arboricola | X. ectaneoides | I | L-43 | AJ969523 | |
| T. arboricola | X. ectaneoides | TN | P-174-II-aA | AJ969565 | |
| T. arboricola | X. ligulata (Körb.) P. James | NZ | P-17-I-ac | AJ969518 | AM261260 |
| T. arboricola | X. ligulata | NZ | P-17-II-a | AJ969519 | |
| T. arboricola | X. ligulata | NZ | P-53-I-a | AJ969528 | |
| T. arboricola | X. ligulata | NZ | P-54-II-a | AJ969530 | |
| T. arboricola | X. parietina (L.) Beltr. | GB | P-18-I-a | AJ969520 | |
| T. arboricola | X. parietina | CY | P-5-I-a-A | AJ969510 | |
| T. arboricola | X. parietina | IS | P-198-II-a | AJ969568 | |
| T. arboricola | X. parietina | IS | P-210-I-a | AJ969570 | |
| T. arboricola | X. parietina | F | P-7-I-a | AJ969512 | |
| T. arboricola | X. parietina | CH | P-320-III-a | AJ969604 | AM261279 |
| T. arboricola | X. polycarpa (Hoffm.) Th.Fr. ex Rieber1 | NZ | P-48-III-a | AJ969526 | |
| T. arboricola | X. sp. | CY | P-5-I-a | AJ969510 | |
| T. arboricola | X. sp. | AUS | P-276-I-ac | AJ969581 | AM261275 |
| T. arboricola | X. hirsuta Eichenberger, Aptroot & Honegger | ZA | P-360-II-ac | AJ969609 | AM261281 |
| T. arboricola | X. turbinata Vain.2 | ZA | P-3-Id | AJ969509 | |
| T. decolorans Ahmadjian | X. candelaria (L.) Th. Fr.3 | IS | P-205-II-ac | AJ969569 | AM261272 |
| T. decolorans |
Xanthomendoza hasseana (Räsänen) Søchting, Kärnefelt & S.Y. Kondr.4 |
USA | P-400-Ia | AM159210 | |
| T. decolorans | X. parietina | AUS | P-10-I-ac | AJ969515 | AM261258 |
| T. decolorans | X. parietina | AUS | P-11-Ia | --- | AM261259 |
| T. decolorans | X. parietina | AUS | L-275-IIc | AJ969580 | AM261274 |
| T. decolorans | X. parietina | USA | L-8c | AJ969513 | AM261256 |
| T. decolorans | X. parietina | USA | L-9c | AJ969514 | AM261257 |
| T. decolorans | X. parietina | S | P-97-I-a | AJ969539 | |
| T. decolorans | X. parietina | F | P-121a-IIIb | AJ969545 | AM261264 |
| T. decolorans | X. parietina | F | P-121-I-b | AJ969549 | AM261266 |
| T. decolorans | X. parietina | F | L-121-I-d | AJ969546 | AM261265 |
| T. decolorans | X. parietina | F | P-121-II-c | AJ969550 | AM261267 |
| T. decolorans | X. parietina | F | P-121-II-g | --- | AM261268 |
| T. decolorans | X. parietina | F | P-144-III-b | AJ969554 | |
| T. decolorans | X. parietina | F | P-144-III-h | AJ969556 | |
| T. decolorans | X. parietina | F | L-145-III-g | AJ969557 | AM261270 |
| T. decolorans | X. parietina | F | P-145-I-f-B | AJ969558 | AM261271 |
| T. decolorans | X. parietina | E | P-280-IIa | AJ969583 | AM261276 |
| T. decolorans | X. parietina | CH | P-319-I-gc | AJ970889 | AM261278 |
| T. decolorans | X. parietina | CH | P-319-II-a | AJ969595 | |
| T. decolorans | X. parietina | CH | P-320-I-d2 | AJ969599 | |
| T. decolorans | X. parietina | CH | P-320-II-b | AJ969600 | |
| T. decolorans | X. parietina | CH | P-320-II-f | AJ969603 | |
| T. decolorans | X. polycarpa (Hoffm.) Rieber1 | USA | P-71-II-b | AJ969535 | AM261262 |
| T. decolorans | X. polycarpa | CH | P-215-I-a | AJ969572 | |
|
T. gelatinosa Ahmadjian ex Archibald |
Xanthomendoza weberi (S.Y. Kondr. & Kärnefelt) L. Lindblom5 |
USA | P-57-I-a | AJ969532 | AM261261 |
Trebouxia species identified according to ITS and rbcL phylogenetic analysis (Nyati et al. submitted);
P indicates photobiont isolated, L indicates whole lichen DNA used for PCR amplification;
both introns present;
T. arboricola isolated from X. turbinata which had an insert of 1090 bases within 1512 group I intron.
Xanthoria polycarpa (syn. Massjukiella polycarpa Fedorenko et al.);
X. turbinata (syn. Xanthodactylon turbinatum (Vain.) C.W. Dodge);
X. candelaria (syn. Massjukiella candelaria Fedorenko et al.);
Xanthomendoza hasseana (syn. Gallowayella hassiana Fedorenko et al.); Xanthomendoza weberi (syn. Honeggeria rosmariae Fedorenko et al.).
Table 2.
ITS and partial LSU accession numbers of reference algal strains used in this study
| Algal species | Straina | LSU accession No. | ITS accession No. | Referenceb |
|---|---|---|---|---|
|
Asterochloris erici (Ahmadjian) Skaloud & Peksa |
UTEX 912 | AF345441 | Piercey-Normore et al. (2001) | |
| A. erici | UTEX 910/IB 342 | AM261248 | This study | |
|
A. excentrica (Archibald) Skaloud & Peksa |
UTEX 1714/IB 345 | AM261249 | This study | |
|
A. glomerata (Warén) Skaloud & Peksa |
UTEX 897 | AF345405 | Piercey-Normore et al. (2001) | |
| A. glomerata | UTEX 894/IB 349 | AM261252 | This study | |
|
A. italiana (Archibald) Skaloud & Peksa |
CCAP 219-5b/IB 358 | AM261253 | This study | |
|
Chlorella angustoellipsoidea N.Hanagata & M.Chihara |
C-87 | D17180 | Aimi et al. (1994) | |
|
Trebouxia anticipata Ahmadjian ex Archibald |
UTEX 903/IB 340 | AM261247 | This study | |
| T. arboricola | 92.011A1 | AJ249481 | Friedl et al. (2000) | |
| T. arboricola | 92.011C3 | Z68703 | Bhattacharya et al. (1996) | |
| T. arboricola c | SAG 219-Ia | Z 68705 | Bhattacharya et al. (1996) | |
|
T. asymmetrica Friedl & Gärtner |
B207 | AF344177 | Beck et al. (2002) | |
|
T. corticola (P.A.Archibald) Gärtner |
UTEX 909 | AJ249566 | Friedl et al. (2000) | |
| T. decolorans | UTEX 901/IB 327 | AM261243 | Unpubl. | Unpublished |
|
T. galapagensis (Hildreth & Ahmadjian) Gärtner |
UTEX 2230 | AJ249567 | Friedl et al. (2000) | |
| T. gelatinosa | 87.072B1 | AJ249575 | Friedl et al. (2000) | |
|
T. higginsiae (Hildreth & Ahmadjian) Gärtner |
UTEX 2232 | AJ249574 | Friedl et al. (2000) | |
| T. impressa Ahmadjian | 87.017E1 | AJ249570 | Friedl et al. (2000) | |
|
T. showmanii (Hildreth & Ahmadjian) Gärtner |
UTEX 2234/IB 337 | AM261246 | AF242470 | Kroken et al. (2000) |
IB: Culture collection of algae at the University of Innsbruck; SAG: Algal culture collection, University of Göttingen; UTEX: algal culture collection, University of Texas
All LSU sequences generated in this study; unpublished ITS sequence kindly provided by T. Friedl and G. Helms
Type strain of the genus Trebouxia
DNA extraction, PCR amplification and sequencing
Genomic DNA from algal isolates or whole lichens was extracted using the GFX PCR, DNA and Gel Band Purification Kit (Amersham Biosciences, Little Chalfont, UK) according to the manufacturer’s protocol. PCR amplification of a partial LSU rDNA fragment (position 660-1100 of T. asymmetrica, Z95380) was done with newly designed primers (Table 3). The PCR reactions were performed in 50 μL reaction volumes containing a reaction mix of 0.2 mM of each of four dNTPs, 1.5 μM of each PCR primer, and 1.25 U of Taq DNA polymerase (Sigma Aldrich, Buchs, SG, Switzerland) and 5 μL of 10X PCR buffer (100 mM Tris-HCl, pH 8.3, 500 mM KCl, 15 mM MgCl2 and 0.01% gelatin). Reactions were performed in a PTC 200 DNA engine (MJ Research Inc., Watertown, MA, USA) with the following conditions: initial denaturation for 3 min at 95°C, followed by 30 cycles of (30 sec at 95°C, 30 sec at 60°C, 1 min at 72°C) and a final extension for 10 min at 72°C. Newly designed intron specific primers were used for PCRs (Table 3), which were carried out with genomic DNA isolated from algal strains and from whole lichen DNA extracts. PCR amplification was carried out with the following settings: 3 min at 95°C, 30 cycles of 30 sec at 95°C, 40 sec at 56°C, and 1 min 20 sec at 72°C with final extension at 72°C for 10 min. Sequencing was carried out using ABI Prism BigDye Terminator Mix V3.0 Cycle Sequencing Kit (Life Technologies, Rotkreuz, Switzerland) following the protocol of the manufacturer and capillary electrophoresis of cycle sequencing products was performed on an Applied BioSystems ABI 3730 DNA Analyzer (Life Technologies, Rotkreuz, Switzerland).
Table 3.
List of newly designed primers used in this study
| Primer | Target | Sequence (5′→3′) | Position | Relative to Accession |
|---|---|---|---|---|
| Intron 1 (fwd) | SSU intron | CTGTCACTAGACTGAGTGC | 208 | AM159503 |
| Intron 2 (rev) | SSU intron | CCAGTTTAGAGGCTCGAATC | 601 | AM159503 |
| Intron 3 (rev) | SSU intron | GGWCCGACTATATCTTAAGC | 1716 | AJ969509 |
| Intron 4 (fwd) | SSU intron | TTGTTGTAAAGGGCTCCACT | 709 | AJ969509 |
| Intron 5 (rev) | SSU intron | ACTCCTGTAAGCTCTCCTTCC | 1756 | AJ969509 |
| LSU fwd | LSU | TTTAACACCCATGAGACGCAAGTAAC | 684 | D17810 |
| LSU rev | LSU | GCCTTAACTCAGCTTTCGGTTCA | 1607 | D17810 |
Sequence alignment, secondary structure prediction, and phylogenetic analysis
Sequence contigs were assembled using SequencherTM 4.2.2 (Gene Codes Corp. Ann Arbor, USA) and ambiguous positions were manually corrected. The final sequences were aligned using ClustalX (Thompson et al. 1997). The resulting alignment was visually checked for any discrepancies and manually corrected on MacClade V5.0 (Maddison & Maddison, 2002). Intron insertion position was determined based on BLAST searches and sequence alignments. The putative secondary structure of the L798 intron was determined using comparisons to existing structures and m-fold (Zuker, 2003) following the guidelines of Cech (1988) and Cech et al. (1994).
Maximum likelihood (ML) analysis was carried out using PAUP 4.0 b10 (Swofford, 2003). Bayesian phylogenetic reconstruction was performed with MrBayes version 3.1.2 (Ronquist & Huelsenbeck 2003, Huelsenbeck & Ronquist 2001). To determine the best models according to the AIC criterion (Posada & Buckley 2004, Sakamoto et al. 1986) for ML and Bayesian phylogenetic analyses, MrModeltest version 2.3 (Nylander, 2004) was applied to each data set. For the LSU and SSU green algal and SSU intron data sets, the General Time Reversible (Tavaré, 1986) (GTR+G) model provided the best fit to the data as determined by AIC in MrModeltest. In contrast, the model best describing the LSU intron data set according to AIC was a version of the General Time Reversible (GTR+I+G, AIC=8411.15) although its AIC was only slightly lower than that of GTR+G model (AIC=8411.28). Using the models determined by AIC in MrModeltest, ML and Bayesian analyses were performed on three data sets: 1) Twenty-eight rDNA sequences of the rDNA region excluding all introns, i.e. the “host” of the intron (data set ‘LSU host’), compiled for comparability with the LSU L798 intron sequences. The host cell phylogeny, as presented in Figure 1, is based on alignments of the complete ITS region, including the 5.8S rRNA gene. 2) The LSU intron, comprising 31 sequences (‘LSU intron’); and 3) thirty-six DNA sequences of the SSU S1512 intron (‘SSU intron’). To assess for statistical support, the ML analysis in PAUP used 100 bootstrap replicates in a heuristic search based on optimality criterion likelihood, with random addition of sequences. Maximum parsimony analysis was performed in PAUP with 100 bootstraps. Tree-bisection-reconnection was used as a branch-swapping algorithm. In MrBayes, analyses were run for 5 Mio generations with four chains sampling the parameter space, sampling trees every 1,000 generations for a total of 5,000 trees, reaching stationarity and receiving effective sample sizes > 3,000 as assessed using Tracer version 1.5 (Rambaut & Drummond 2007). Of the sampled trees, 10% were discarded as burn-in. TreeAnnotator version 1.6.0, part of the Beast software (Drummond et al. 2013), was used to summarize trees and create a maximum clade credibility tree which was visualized using FigTree version 1.4.1 (Rambaut 2008).
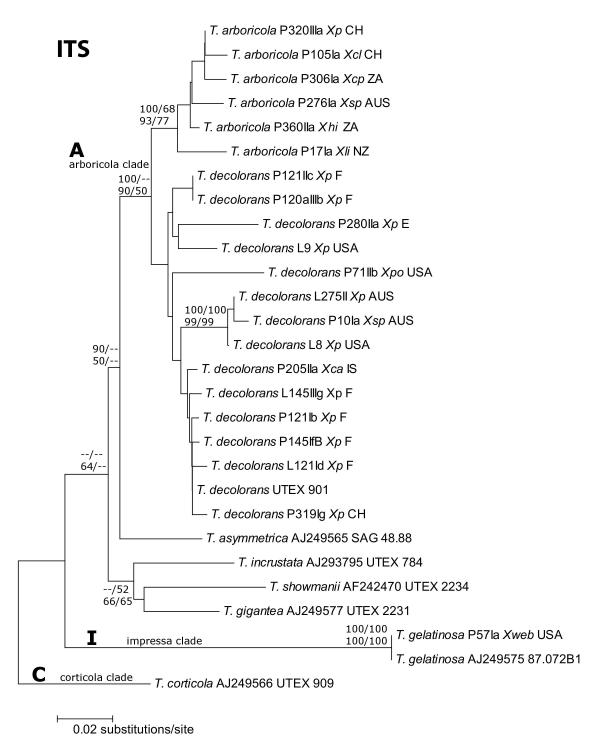
Fig. 1.
Phylogeny of Trebouxia s. lat.. Sequences obtained from GenBank are indicated in Table 3. Support values shown at a respective node are posterior probabilities of nodes from Bayesian analysis in MrBayes (first row, first number), values from 100 bootstrap replicates in a maximum likelihood analysis in Paup (first row, second number), values from 500 bootstrap replicates in a neighbor-joining analysis in MEGA (second row, first number) and maximum parsimony support values from 100 bootstraps generated with PAUP (second row, second number). Clades A, I and C represent ‘arboricola’, ‘impressa’ and ‘corticola’, respectively, as proposed by Helms (2003). The identity of photobionts was inferred on the basis of ITS and rbcL phylogenetic analyses including reference strains (Nyati, Scherrer, Werth and Honegger, in press). Abbreviations used: Xca: Xanthoria candelaria, Xcl: X. calcicola, Xcp: X. capensis, Xec: X. ectaneoides, Xh: X. hirsuta, Xli: X. ligulata, Xp: X. parietina, Xpo: X. polycarpa, Xtu: X. turbinata, Xsp: Xanthoria sp., Xweb: Xanthomendoza weberi.. Phylogeny of the nuclear ribosomal internal transcribed spacer region (complete ITS region, including the 5.8S rRNA gene) as inferred using Maximum Likelihood (ML) analysis. Asterochloris sequences not used in analyses due to very high sequence divergence.
To compare whether partitioned rDNA-host and intron data sets were homogeneous, incongruence length difference tests (Farris et al. 1995) were performed on two combined data sets in PAUP, using 1000 replicates and a heuristic search strategy, with parsimony as optimality criterion. The first combined data set contained 23 sequences of the L798 intron with corresponding rDNA host sequences (‘Combined LSU intron’). The second combined data set contained 33 DNA sequences of the S1512 intron with their corresponding rDNA host sequences (‘Combined SSU intron’).
Missing characters, gaps, and ambiguous sites were excluded in all phylogenetic analyses. For the visualization of phylogenetic results, distance trees were constructed by the neighbor-joining algorithm implemented in MEGA version 5 (Tamura et al. 2007) and annotated with support values from Bayesian and ML analyses as well as with support values from 500 bootstraps using the neighbor-joining algorithm in MEGA.
RESULTS AND DISCUSSION
Trebouxia L798 group I intron
In the current investigation 28 LSU rDNA group I introns were found, which are all inserted at position L798 and belong to group IB introns (Fig. 2). In a secondary structure based alignment of Trebouxia LSU introns, the sequence of Chlorella angustoellipsoidea C-87 was included (Aimi et al. 1994) because this intron was also 445 base pairs (bp) in length and had all the characteristic RNA folding (P1-P10) properties of group IB introns (Burke et al. 1987). The flanking exon regions were also highly conserved.
Fig. 2.
Putative secondary structure of the group I intron within the large subunit rRNA precursor of Trebouxia decolorans UTEX 901 drawn according to the conventions of Cech et al. (1994). Arrows point to the 5′ and 3′ splice junctions of this L798 group I intron. Also shown are the locations of the pairing segments P1-P10. The solid lines are used to position secondary structure elements that are believed to interact in close proximity.
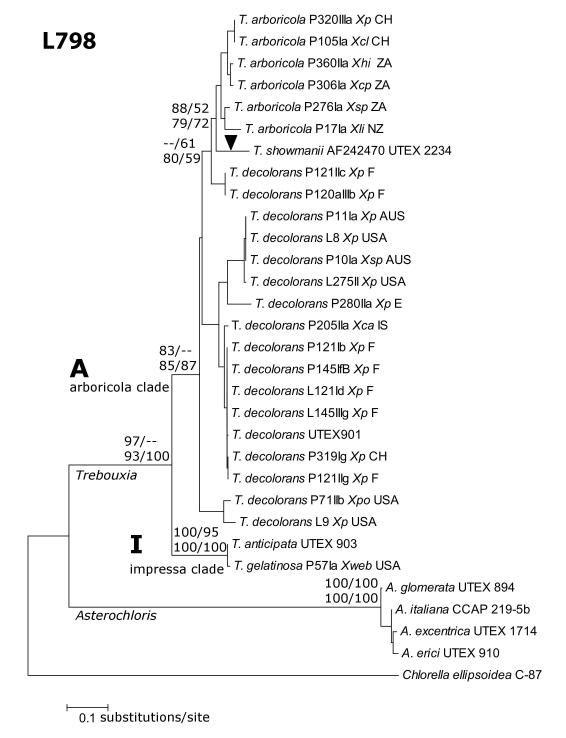
The L798 group I intron phylogeny (Fig 3) showed a similar grouping of taxa into clades as the ITS host cell tree (Fig. 1), indicating vertical inheritance of the intron. The only exception was the intron of T. showmanii (Figs. 1 and 3), which was placed in the ‘arboricola’ clade, indicating the possibility of lateral transfer. A comparison of the conserved core catalytic sequences (P, Q, R, S) revealed high similarity with multiple nucleotide substitutions between Trebouxia subclades and the Asterochloris clade (Table 4). Moreover, an incongruent length difference test rejected the hypothesis of homogeneity among intron and host data sets (p=0.005). Due to the conserved secondary structures among the LSU group I intron of representatives of the genera Trebouxia and Asterochloris (Microthamniales) and of Chlorella angustoellipsoidea (Chlorellales; Table 4), we hypothesize that this intron was present in the common ancestor of the genera Trebouxia and Asterochloris.
Fig. 3.
Phylogeny of the LSU 798 group I intron in representatives of Trebouxia s. str. and Asterochloris Tscherm.-Woess. An intron sequence of Chlorella angustoellipsoidea strain C-87 was obtained from GenBank (accession D17810) and used as outgroup taxon. Arrowhead indicates potential site for lateral intron transfer. All the support values were generated as in Figure 2.
Table 4.
Comparison of catalytic core (PQRS) sequences of LSU 798 group I introns in representatives of the genera Trebouxia s. str. and Asterochloris. The identity of photobionts of the investigated Xanthoria and Xanthomendoza species was inferred on the basis of ITS and rbcL analyses. The positions of P, Q, R, S regions are based on the reference sequence of the Chlorella angustoellipsoidea strain C-87 intron (accession number D17180).
| P | Q | R | S | |
|---|---|---|---|---|
| T. decolorans | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| L145IIIg Xp F | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| L121Id Xp F | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P121IIg Xp F | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P145IdfB Xp F | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P319Ig Xp CH | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P120aIIIb Xp F | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P121II cd Xp F | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P280IIa Xp E | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P205IIa Xca IS | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P121Ib Xp F | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P71IIb Xpo USA | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAGA | AAGAUAUAGUCG |
| L9 Xp USA | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUGUAGUCG |
| L8 Xp USA | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAAAUAUAGUCG |
| L275II Xp USA | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAAAUAUAGUCG |
| P11Ia Xp AUS | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAAAUAUAGUCG |
| P10Ia Xsp AUS | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAAAUAUAGUCG |
| P306Ia Xcp ZA | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P360IIa Xhi ZA | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P105Ia Xcl CH | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P276Ia Xsp ZA | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P320IIIa Xp CH | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P17Ia Xli NZ | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| T. showmanii | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUACA | AAGAUAUAGUCG |
| T. anticipata | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| P 57 Ia Xweb U | AAUUGCGGGGAC | AAUCCGCAGC | GUUCACAGACUAAA | AAGAUAUAGUCG |
| T. glomerata | AAUUGCCGGGAC | AAUCGGCAGC | GUUCACAGACUAGA | AAGGUAUAGUCG |
| T. italiana | AAUUGCCGGGAC | AAUCGGCAGC | GUUCACAGACUAGA | AAGAUAUAGUCG |
| T. excentrica | AAUUGCCGGGAC | AAUCGGCAGC | GUUCACAGACUAGA | AAGAUAUAGUCG |
| T. erici | AAUUGCCGGGAC | AAUCGGCAGC | GUUCACAGACUAGA | AAGAUAUAGUCG |
| C. angustoellipsoidea | AAUUGCGGGAAA | AAUCCGCAGC | GUUCACAGACUAGA | AAGGUAUAGUCG |
Trebouxia S1512 group I intron
Intron specific primers were used for PCR amplification reactions, which were carried out with genomic DNA isolated from algal strains and from whole lichen DNA extracts. Several different primer combinations were used to exclude PCR bias. These tests were done to infer the frequencies of introns in the investigated algal isolates. Of the 124 nrDNA sequences generated for Trebouxia photobionts of Xanthoria s. lat and Xanthomendoza spp., 32 contained a group I intron. The intron-harboring Trebouxia photobionts belonged to the “arboricola” clade A as described by Helms (2003). In all photobiont samples except for one, the intron varied between 434 bp and 530 bp in length. The 460 bp intron of the T. arboricola photobiont of the African Xanthoria turbinata (accession No. AJ969509) had an additional insertion of 1090 bp at a conserved site, making the entire intron sequence 1550 bp in length. Although this intron is very long, it does encode an ORF for a homing endonuclease as assessed by blast searches (Altschul et al. 1997). No similarities were found between this longer intron and complex nested inserts found in 18S rDNA of ascomycetes, where putative spliceosomal introns were inserted within group I introns (DePriest & Been 1992, DePriest 2004, Myllys et al. 1999)
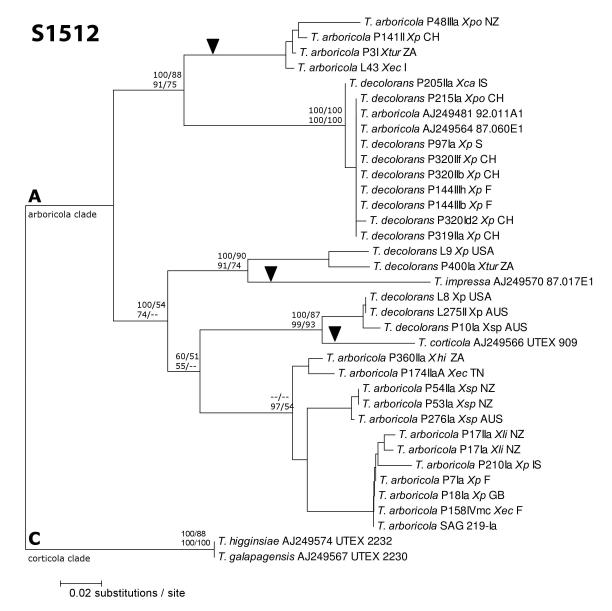
In comparison with published data (Bhattacharya et al. 1996b) all insertions at position 1512 were identified as group I introns. Twenty-nine newly generated intron sequences together with available data were analyzed using phylogenetic methods (Figs. 3 and 4) and compared with the ITS phylogeny (Fig. 1) to identify potential cases of lateral transfer (Bhattacharya et al. 1996b, Friedl et al. 2000). The ITS and S1512 group I intron phylogenies showed a similar grouping of clades (Figs. 1 and 4), indicating overall vertical inheritance. However, incongruence length difference tests suggested that the data sets were not homogeneous (p=0.001). Indeed, three interesting incongruities were found among host and intron phylogenies (marked with arrowheads in Fig. 4): 1) the intron sequence of Trebouxia corticola (UTEX 909, a putatively free-living alga from tree bark in Webster, MA, USA isolated by V. Ahmadjian in 1959), a representative of the “corticola” clade C according to (Helms, 2003), falls near three intron sequences from clade A. These Trebouxia decolorans photobionts were identified from X. parietina thalli from Barossa valley (AUS) and Sonoma County (California, USA) and from an unnamed Xanthoria species from Tasmania (AUS). 2) The intron sequence of Trebouxia impressa (Friedl et al. 2000), a representative of the “impressa” clade I according to Helms (2003), falls near two T. decolorans intron sequences from clade A. These Trebouxia decolorans photobionts were found in thalli of X. parietina and Xanthomendoza hasseana in California. 3) Intron sequences of T. arboricola isolates from X. ectaneoides (Sicily, Italy), X. parietina (Avenches, Switzerland), X. polycarpa (Otago, New Zealand), and X. turbinata (Port Nolloth, South Africa) formed a separate group outside the group of samples belonging to T. decolorans, receiving high support in all three analyses (Fig 4). All these incongruities are likely to have resulted from intron lateral transfers.
Fig. 4.
Phylogeny of the SSU 1512 group I intron in representatives of Trebouxia s. str. All the support values were generated as in Figure 2.
LSU and SSU group I intron distribution in Trebouxia and Asterochloris spp
Introns in both SSU and LSU rDNA were found in only nine Trebouxia isolates. The distribution of these taxa in the photobionts of Xanthoria s. lat. (Table 1) and in the host tree (Table 2) follows neither a geographic nor a taxonomic pattern. Trebouxia isolates from thalli of Xanthoria parietina from the Northern Hemisphere are however over-represented in this analysis, and only one sample was studied from some of the other species. In a survey of the genetic diversity among Trebouxia photobionts of Teloschistaceae (n = 124) the S1512 group I intron was found in 50% of T. arboricola, but in only 23% of T. decolorans samples (Nyati et al. submitted). Friedl et al. (2000) found a S1512 intron in 28% of 85 algal strains that were tested. In a PCR-based screening of sterile cultured isolates of T. arboricola or T. decolorans, respectively, from populations of Xanthoria parietina and X. ectaneoides from maritime, coastal, rural, and urban sites (for details see Itten & Honegger 2010) a very unequal distribution of S1512 group I introns was observed among populations, ranging from none to 87% (Table 5). The percentage distribution of introns was assessed by combining sequences obtained from several photobiont isolates from within a single lichen thallus as well as from a number of different lichen thalli within a population. This result may reflect the dynamic nature of intron gain and loss. Very high genetic diversity by RADP-PCR fingerprinting has been observed from the axenically cultured mycobiont isolates from the same populations (Itten & Honegger 2010). The Trebouxia photobionts of Xanthoria ectaneoides or X. parietina populations with the highest and lowest percentages of S1512 group I introns were collected less than 1 km apart from each other. The first was found on granite rock in the supralittoral fringe of a tiny island (Île verte), the other on shrubs in the old port of Roscoff in Brittany (NW France; Table 5). However, we cannot exclude that recombination could redistribute introns among individual algal cells in free-living populations of Trebouxia, followed by gene flow, which would lead to the same pattern of intron distribution in populations.
Table 5.
Presence of SSU1512 group I intron in axenically cultured Trebouxia isolates from five Xanthoria populations collected in Europe
| Photobiont | Voucher No. |
Mycobiont | Collecting site |
Substrate | Introna/ isolates, % |
|---|---|---|---|---|---|
| Trebouxia arboricola | 158 | X. ectaneoides | maritime, F | saxicolous | 20/23, 87% |
| T. decolorans | 120-121 | X. parietina | rural, F | epiphytic | 1/25, 4% |
| T. decolorans | 144-145 | X. parietina | rural, F | epiphytic | 6/63, 9.5% |
| T. decolorans | 164 | X. parietina | coastal, F | epiphytic | 0/12, 0% |
|
T. arboricola,
T. decolorans |
319-320 | X. parietina | urban, CHb | epiphytic, saxicolous |
18/47, 38% |
Presence of intron tested with PCR assays;
All T. decolorans isolates were photobiont of epiphytic samples, while T. arboricola was identified from saxicolous specimens. F, France. CH, Switzerland. Details about these Xanthoria populations are found in Itten & Honegger (2010).
Introns of lichen photobionts and mycobionts
The origin of S1512 group I introns in chlorophytes is still a matter of debate. Based on the observation of a close phylogenetic relationship of the S1512 group I intron lineage in chlorophytes to viral introns found in Chlorella spp., the viruses were hypothesized to be either the source or at least the vector, which facilitates the spread of group I introns among eukaryotes (Bhattacharya et al. 1996b, Aimi et al. 1994).
Horizontal transfer of group I introns is known from many biological systems. Examples of interkingdom transfers are: 1) plant parasitic fungus to host plant (Nishida & Sugiyama 1995, Vaughn et al. 1995, Sanchez-Puerta et al. 2008); 2) fungus to green alga (Lindstrom & Pistolic 2005); 3) fungus to red alga (Müller et al. 2005); 4) red alga to brown alga (Bhattacharya et al. 2001); 5) algae to amoebae (Turmel et al. 1995); 6) between eubacteria and chloroplasts (Kuhsel et al. 1990). Inter-organellar transfer of group I introns has been shown from mitochondria to the nucleus (Curtis & Archibald 2010), and from mitochondria to the chloroplast (Pombert et al. 2005). Interfamilial transfers among lichen-forming ascomycetes within the Physciaceae were described by Simon et al. (2005). Based on the close relationship of introns in green algal lichen photobionts with bacterial and fungal introns, del Campo et al. (2009) proposed lichen thalli to be potential sites of horizontal transfer of introns. Indeed, already in the Lower Devonian (approx. 415 Myr ago) were the thalli of lichen-forming fungi microcosms, built up by the quantitatively predominant lichen-forming fungus and its photoautotrophic green algal or cyanobacterial partner, but comprising also large numbers of epi- and endolichenic bacteria and non-lichenized fungi, all in close physical contact with the lichen mycobiont and partly also with the photobiont (Honegger et al. 2013), as is the case in extant lichens (Honegger 2012), However, no direct evidence has yet been reported demonstrating SSU rDNA group I intron movement between the fungal and algal partners in the lichen symbiosis.
Only few insertion positions (S516, S943, S1046, and S1506) for group I introns are found in both green algae and ascomycetes (Gargas et al. 1995b). The SSU intron at position S1512, which is present on the surface of the mature ribosome in tertiary structure, is restricted to green algae (Gargas et al. 1995a, Gargas et al. 1995b); it has never been found in fungi or other organisms. DePriest and Been (1992) concluded that group I introns of lichen algae do not originate from their respective fungal partners or vice versa. Bhattacharya et al. (2002) demonstrated that there is no evolutionary relationship between group I introns of lichen-forming ascomycetes and their green algal partners; thus horizontal transfer can be excluded. One process that might lead to a similar pattern of intron distribution as observed in our study is photobiont switching (Piercey-Normore & DePriest 2001, Nelsen & Gargas 2008, Nelsen & Gargas 2009). Photobiont switches could occur if a developing germling incorporates either a free-living alga, or an alga that has been released from a thallus e.g. after being digested by invertebrates. Algal cells have been shown to survive the gut passage of mites (Meier et al. 2002) and snails (Boch et al. 2011), providing an opportunity for algal uptake by developing germlings. Friedl et al. (2000) suggested that lichenization might facilitate the spread of 1512 introns among algal strains that coexist in fungal thalli. However, it is difficult to imagine how intron gain might proceed within the lichen thallus. In morphologically advanced, foliose, or fruticose lichens the algal cells have physical contact only with sister cells derived from the same mother cell, but each of them is in direct contact with the fungal partner via the appressorial or haustorial complex (Honegger, 1991). A mycobiont-derived, water-repellent wall surface layer composed of hydrophobins and/or other hydrophobic wall surface components (Scherrer et al. 2000), ensheaths the algal and fungal surfaces in the thalline interior (Honegger 1991, Honegger 2001). During the regular wetting and drying cycles solutes are passively exchanged underneath this hydrophobic sealing via the apoplastic continuum of both partners, but it seems unlikely that mobile genetic elements would be transmitted through this process. It is conceivable that intron gain occurs while lichen photobionts are not symbiotic, but free-living and in contact with cells of other species or strains. The example of the S1512 group I intron of the type strain of Trebouxia corticola, as reported in this study, is consistent with this idea. This strain was isolated from putatively free-living cells, not from a lichen, and its intron is similar to that in Trebouxia decolorans isolates from a different clade, the “arboricola” clade A (i.e., Helms 2003). According to the literature, Trebouxia arboricola, the type species of the genus and photobiont of numerous lichen-forming ascomycetes, including Teloschistaceae, is also a common and widespread free-living alga (Rindi & Guiry 2003, John et al. 2002, Ettl & Gärtner 1995, Beck et al. 1998), despite contradictory claims (Ahmadjian 1988). The mechanisms underlying intron gain in lichen photobionts merit therefore a thorough investigation in future studies.
ACKNOWLEDGEMENTS
Our sincere thanks go to Prof. Georg Gärtner, Innsbruck, for generous gifts of reference strains, and to the Swiss National Science Foundation for financial support (grant Nr. 31-103860/1 to R.H.). D.B. acknowledges generous support from the National Science Foundation and the National Institutes of Health (grants EF 04-31117 and R01 ES013679, respectively).
Abbreviations
- LSU
large subunit
- SSU
small subunit
- ITS
internal transcribed spacer
Contributor Information
Shyam Nyati, Institute of Plant Biology, University of Zurich, Zollikerstrasse 107, 8008 Zurich, Switzerland Department of Radiation Oncology, University of Michigan, 109 Zina Pitcher Place, Ann Arbor, Michigan 48109, United States.
Debashish Bhattacharya, Department of Ecology, Evolution and Natural Resources and Institute of Marine and Coastal Science, Rutgers University, 59 Dudley Road, New Brunswick, New Jersey 08901, USA.
Silke Werth, Faculty of Life- and Environmental Sciences, University of Iceland, Sturlugata 7, 101 Reykjavík, Iceland.
Rosmarie Honegger, Institute of Plant Biology, University of Zurich, Zollikerstrasse 107, 8008 Zurich, Switzerland.
References
- Ahmadjian V. The lichen alga Trebouxia - does it occur free-living. Plant. Syst. Evol. 1988;158:243–47. [Google Scholar]
- Aimi T, Yamada T, Yamashita M, Murooka Y. Characterization of the nuclear large subunit ribosomal RNA encoding gene and the group-I self splicing intron from Chlorella ellipsoidea C-87. Gene. 1994;145:139–44. doi: 10.1016/0378-1119(94)90337-9. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic acids res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachar D, Guillou L, Christen R. Detection of introns in eukaryotic small subunit ribosomal RNA gene sequences. Dataset Papers in Biology. 2013;2013:12. [Google Scholar]
- Beck A, Friedl T, Rambold G. Selectivity of photobiont choice in a defined lichen community: inferences from cultural and molecular studies. New Phytol. 1998;139:709–20. [Google Scholar]
- Bhattacharya D, Cannone JJ, Gutell RR. Group I intron lateral transfer between red and brown algal ribosomal RNA. Curr. Genet. 2001;40:82–90. doi: 10.1007/s002940100227. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Damberger S, Surek B, Melkonian M. Primary and secondary structure analyses of the rDNA group-I introns of the Zygnematales (Charophyta) Curr. Genet. 1996a;29:282–86. doi: 10.1007/BF02221559. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Friedl T, Damberger S. Nuclear-encoded rDNA group I introns: Origin and phylogenetic relationships of insertion site lineages in the green algae. Mol. Biol. Evol. 1996b;13:978–89. doi: 10.1093/oxfordjournals.molbev.a025666. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Friedl T, Helms G. Vertical evolution and intragenic spread of lichen-fungal group I introns. J. Mol. Evol. 2002;55:74–84. doi: 10.1007/s00239-001-2305-x. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Surek B, Rusing M, Damberger S, Melkonian M. Group I introns are inherited through common ancestry in the nuclear encoded ribosomal RNA of Zygnematales (Charophyceae) Pro. Nat. Acad. Sci. USA. 1994;91:9916–20. doi: 10.1073/pnas.91.21.9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Weber K, An SS, Berning-Koch W. Actin phylogeny identifies Mesostigma viride as a flagellate ancestor of the land plants. J. Mol. Evol. 1998;47:544–50. doi: 10.1007/pl00006410. [DOI] [PubMed] [Google Scholar]
- Boch S, Prati D, Werth S, Rüetschi J, Fischer M. Lichen endozoochory by snails. PLoS ONE. 2011;6:e18770. doi: 10.1371/journal.pone.0018770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JM, Belfort M, Cech TR, Davies RW, Scweyen RJ, Shub DA, Szostak JW, Tabak HF. Structural conventions for group I introns. Nucleic acids res. 1987;15:7217–21. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR. Conserved sequences and structures of Group-I introns - building an active-site for RNA catalysis - a review. Gene. 1988;73:259–71. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- Cech TR, Damberger SH, Gutell RR. Representation of the secondary and tertiary structure of Group-I Introns. Nat. Struct. Biol. 1994;1:273–80. doi: 10.1038/nsb0594-273. [DOI] [PubMed] [Google Scholar]
- Curtis BA, Archibald JM. A spliceosomal intron of mitochondrial DNA origin. Curr. Biol. 2010;20:R919–20. doi: 10.1016/j.cub.2010.09.038. [DOI] [PubMed] [Google Scholar]
- del Campo EM, Casano LM, Gasulla F, Barreno E. Presence of multiple group I introns closely related to bacteria and fungi in plastid 23S rRNAs of lichen-forming Trebouxia. Int. microbiol.: J. Spanish Soc. Microbiol. 2009;12:59–67. [PubMed] [Google Scholar]
- Del Campo EM, Casano LM, Gasulla F, Barreno E. Suitability of chloroplast LSU rDNA and its diverse group I introns for species recognition and phylogenetic analyses of lichen-forming Trebouxia algae. Mol. Phylogenet. Evol. 2010;54:437–44. doi: 10.1016/j.ympev.2009.10.024. [DOI] [PubMed] [Google Scholar]
- DePriest PT. Small subunit rDNA variation in a population of lichen fungi due to optional group I introns. Gene. 1993;134:67–74. doi: 10.1016/0378-1119(93)90175-3. [DOI] [PubMed] [Google Scholar]
- DePriest PT. Early molecular investigations of lichen-forming symbionts: 1986-2001. Annu. Rev. Microbiol. 2004;58:273–301. doi: 10.1146/annurev.micro.58.030603.123730. [DOI] [PubMed] [Google Scholar]
- DePriest PT, Been MD. Numerous group I introns with variable distributions in the ribosomal DNA of a lichen fungus. J.Mol. Biol. 1992;228:315–21. doi: 10.1016/0022-2836(92)90819-6. [DOI] [PubMed] [Google Scholar]
- Drummond A, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2013 doi: 10.1093/molbev/mss075. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations - a Review. Gene. 1989;82:91. doi: 10.1016/0378-1119(89)90034-6. &. [DOI] [PubMed] [Google Scholar]
- DEL CAMPO EM, GIMENO J, DE NOVA JPG, CASANO LM, GASULLA F, GARCÍA-BREIJO F, REIG-ARMIÑANA J, BARRENO2 E. South European populations of Ramalina farinacea (L.) Ach. share different Trebouxia algae. In: T. H. N. I., editor. Biology of Lichens – Symbiosis, Ecology, Environm. Monitoring, Systematics, Cyber Applications. J. Cramer in der Gebrüder Borntraeger Verlagsbuchhandlung; Stuttgart: 2010. pp. 247–56. [Google Scholar]
- Einvik C, Elde M, Johansen S. Group I twintrons: genetic elements in myxomycete and schizopyrenid amoeboflagellate ribosomal DNAs. J. Biotechnol. 1998;64:63–74. doi: 10.1016/s0168-1656(98)00104-7. [DOI] [PubMed] [Google Scholar]
- Ettl H, Gärtner G. Syllabus der Boden-, Luft-und Flechtenalgen. Gustav Fischer; Stuttgart, Germany: 1995. [Google Scholar]
- Farris JS, Källersjö M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1995;10:315–19. [Google Scholar]
- Friedl T, Besendahl A, Pfeiffer P, Bhattacharya D. The distribution of group I introns in lichen algae suggests that lichenization facilitates intron lateral transfer. Mol. Phylogenet. Evol. 2000;14:342–52. doi: 10.1006/mpev.1999.0711. [DOI] [PubMed] [Google Scholar]
- Gargas A, Depriest PT, Grube M, Tehler A. Multiple origins of lichen symbioses in fungi suggested by SSU rDNA phylogeny. Science. 1995a;268:1492–95. doi: 10.1126/science.7770775. [DOI] [PubMed] [Google Scholar]
- Gargas A, Depriest PT, Taylor JW. Positions of multiple insertions in SSU rDNA of lichen-forming fungi. Mol. Biol. Evol. 1995b;12:208–18. doi: 10.1093/oxfordjournals.molbev.a040199. [DOI] [PubMed] [Google Scholar]
- Gutierrez G, Blanco O, Divakar PK, Lumbsch HT, Crespo A. Patterns of group I intron presence in nuclear SSU rDNA of the Lichen family Parmeliaceae. J. Mol. Evol. 2007;64:181–95. doi: 10.1007/s00239-005-0313-y. [DOI] [PubMed] [Google Scholar]
- Harris LB, Rogers SO. Evolution of small putative group I introns in the SSU rRNA gene locus of Phialophora species. BMC Res. Notes. 2011;4:258. doi: 10.1186/1756-0500-4-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen P, Simon DM, Bhattacharya D. The natural history of group I introns. Trends Genet. 2005a;21:111–19. doi: 10.1016/j.tig.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Haugen P, Wikmark O-G, Vader A, Coucheron DH, Sjøttem E, Johansen SD. The recent transfer of a homing endonuclease gene. Nucleic Acids Res. 2005b;33:2734–41. doi: 10.1093/nar/gki564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms G. PhD thesis. University of Göttingen; 2003. Taxonomy and symbiosis in associations of Physciaceae and Trebouxia; p. 156. [Google Scholar]
- Honegger R. Functional aspects of the lichen symbiosis. Ann. Rev. Plant Physiol. Mol. Biol. 1991;42:553–78. [Google Scholar]
- Honegger R. The symbiotic phenotype of lichen-forming ascomycetes. In: Hock B, editor. Fungal associations. Springer; Berlin: 2001. pp. 165–88. [Google Scholar]
- Honegger R. The impact of different long-term storage conditions on the viability of lichen-forming ascomycetes and their green algal photobiont, Trebouxia spp. Plant Biol. 2003;5:324–30. [Google Scholar]
- Honegger R. Differential gene expression within the cyanobacterial cell population of a lichen thallus. New Phytol. 2012;196:657–60. doi: 10.1111/j.1469-8137.2012.04361.x. [DOI] [PubMed] [Google Scholar]
- Honegger R, Edwards D, Axe L. The earliest records of internally stratified cyanobacterial and algal lichens from the Lower Devonian of the Welsh Borderland. New Phytol. 2013;197:264–75. doi: 10.1111/nph.12009. [DOI] [PubMed] [Google Scholar]
- Hoshina R, Imamura N. Phylogenetically close group I introns with different positions among Paramecium bursaria photobionts imply a primitive stage of intron diversification. Mol. Biol. Evol. 2009;26:1309–19. doi: 10.1093/molbev/msp044. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–55. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Itten B, Honegger R. Population genetics in the homothallic lichen-forming ascomycete Xanthoria parietina. Lichenologist. 2010;42:751–61. [Google Scholar]
- Johansen S, Muscarella DE, Vogt VM. Insertion elements in ribosomal DNA. CRC Press; 1996. pp. 89–110. [Google Scholar]
- John DM, Whitton BA, Brook AJ. The freshwater algal flora of the British Isles: An identification guide to freshwater and terrestrial algae. Cambridge University Press; Cambridge, U.K.: 2002. [Google Scholar]
- Kuhsel MG, Strickland R, Palmer JD. An ancient Group-I intron shared by eubacteria and chloroplasts. Science. 1990;250:1570–73. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- Lindstrom SC, Pistolic J. Detection of a Group I (IE) fungal intron in the green algal genus Urospora (Ulvophyceae) J. Phycol. 2005;41:359–65. [Google Scholar]
- Maddison WP, Maddison DR. MacClade. Sinauer Associates; Sundarland, Massachusetts, USA: 2002. [Google Scholar]
- McManus HA, Lewis LA, Fucikova K, Haugen P. Invasion of protein coding genes by green algal ribosomal group I introns. Mol. Phylogenet. Evol. 2012;62:109–16. doi: 10.1016/j.ympev.2011.09.027. [DOI] [PubMed] [Google Scholar]
- Meier FA, Scherrer S, Honegger R. Faecal pellets of lichenivorous mites contain viable cells of the lichen-forming ascomycete Xanthoria parietina and its green algal photobiont, Trebouxia arboricola. Biol. J. Linnean Soc. 2002;76:259–68. [Google Scholar]
- Michel F, Westhof E. Modeling of the 3 dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Müller KM, Ellenor DW, Sherwood AR, Sheath RG, Cannone JJ, Gutell RR. Evidence for lateral transfer of an IE intron between fungal and red algal small subunit rRNA genes. J. Phycol. 2005;41:380–90. [Google Scholar]
- Myllys L, Kallersjo M, Tehler A. Variable sizes of introns in the SSU rDNA in three species of Roccella (Arthoniales, Euascomycetes) Curr. Genet. 1999;36:79–85. doi: 10.1007/s002940050475. [DOI] [PubMed] [Google Scholar]
- Nelsen MP, Gargas A. Dissociation and horizontal transmission of codispersing lichen symbionts in the genus Lepraria (Lecanorales: Stereocaulaceae) New Phytol. 2008;177:264–75. doi: 10.1111/j.1469-8137.2007.02241.x. [DOI] [PubMed] [Google Scholar]
- Nelsen MP, Gargas A. Symbiont flexibility in Thamnolia vermicularis (Pertusariales: Icmadophilaceae) Bryologist. 2009;112:404–17. [Google Scholar]
- Nielsen H, Johansen SD. Group I introns: Moving in new directions. RNA Biol. 2009;6:375–83. doi: 10.4161/rna.6.4.9334. [DOI] [PubMed] [Google Scholar]
- Nishida H, Sugiyama J. A common group-I intron between a plant-parasitic fungus and its host. Mol. Biol. Evol. 1995;12:883–86. doi: 10.1093/oxfordjournals.molbev.a040264. [DOI] [PubMed] [Google Scholar]
- Nyati S, Scherrer S, Werth S, Honegger R. Green algal photobiont diversity (Trebouxia spp.) in representatives of Teloschistaceae (Lecanoromycetes, lichen-forming ascomycetes) The Lichenologist. submitted. [Google Scholar]
- Nylander JAA. Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest v2. Program distributed by the author. Available from http://www.abc.se/~nylander/ [Google Scholar]
- Piercey-Normore MD, DePriest PT. Algal switching among lichen symbioses. Am. J. Bot. 2001;88:1490–98. [PubMed] [Google Scholar]
- Pombert JF, Keeling PJ. The mitochondrial genome of the entomoparasitic green alga helicosporidium. PLoS One. 2010;5:e8954. doi: 10.1371/journal.pone.0008954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombert JF, Otis C, Lemieux C, Turmel M. The chloroplast genome sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) reveals unusual structural features and new insights into the branching order of chlorophyte lineages. Mol. Biol. Evol. 2005;22:1903–18. doi: 10.1093/molbev/msi182. [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Rambaut A. FigTree version 1.2. 2008 Available from http://tree.bio.ed.ac.uk/software/figtree/
- Rambaut A, Drummond A. Tracer v 1.4. 2007 Available from http://beast.bio.ed.ac.uk/Tracer.
- Reeb V, Haugen P, Bhattacharya D, Lutzoni F. Evolution of Pleopsidium (lichenized ascomycota) S943 group I introns and the phylogeography of an intron-encoded putative homing endonuclease. J. Mol. Evol. 2007;64:285–98. doi: 10.1007/s00239-005-0179-z. [DOI] [PubMed] [Google Scholar]
- Rindi F, Guiry MD. Composition and distribution of subaerial algal assemblages in Galway City, western Ireland. Cryptogam. Algol. 2003;24:245–67. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–74. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ruoff B, Johansen S, Vogt VM. Characterization of the self-splicing products of a mobile intron from the nuclear rDNA of Physarum polycephalum. Nucleic Acids Res. 1992;20:5899–906. doi: 10.1093/nar/20.22.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y, Ishiguro M, Kitagawa G. Akaike Information Criterion Statistics. D. Reidel; Dordrecht: 1986. [Google Scholar]
- Sanchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD. Frequent, phylogenetically local horizontal transfer of the cox1 group I Intron in flowering plant mitochondria. Mol. Biol. Evol. 2008;25:1762–77. doi: 10.1093/molbev/msn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer S, De Vries O, Dudler R, Wessels J, Honegger R. Interfacial self-assembly of fungal hydrophobins of the lichen-forming ascomycetes Xanthoria parietina and X. ectaneoides. Fungal Genet. Biol. 2000;30:81–93. doi: 10.1006/fgbi.2000.1205. [DOI] [PubMed] [Google Scholar]
- Simon D, Moline J, Helms G, Friedl T, Bhattacharya D. Divergent histories of rDNA group I introns in the lichen family Physciaceae. J. Mol. Evol. 2005;60:434–46. doi: 10.1007/s00239-004-0152-2. [DOI] [PubMed] [Google Scholar]
- Sogin M, Ingold A, Karlok M, Nielsen H, Engberg J. Phylogenetic evidence for the acquisition of ribosomal RNA introns subsequent to the divergence of some of the major Tetrahymena groups. EMBO J. 1986;5:3625–30. doi: 10.1002/j.1460-2075.1986.tb04691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4 Sinauer Associates; Sundarland, Massachusetts, USA: 2003. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–99. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lectures on Mathematics in the Life Sciences (American Mathematical Society) 1986;17:57–86. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Cote V, Otis C, Mercier JP, Gray MW, Lonergan KM, Lemieux C. Evolutionary transfer of ORF containing Group-I introns between different subcellular compartments (chloroplast and mitochondrion) Mol. Biol. Evol. 1995;12:533–45. doi: 10.1093/oxfordjournals.molbev.a040234. [DOI] [PubMed] [Google Scholar]
- Vaughn JC, Mason MT, Sperwhitis GL, Kuhlman P, Palmer JD. Fungal origin by horizontal transfer of a plant mitochondrial Group-I intron in the chimeric CoxI gene of Peperomia. J. Mol. Evol. 1995;41:563–72. doi: 10.1007/BF00175814. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]