Abstract
Although microbial culture remains the gold standard for diagnosis of many ocular infections, the technique is limited by low yield, inability to detect certain organisms, and potentially long delays to results. DNA-based molecular diagnostic techniques use detection of specific nucleic acid sequences as evidence for presence of suspected pathogens. The polymerase chain reaction (PCR) is a powerful molecular biology technique that allows for detection of fewer than 10 copies of pathogen genome. Recent technical advances in PCR have permitted quantitation of pathogen load using quantitative PCR (qPCR), and have permitted multiplexing of primer sets. Use of pan-bacterial and pan-fungal primers for ribosomal DNA sequences has allowed diagnosis of bacterial and fungal infections using molecular techniques. In this review, we highlight recent advances in the application of PCR to the diagnosis of anterior segment and posterior segment ocular infectious diseases.
Keywords: Ocular infection, uveitis, polymerase chain reaction, Acanthamoeba, keratitis, retinitis, Toxoplasmosis, glaucomatocyclitic crisis
Introduction
Microbial culture has been the mainstay of diagnosis of infectious disease since the first pure bacterial cultures were produced by Koch in the 1880’s. However, culture techniques suffer from a number of limitations that reduce their utility. Culture techniques are relative insensitive. Yields for clearly infectious processes such as postoperative endophalmitis (70% [1]) and corneal ulcer (~55–60% [2,3]) are relatively poor. Cultures can be very slow, taking up to 2 weeks for fastidious organisms such as Propionibacterium acnes. Culture for certain organisms, such as Acanthamoeba require complicated conditions and suffer from low yield. Viruses are difficult to culture; diagnosis often depends on clinical presentation, cytopathologic changes, or serologic evidence of prior infection.
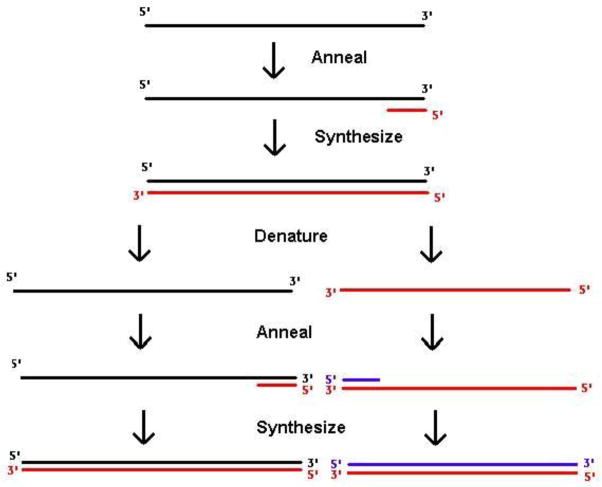
The polymerase chain reaction (PCR) is a molecular biologic technique for detection and analysis of specific DNA sequences. In PCR (Figure 1), two short DNA oligonucleotides complementary to sequence in the organism of interest are used to amplify the intervening DNA using a thermostable DNA polymerase. The resulting DNA fragment may be analyzed by gel electrophoresis to document the presence of DNA from the suspected pathogen, or sequenced to confirm its presence.
Figure 1.
Schematic of the polymerase chain reaction. DNA to be detected (black) is incubated with oligonucleotide primers (red and blue) complementary to sequences in the suspected pathogen. Hybridization of primers allows DNA synthesis by a thermostable polymerase. PCR is an exponential process; after N rounds, 2N copies of the starting material will be generated in theory.
The original PCR technique has had significant utility in the diagnosis of ocular infectious disease, particularly for posterior uveitis such as acute retinal necrosis and ocular toxoplasmosis. However, the original PCR technique had its own significant drawbacks. The presence of inhibitors of DNA polymerase in biopsy samples produced false negative results [4,5]. The high sensitivity of PCR made false-positive detection of commensal DNA common [6]. Samples needed to be run independently, limiting the number of organisms that could be tested from sparse clinical material. Bacterial and fungal diseases were difficult to diagnose by PCR because of the large number of primer sets potentially required to account for all organisms in a differential diagnosis.
Technical advances in PCR over the last decade have largely eliminated these limitations. Techniques borrowed from forensic science have allowed purification of DNA free of inhibitors from minute samples [7]. Real-time, or quantitative PCR (qPCR) uses real-time detection of fluorescent amplification products to allow quantitation of starting material [8]. Multiplex PCR allows running of multiple primer sets simultaneously, which permits more organisms to be tested from a single starting biopsy [9]. Conserved sequences within bacteria (16S) and fungi (5.8S/18S/28S) allow for pan-bacterial and pan-fungal PCR to be performed from single primer sets [3]. With these improvements, the utility of PCR has increased and application has widened. In this review, we highlight recent uses of PCR in anterior segment and posterior segment ocular infectious disease.
Utility of PCR in Evaluation of Anterior Segment Disease
PCR in Microbial Keratitis
Several recent studies support the use of PCR in the diagnosis of microbial keratitis. PCR offers speed and sensitivity relative to culture; however, a disadvantage of PCR is its high rate of false positive error from commensal contaminants or dead bacteria. PCR also offers the ability to rapidly differentiate bacterial and fungal ulcers, which cannot readily be achieved by clinical inspection [10]. Eleinen et al. [11] studied 88 patients with infectious corneal ulcers, performing corneal scrapings and subjecting the samples to potassium hydroxide wet mount, Gram stain, bacterial culture, fungal culture, and broad-range PCR with primer pairs targeted to the 16S (bacterial) and 18S (fungal) rRNA genes. For bacterial keratitis, the sensitivity of culture was found to be 57.58%, while PCR sensitivity was 87.88%. For fungal keratitis, the sensitivity of potassium hydroxide wet mount was 65.91%, culture was 59.09%, and PCR was 90.91%. In a prospective cohort study, Kim et al. [3] evaluated a total of 108 consecutive corneal ulcers by culture and pan-bacterial and pan-fungal PCR, as well as 87 air swab controls. Microbial culture identified an organism in 56% of cases. The majority of the culture-positive ulcers were found to be PCR-positive, but the yield was higher in fungal (95%) vs. bacterial (76%) cases. There was a high concordance of PCR speciation with culture results for fungal ulcers, with 89% agreement. There was a lower concordance rate for bacterial ulcers, however, with only 63% agreement. This was felt to be secondary to detection of normal ocular flora, which are primarily bacterial and rarely fungal. In culture-negative ulcers, potential bacterial or fungal pathogens were identified in 88% by PCR. In another prospective study, Enbong et al. [2] studied 30 cases of presumed fungal keratitis. Positivity for fungus was found in 20% of potassium hydroxide wet mounts, 40% of cultures, and 93.3% of PCR. There was a high concordance between fungal culture and PCR of 91.7%. PCR was also found to be positive for fungus in 88.9% of culture-negative cases. Tananuvat et al. [12] similarly reported a sensitivity of 90.9% and a specificity of 94.7% of fungal PCR in microbial keratitis. In aggregate, these results suggest PCR techniques have high sensitivity for detection of bacterial and particularly fungal pathogens and is particularly useful for diagnosis of fungal keratitis.
Several recent studies have looked at methods to improve diagnosis of fungal keratitis by PCR. Kuo et al. [13] used a dot hybridization assay in order to speciate fungal ribosomal DNA quickly. The sensitivity of detecting fungal keratitis by this method was 100%, and the specificity was 96.7%. Goldschmidt et al. [14] similarly described a real-time PCR high-resolution melting analysis that is able to detect yeasts and filamentous fungi, differentiate them from each other, and discriminate amongst different species of yeast, all in one run. This method was found to be 100% sensitive and specific for culture-positive fungal keratitis and positive in 7 out of 10 culture-negative cases of suspected fungal keratitis.
PCR in Acanthamoeba Keratitis
Acanthamoeba is a free-living protozoan parasite which is ubiquitous in the environment and is able to cause keratitis in immune-competent individuals. Acanthamoeba is particularly difficult to diagnose, as its clinical signs can be confused with herpetic or fungal keratitis, and the use of direct microscopy and culture can often miss the diagnosis due to poor sensitivity. PCR, however, has demonstrated a much higher sensitivity than direct microscopy and culture in making the diagnosis of Acanthamoeba keratitis. Lehmann et al. [15], demonstrated that PCR was not only able to detect Acanthamoeba in 84% of corneal epithelial samples (compared with 53% for culture), but was also able to detect it from tear samples in 66% of cases (although with the caveat that presence of topical anesthetics may reduce sensitivity of PCR [16], and DNA preparation [17, 18] and amplification [19–21] techniques should be optimized). Other series have described the diagnosis of Acanthamoeba keratitis by PCR in culture-negative cases, as well as the utility of PCR in confirming confocal microscopy findings [22,23]. PCR may also be used to quantify the copy number of Acanthamoeba in a sample [24–26]. Ikeda et al. [25] studied 29 eyes with Acanthamoeba keratitis and found that the Acanthamoeba copy number at the first visit correlated with the clinical stage of the keratitis, and both the copy number and stage were significant risk factors for a poor outcome. Also, in cases with a poor outcome, the DNA copy number was not reduced by more than 90% after 1 month of treatment.
PCR in viral conjunctivitis and keratitis
Viral infections of the ocular surface, such as conjunctivitis and keratitis, are usually diagnosed clinically, as confirmatory testing by the gold standard of viral isolation with tissue culture often has low-yield and is time-consuming. However, not all cases of ocular surface viral infections are easy to diagnose clinically and may be confused with bacterial infections.
Elnifro et al. [27] tested a multiplex PCR for the detection of adenovirus, herpes simplex virus (HSV), and chlamydia trachomatis in 805 conjunctival swabs from patients with suspected viral or chlamydial conjunctivitis. This multiplex PCR was prospectively compared to the corresponding uniplex PCRs, virus isolation, and an immunoassay technique. The multiplex PCR was found to have a sensitivity of 98% for adenovirus, 92% for HSV, and 100% for chlamydia. The sensitivity of adenoviral isolation by cell culture was 53%, HSV isolation by cell culture was 94.5%, and chlamydial immune dot blot test was 71%. Another comparison of the techniques of adenoviral isolation by cell culture, direct detection of adenoviral antigens by direct fluorescent assay (DFA), and adenoviral PCR in conjunctival swabs during an outbreak of epidemic keratoconjunctivitis (EKC). The sensitivity of adenoviral isolation was 51.1%, DFA was 48.8%, and PCR was 62.1%. Although the DFA was the most rapid test, PCR was the most sensitive and could yield results in approximately 24 hours [28]. Weitgasser et al. [29] evaluated conjunctival swabs from 15 patients with adenoviral conjunctivitis and found that 14 of the samples were positive when screened by direct immunofluorescence and all were positive by PCR. Not only is PCR sensitive in diagnosing viral conjunctivitis, it has also been demonstrated as capable of identifying the serotypes of adenoviruses that cause keratoconjunctivitis [30,31].
PCR has also been shown to be valuable in diagnosing HSV keratitis, with the sensitivity from corneal scrapings reported to be from 70–100% and specificity ranging from 67.9% to greater than 95% [32–34]. Although the sensitivity of PCR in detecting HSV is higher with corneal scrapings compared to tear samples, tear samples should be considered in cases where corneal scraping is not feasible, such as in stromal keratitis [35–37]. HSV PCR may be considered in cases of anterior segment inflammation of unclear etiology. In one study, 17% of cases of anterior segment inflammation in which HSV was not suspected showed HSV positivity by PCR [38].
The quantification of HSV by PCR has also been shown to have useful applications. Although previous studies have suggested that active viral replication is not related to persistent epithelial defects and progressive HSV keratitis, Fukuda et al demonstrated high HSV DNA copy numbers by PCR in persistent epithelial defects [39]. Another group used HSV DNA copy number to identify patients with HSV keratitis not responding to treatment due to acyclovir resistance [40].
HSV PCR has also been used to test corneal explants of transplant recipients, as well as the corneoscleral rim of donors. This has not only helped diagnose HSV in failed corneal grafts where HSV was not suspected, but also has identified patients at risk of developing HSV keratitis by donor-to-host transmission [41–44].
One question that has been posed is whether the asymptomatic shedding of HSV on the ocular surface can lead to false-positive results by PCR. Kaufman et al studied 50 asymptomatic patients and found that 98% of them shed HSV DNA in their tears or saliva over a 30-day course [45]. For this reason, Leigh et al retrospectively evaluated 206 corneal samples submitted for HSV PCR analysis, and 11.2% tested positive for HSV. Of these positive samples, 95.7% of them met clinical criteria for HSV keratitis, suggesting that asymptomatic ocular shedding of HSV does not contribute to a significant rate of false positive results [46]. Such studies highlight the utility of quantitative PCR to determine viral load.
Although HSV is a common cause of keratitis, another study investigated keratitis of unknown etiology and found a small percentage to be caused by CMV. Kandori et al. [47] evaluated 78 patients with epithelial, stromal, and endothelial keratitis of unknown etiology by CMV PCR and found that 24.1% of cases of corneal edema (endothelial keratitis) were caused by CMV, and 62.5% of these eyes had clinical improvement with appropriate treatment of the CMV. None of the cases of epithelial and stromal keratitis tested positive for CMV.
PCR in Glaucoumatocyclitic Crisis (Posner-Schlossman Syndrome)
Glaucomatocyclitic crisis is characterized by episodic anterior uveitis with raised intraocular pressure that is out of proportion to the degree of anterior chamber reaction. Recent evidence suggests that at least a subset of this disorder may be viral in etiology [48,49]. Chee et al. [50] found that 22.8% of patients with anterior uveitis associated with elevated intraocular pressure were positive for CMV by PCR analysis of aqueous samples. Of these CMV-positive patients, 75% had presented with clinical signs consistent with glaucomatocyclitic crisis, and 20.8% presented as Fuchs heterchromic iridocyclitis. Another study by Chee and Jap [51] evaluated aqueous samples of 103 eyes with presumed glaucomatocyclitic crisis or Fuchs heterochromic iridocyclitis by PCR for CMV. Of the patients with presumed glaucomatocyclitic crisis, 52.2% were CMV positive, and of the patients with Fuchs heterchromic iridocyclitis, 41.7% were CMV positive. They evaluated the clinical characteristics of these patients to determine if there were any differences between the CMV-positive and CMV-negative patients; however, no clinical differences could be discerned.
Another prospective study of 53 patients with glaucoumatocyclitic crisis found that 14 had aqueous samples positive for CMV; the remainder were CMV negative. Although all of the patients with glaucomatocyclitic crisis had elevated aqueous chemokine concentration, there was no difference in cytokine expression between the CMV-positive and CMV-negative patients. The authors suggested that further testing is needed to determine if CMV is truly the cause of the inflammation in this disorder [52]. A smaller study evaluated aqueous samples of 7 patients with glaucomatocyclitic crisis and found 5 of these samples to be positive for CMV by PCR. Two of these patients were treated with oral antiviral therapy, and relapses occurred in these cases after cessation of treatment, suggesting that CMV is a potential cause of glaucomatocyclitic crisis and that oral valganciclovir may be an effective treatment [53].
Utility of molecular techniques for posterior segment disease
A common indication for performing diagnostic PCR for posterior segment disease is in the presence of significant media opacity from dense vitritis or cataract and other situations where diagnoses may be challenging such as between viral retinitis and ocular toxoplasmosis.
Viral Retinitis
Historically, posterior segment uveitis was the initial condition where PCR was first used in ophthalmic disease. Knox et al. [54] performed PCR on 38 vitreous samples from 37 patients with atypical posterior uveitis. They identified viral DNA in 24 eyes and, of significance, none of the PCR negative eyes had a clinical course that was consistent with a viral retinitis. Since that time, the use of PCR for diagnosis has become relatively routine. A small sample of aqueous (as little as 10 microliters) from the anterior chamber can be used to detect HSV, VZV, CMV and toxoplasma gondii DNA [55,56]. Additionally quantitative PCR may be useful in monitoring disease and treatment response [57]. PCR has sensitivities greater than 90% for VZV, HSV and CMV and specificities greater than 95% for these viruses [58–63] with faster results than viral culture. PCR is also useful for subtypes of viral infections, for example ARN can be caused by HSV type 1, HSV type 2 or VZV. PCR can also be used in the management of resistant viral strains. CMV can acquire ganciclovir resistance such as in the UL97 polymerase gene, which will affect management decisions. Such mutations are readily detected by PCR [64–67].
Ocular Toxoplasmosis
Atypical toxoplasmosis can be difficult to diagnose based on clinical examination alone, as it can mimic other infectious retinitides particularly in immunosuppressed patients [68,69] [70]. The highly repeated B2 gene of Toxoplasma gondii confers high sensitivity to PCR for this parasite [71,72]. PCR for toxoplasmosis is most useful from vitreous biopsy but some organisms may be found in anterior chamber as well [72,73]. Recent advances have permitted quantitative PCR for ocular toxoplasmosis, demonstrating that many cases have high copy numbers (> 106/mL) [74]. For atypical toxoplasmosis, PCR of the aqueous may be complementary with Goldmann-Witmer coefficient (GWC) analysis [56, 75]. Several studies have investigated combined PCR and GWC for the diagnosis of toxoplasmic retinochoroiditis and found that PCR and GWC together had 80–93% sensitivity for detection with specificity of ~93% [76, 77]. The results of PCR in vitreous samples are more likely to be positive if the same is taken near the onset of symptoms, the eye is inflamed, and the patient has not received any anti-Toxoplasma medication, while GWC takes longer to become positive but also remains positive longer. Yields of PCR-amplifiable DNA become poor as the eye becomes quiet [78].
PCR has been used to demonstrate that there are at least three major strain subtypes of Toxoplasma gondii [79–81]. Each subtype has different susceptibility to antibiotic treatment and virulence in animal models. There is still controversy as to which strains are most commonly associated with ocular toxoplasmosis [82,83]. It is possible that knowledge of the strain (obtainable only by PCR) may help guide therapy in the future.
Endophthalmitis
Endophthalmitis is a condition where PCR results can immediately impact disease management. In the Endophthalmitis Vitrectomy Study, 30% of cases were culture negative [84]. In the initial paper examining use of PCR in post-operative endophthalmitis, Okrhavi et al. [85] demonstrated detectable 16S bacterial sequences in all culture-negative samples. Therese et al. [86] showed that PCR correlated with 100% of culture positive and 44% of culture negative specimens. In this study, 1/3 of the culture negative and PCR negative to eubacterial 16S primers were positive for fungal sequence by PCR.
The French Institutional Endophthalmitis Study (FRIENDS) group evaluated 16S PCR compared to conventional cultures to detect and identify bacterial agent causing acute postcataract endophthalmitis in 100 patients [87]. This group found minimal difference in detection between cultures and PCR when comparing both aqueous and vitreous. However, after treatment, PCR still was able to detect bacteria in 70% compared to 9% by culture, likely due to persistence of DNA from dead organisms.
Due to low pathogen levels and fastidiousness, definitive diagnosis of delayed onset endophthalmitis can be difficult. Lohmann et al. [88] collected 25 samples with the clinical diagnosis of delayed onset postoperative infectious endophthalmitis from the aqueous and vitreous. For aqueous samples, diagnostic culture yield was 0% but was 84% for PCR, principally Propionibacterium acnes. For vitreous samples, the pathogen was identified in 92% of eyes by PCR compared to 24% by diagnostic culture. PCR is also useful in suspected fungal endophthalmitis, as fungi may require up to 3 weeks for growth. In one large study, 24 of 43 intraocular specimens were positive for fungal disease by microscopy and culture while 32 were positive by PCR. PCR increased the sensitivity of detection by 18.6% [89].
Ocular Tuberculosis
Ocular involvement occurs in 1–2% of patients with tuberculosis. Mycobacteria have a wide clinical spectrum which can make diagnosis difficult. PCR has been used to detect Mycobacterium tuberculosis genome in ocular fluids [90–92]. However, Arora et al. [91] reported that PCR was quite specific but was positive in only 37% of cases of presumed TB and the positivity related to the load of organisms. The low sensitivity of PCR from ocular fluids in M. tuberculosis could be due to low bacterial load in the fluid and thick cell wall of the bacterium [93]. Recently, PCR has been used to demonstrate the presence of M. tuberculosis DNA in a majority of samples from patients with Eales disease, often at high copy number [94].
Integrated use of PCR for broad-spectrum testing in anterior and posterior segment disease
With recent technical advances, as well as increased scope of application, PCR has become a useful tool in the clinic for diagnosis of ocular infectious disease. Harper et al. [55] reviewed their clinical use of PCR in 143 aqueous and vitreous samples, finding PCR could identify a pathogen is 77 of 95 patients. This group found their positive predictive value was 98.7% and negative predictive value was 67.9%, suggesting high utility for PCR. Indeed, clinical treatment was changed for 26 of the 95 patients on the basis of PCR testing. Thompson et al. [95] reviewed a large series of ocular samples tested by PCR over a decade. A total of 3,056 samples were tested; PCR results were positive more often than culture for herpes simplex, varicella zoster, Chlamydia, and Acanthamoeba. Recently, Sugita et al. [96] created a generalizable, comprehensive protocol for testing for most known viruses, Toxoplasmosis, bacteria, and fungi from single samples. This group found the positive predictive value of testing to be 98.6%, and negative predictive value of 92.4%.
Future directions
One drawback of current PCR techniques is the necessity of ‘knowing what you are looking for’. PCR cannot identify unexpected organisms in the way that culture can. Deep DNA sequencing for pathogen detection allows detection of all foreign DNA within a sample, and may allow discovery of novel pathogens [97,98]. Techniques utilizing biome representative sequencing [99] in particular have the potential to rapidly and cheaply identify all known and unknown organism in a biopsy. Such techniques will likely expand the range of molecular biology in the diagnosis of ocular inflammatory and infectious disease in the future.
Conclusion
PCR is now an extremely useful test for the diagnosis of ocular inflammatory disease, and may be considered (along with culture) as a first-line test for many infectious conditions in both anterior and posterior segments.
Acknowledgments
This work supported by NIH grant 1R01EY022038 to RVG, the Burroughs-Wellcome Translational Scientist Award to RVG, and an unrestricted grant from Research to Prevent Blindness.
Footnotes
Conflict of Interest
Parisa Taravati, Deborah Lam, Russell N. Van Gelder declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Contributor Information
Parisa Taravati, Email: taravati@uw.edu.
Deborah Lam, Email: deblam@uw.edu.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Results of the Endophthalmitis Vitrectomy Study. A randomized trial of immediate vitrectomy and of intravenous antibiotics for the treatment of postoperative bacterial endophthalmitis. Endophthalmitis Vitrectomy Study Group. Arch Ophthalmol. 1995;113:1479–1496. [PubMed] [Google Scholar]
- 2.Embong Z, Wan Hitam WH, Yean CY, et al. Specific detection of fungal pathogens by 18S rRNA gene PCR in microbial keratitis. BMC Ophthalmol. 2008;8:7. doi: 10.1186/1471-2415-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim E, Chidambaram JD, Srinivasan M, et al. Prospective comparison of microbial culture and polymerase chain reaction in the diagnosis of corneal ulcer. Am J Ophthalmol. 2008;146:714–723. 723, e711. doi: 10.1016/j.ajo.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Wiedbrauk DL, Werner JC, Drevon AM. Inhibition of PCR by aqueous and vitreous fluids. J Clin Microbiol. 1995;33:2643–2646. doi: 10.1128/jcm.33.10.2643-2646.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milman T, Mirani N, Gibler T, et al. Actinomyces israelii endogenous endophthalmitis. Br J Ophthalmol. 2008;92:427–428. doi: 10.1136/bjo.2007.123596. [DOI] [PubMed] [Google Scholar]
- 6.Morris DJ, Bailey AS, Cooper RJ, et al. Polymerase chain reaction for rapid detection of ocular adenovirus infection. J Med Virol. 1995;46:126–132. doi: 10.1002/jmv.1890460208. [DOI] [PubMed] [Google Scholar]
- 7.Hedman J, Radstrom P. Overcoming inhibition in real-time diagnostic PCR. Methods Mol Biol. 2013;943:17–48. doi: 10.1007/978-1-60327-353-4_2. [DOI] [PubMed] [Google Scholar]
- 8.Dworkin LL, Gibler TM, Van Gelder RN. Real-time quantitative polymerase chain reaction diagnosis of infectious posterior uveitis. Arch Ophthalmol. 2002;120:1534–1539. doi: 10.1001/archopht.120.11.1534. [DOI] [PubMed] [Google Scholar]
- 9.Dabil H, Boley ML, Schmitz TM, Van Gelder RN. Validation of a diagnostic multiplex polymerase chain reaction assay for infectious posterior uveitis. Arch Ophthalmol. 2001;119:1315–1322. doi: 10.1001/archopht.119.9.1315. [DOI] [PubMed] [Google Scholar]
- 10.Dalmon C, Porco TC, Lietman TM, et al. The clinical differentiation of bacterial and fungal keratitis: a photographic survey. Invest Ophthalmol Vis Sci. 2012;53:1787–91. doi: 10.1167/iovs.11-8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11❖.Eleinen KG, Mohalhal AA, Elmekawy HE, et al. Polymerase chain reaction-guided diagnosis of infective keratitis - a hospital-based study. Curr Eye Res. 2012;37:1005–1011. doi: 10.3109/02713683.2012.698357. In this study, the authors subjected corneal scrapings from 88 patients to routine culture, Gram stain, and KOH stain as well as 16S and 18S PCR. The majority of samples were positive for fungal or bacterial pathogens. Results were available within 4–8 hours compared with days for culture. [DOI] [PubMed] [Google Scholar]
- 12❖.Tananuvat N, Salakthuantee K, Vanittanakom N, et al. Prospective comparison between conventional microbial work-up vs PCR in the diagnosis of fungal keratitis. Eye (Lond) 2012;26:1337–1343. doi: 10.1038/eye.2012.162. The authors tested 30 samples of suspected fungal keratitis by PCR and culture. 28 samples were positive by PCR compared with 40% by culture and 20% by smear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo MT, Chang HC, Cheng CK, et al. A highly sensitive method for molecular diagnosis of fungal keratitis: a dot hybridization assay. Ophthalmology. 2012;119:2434–2442. doi: 10.1016/j.ophtha.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 14.Goldschmidt P, Degorge S, Benallaoua D, et al. New strategy for rapid diagnosis and characterization of keratomycosis. Ophthalmology. 2012;119:945–950. doi: 10.1016/j.ophtha.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann OJ, Green SM, Morlet N, et al. Polymerase chain reaction analysis of corneal epithelial and tear samples in the diagnosis of Acanthamoeba keratitis. Invest Ophthalmol Vis Sci. 1998;39:1261–1265. [PubMed] [Google Scholar]
- 16.Goldschmidt P, Rostane H, Saint-Jean C, et al. Effects of topical anaesthetics and fluorescein on the real-time PCR used for the diagnosis of Herpesviruses and Acanthamoeba keratitis. Br J Ophthalmol. 2006;90:1354–1356. doi: 10.1136/bjo.2006.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laummaunwai P, Ruangjirachuporn W, Boonmars T. A simple PCR condition for detection of a single cyst of Acanthamoeba species. Parasitol Res. 2012;110:1569–1572. doi: 10.1007/s00436-011-2662-3. [DOI] [PubMed] [Google Scholar]
- 18.Goldschmidt P, Degorge S, Saint-Jean C, et al. Resistance of Acanthamoeba to classic DNA extraction methods used for the diagnosis of corneal infections. Br J Ophthalmol. 2008;92:112–115. doi: 10.1136/bjo.2007.125898. [DOI] [PubMed] [Google Scholar]
- 19.Yera H, Zamfir O, Bourcier T, et al. Comparison of PCR, microscopic examination and culture for the early diagnosis and characterization of Acanthamoeba isolates from ocular infections. Eur J Clin Microbiol Infect Dis. 2007;26:221–224. doi: 10.1007/s10096-007-0268-6. [DOI] [PubMed] [Google Scholar]
- 20.Boggild AK, Martin DS, Lee TY, et al. Laboratory diagnosis of amoebic keratitis: comparison of four diagnostic methods for different types of clinical specimens. J Clin Microbiol. 2009;47:1314–1318. doi: 10.1128/JCM.00173-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldschmidt P, Degorge S, Benallaoua D, et al. New tool for the simultaneous detection of 10 different genotypes of Acanthamoeba available from the American Type Culture Collection. Br J Ophthalmol. 2009;93:1096–1100. doi: 10.1136/bjo.2008.144873. [DOI] [PubMed] [Google Scholar]
- 22.Kandori M, Inoue T, Takamatsu F, et al. Two cases of acanthamoeba keratitis diagnosed only by real-time polymerase chain reaction. Cornea. 2010;29:228–231. doi: 10.1097/ICO.0b013e3181a39020. [DOI] [PubMed] [Google Scholar]
- 23.Mathers WD, Nelson SE, Lane JL, et al. Confirmation of confocal microscopy diagnosis of Acanthamoeba keratitis using polymerase chain reaction analysis. Arch Ophthalmol. 2000;118:178–183. doi: 10.1001/archopht.118.2.178. [DOI] [PubMed] [Google Scholar]
- 24.Itahashi M, Higaki S, Fukuda M, et al. Utility of real-time polymerase chain reaction in diagnosing and treating acanthamoeba keratitis. Cornea. 2011;30:1233–1237. doi: 10.1097/ICO.0b013e3182032196. The authors studied six cases of presumed Acanthamoeba keratis. [DOI] [PubMed] [Google Scholar]
- 25❖.Ikeda Y, Miyazaki D, Yakura K, et al. Assessment of real-time polymerase chain reaction detection of Acanthamoeba and prognosis determinants of Acanthamoeba keratitis. Ophthalmology. 2012;119:1111–1119. doi: 10.1016/j.ophtha.2011.12.023. The authors studied 29 eyes with Acanthamoeba keratitis and 75 with bacterial keratitis. The detection of amoebic DNA was 50 times more sensitive by real-time PCR. Copy number of Acanthamoeba at presentation was correlated with outcome. [DOI] [PubMed] [Google Scholar]
- 26.Khairnar K, Tamber GS, Ralevski F, Pillai DR. Comparison of molecular diagnostic methods for the detection of Acanthamoeba spp. from clinical specimens submitted for keratitis. Diagn Microbiol Infect Dis. 2011;70:499–506. doi: 10.1016/j.diagmicrobio.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Elnifro EM, Cooper RJ, Klapper PE, et al. Multiplex polymerase chain reaction for diagnosis of viral and chlamydial keratoconjunctivitis. Invest Ophthalmol Vis Sci. 2000;41:1818–1822. [PubMed] [Google Scholar]
- 28.Percivalle E, Sarasini A, Torsellini M, et al. A comparison of methods for detecting adenovirus type 8 keratoconjunctivitis during a nosocomial outbreak in a Neonatal Intensive Care Unit. J Clin Virol. 2003;28:257–264. doi: 10.1016/s1386-6532(03)00011-8. [DOI] [PubMed] [Google Scholar]
- 29.Weitgasser U, Haller EM, El-Shabrawi Y. Evaluation of polymerase chain reaction for the detection of adenoviruses in conjunctival swab specimens using degenerate primers in comparison with direct immunofluorescence. Ophthalmologica. 2002;216:329–332. doi: 10.1159/000066175. [DOI] [PubMed] [Google Scholar]
- 30.Tabbara KF, Omar N, Hammouda E, et al. Molecular epidemiology of adenoviral keratoconjunctivitis in Saudi Arabia. Mol Vis. 2010;16:2132–2136. [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura M, Hirano E, Kowada K, et al. Surveillance of adenovirus D in patients with epidemic keratoconjunctivitis from Fukui Prefecture, Japan, 1995–2010. J Med Virol. 2012;84:81–86. doi: 10.1002/jmv.22252. [DOI] [PubMed] [Google Scholar]
- 32.El-Aal AM, El Sayed M, Mohammed E, et al. Evaluation of herpes simplex detection in corneal scrapings by three molecular methods. Curr Microbiol. 2006;52:379–382. doi: 10.1007/s00284-005-0289-2. [DOI] [PubMed] [Google Scholar]
- 33.Subhan S, Jose RJ, Duggirala A, et al. Diagnosis of herpes simplex virus-1 keratitis: comparison of Giemsa stain, immunofluorescence assay and polymerase chain reaction. Curr Eye Res. 2004;29:209–213. doi: 10.1080/02713680490504911. [DOI] [PubMed] [Google Scholar]
- 34.Farhatullah S, Kaza S, Athmanathan S, et al. Diagnosis of herpes simplex virus-1 keratitis using Giemsa stain, immunofluorescence assay, and polymerase chain reaction assay on corneal scrapings. Br J Ophthalmol. 2004;88:142–144. doi: 10.1136/bjo.88.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35❖.Satpathy G, Mishra AK, Tandon R, et al. Evaluation of tear samples for Herpes Simplex Virus 1 (HSV) detection in suspected cases of viral keratitis using PCR assay and conventional laboratory diagnostic tools. Br J Ophthalmol. 2011;95:415–418. doi: 10.1136/bjo.2010.191049. The authors compare direct HSV antigen detection, viral culture, and PCR from tears and corneal scrapings. They find that PCR from corneal scrapings was most sensitive (36%) but fair yield was detected in tears (13.9%) [DOI] [PubMed] [Google Scholar]
- 36.Khodadoost MA, Sabahi F, Behroz MJ, et al. Study of a polymerase chain reaction-based method for detection of herpes simplex virus type 1 DNA among Iranian patients with ocular herpetic keratitis infection. Jpn J Ophthalmol. 2004;48:328–332. doi: 10.1007/s10384-004-0081-z. [DOI] [PubMed] [Google Scholar]
- 37.Pramod NP, Thyagarajan SP, Mohan KV, Anandakannan K. Polymerase chain reaction in the diagnosis of herpetic keratitis: experience in a developing country. Can J Ophthalmol. 2000;35:134–140. doi: 10.1016/s0008-4182(00)80006-x. [DOI] [PubMed] [Google Scholar]
- 38.Kakimaru-Hasegawa A, Kuo CH, Komatsu N, et al. Clinical application of real-time polymerase chain reaction for diagnosis of herpetic diseases of the anterior segment of the eye. Jpn J Ophthalmol. 2008;52:24–31. doi: 10.1007/s10384-007-0485-7. [DOI] [PubMed] [Google Scholar]
- 39.Fukuda M, Deai T, Higaki S, et al. Presence of a large amount of herpes simplex virus genome in tear fluid of herpetic stromal keratitis and persistent epithelial defect patients. Semin Ophthalmol. 2008;23:217–220. doi: 10.1080/08820530802111366. [DOI] [PubMed] [Google Scholar]
- 40.Hlinomazova Z, Loukotova V, Horackova M, Sery O. The treatment of HSV1 ocular infections using quantitative real-time PCR results. Acta Ophthalmol. 2012;90:456–460. doi: 10.1111/j.1755-3768.2010.01933.x. [DOI] [PubMed] [Google Scholar]
- 41.van Gelderen BE, Van der Lelij A, Treffers WF, van der Gaag R. Detection of herpes simplex virus type 1, 2 and varicella zoster virus DNA in recipient corneal buttons. Br J Ophthalmol. 2000;84:1238–1243. doi: 10.1136/bjo.84.11.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimomura Y, Deai T, Fukuda M, et al. Corneal buttons obtained from patients with HSK harbor high copy numbers of the HSV genome. Cornea. 2007;26:190–193. doi: 10.1097/ICO.0b013e31802eaee6. [DOI] [PubMed] [Google Scholar]
- 43.Remeijer L, Duan R, van Dun JM, et al. Prevalence and clinical consequences of herpes simplex virus type 1 DNA in human cornea tissues. J Infect Dis. 2009;200:11–19. doi: 10.1086/599329. [DOI] [PubMed] [Google Scholar]
- 44.Robert PY, Adenis JP, Denis F, et al. Herpes simplex virus DNA in corneal transplants: prospective study of 38 recipients. J Med Virol. 2003;71:69–74. doi: 10.1002/jmv.10454. [DOI] [PubMed] [Google Scholar]
- 45.Kaufman HE, Azcuy AM, Varnell ED, et al. HSV-1 DNA in tears and saliva of normal adults. Invest Ophthalmol Vis Sci. 2005;46:241–247. doi: 10.1167/iovs.04-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leigh JF, Acharya N, Cevallos V, Margolis TP. Does asymptomatic shedding of herpes simplex virus on the ocular surface lead to false-positive diagnostic PCR results? Br J Ophthalmol. 2008;92:435–436. doi: 10.1136/bjo.2007.122150. [DOI] [PubMed] [Google Scholar]
- 47.Kandori M, Inoue T, Takamatsu F, et al. Prevalence and features of keratitis with quantitative polymerase chain reaction positive for cytomegalovirus. Ophthalmology. 2010;117:216–222. doi: 10.1016/j.ophtha.2009.06.059. [DOI] [PubMed] [Google Scholar]
- 48.Teoh SB, Thean L, Koay E. Cytomegalovirus in aetiology of Posner-Schlossman syndrome: evidence from quantitative polymerase chain reaction. Eye (Lond) 2005;19:1338–1340. doi: 10.1038/sj.eye.6701757. [DOI] [PubMed] [Google Scholar]
- 49.Kongyai N, Sirirungsi W, Pathanapitoon K, et al. Viral causes of unexplained anterior uveitis in Thailand. Eye (Lond) 2012;26:529–534. doi: 10.1038/eye.2011.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chee SP, Bacsal K, Jap A, et al. Clinical features of cytomegalovirus anterior uveitis in immunocompetent patients. Am J Ophthalmol. 2008;145:834–840. doi: 10.1016/j.ajo.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 51.Chee SP, Jap A. Presumed fuchs heterochromic iridocyclitis and Posner-Schlossman syndrome: comparison of cytomegalovirus-positive and negative eyes. Am J Ophthalmol. 2008;146:883–889. e881. doi: 10.1016/j.ajo.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 52❖.Li J, Ang M, Cheung CM, et al. Aqueous cytokine changes associated with Posner-Schlossman syndrome with and without human cytomegalovirus. PLoS One. 2012;7:e44453. doi: 10.1371/journal.pone.0044453. The authors examine cytokine profiles of the aqueous in 53 patients with glaucomatocyclitic crisis, of whom 13 were positive for CMV DNA by PCR. The authors found no difference in cytokines between PCR-positive and – negative cases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodier-Bonifas C, Cornut PL, Billaud G, Lina B, et al. Cytomegalovirus research using polymerase chain reaction in Posner-Schlossman syndrome. J Fr Ophtalmol. 2011;34:24–29. doi: 10.1016/j.jfo.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 54.Knox CM, Chandler D, Short GA, Margolis TP. Polymerase chain reaction-based assays of vitreous samples for the diagnosis of viral retinitis. Use in diagnostic dilemmas. Ophthalmology. 1998;105:37–44. doi: 10.1016/s0161-6420(98)71127-2. discussion 44–35. [DOI] [PubMed] [Google Scholar]
- 55❖.Harper TW, Miller D, Schiffman JC, Davis JL. Polymerase chain reaction analysis of aqueous and vitreous specimens in the diagnosis of posterior segment infectious uveitis. Am J Ophthalmol. 2009;147:140–147. e142. doi: 10.1016/j.ajo.2008.07.043. This group reviewed their clinical use of PCR in 143 aqueous and vitreous samples, finding PCR could identify a pathogen is 77 of 95 patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rothova A, de Boer JH, Ten Dam-van Loon NH, et al. Usefulness of aqueous humor analysis for the diagnosis of posterior uveitis. Ophthalmology. 2008;115:306–311. doi: 10.1016/j.ophtha.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 57.Yin PD, Kurup SK, Fischer SH, et al. Progressive outer retinal necrosis in the era of highly active antiretroviral therapy: successful management with intravitreal injections and monitoring with quantitative PCR. J Clin Virol. 2007;38:254–259. doi: 10.1016/j.jcv.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 58.Abe T, Sato M, Tamai M. Correlation of varicella-zoster virus copies and final visual acuities of acute retinal necrosis syndrome. Graefes Arch Clin Exp Ophthalmol. 1998;236:747–752. doi: 10.1007/s004170050153. [DOI] [PubMed] [Google Scholar]
- 59.Abe T, Tsuchida K, Tamai M. A comparative study of the polymerase chain reaction and local antibody production in acute retinal necrosis syndrome and cytomegalovirus retinitis. Graefes Arch Clin Exp Ophthalmol. 1996;234:419–424. doi: 10.1007/BF02539407. [DOI] [PubMed] [Google Scholar]
- 60.Ganatra JB, Chandler D, Santos C, et al. Viral causes of the acute retinal necrosis syndrome. Am J Ophthalmol. 2000;129:166–172. doi: 10.1016/s0002-9394(99)00316-5. [DOI] [PubMed] [Google Scholar]
- 61.McCann JD, Margolis TP, Wong MG, et al. A sensitive and specific polymerase chain reaction-based assay for the diagnosis of cytomegalovirus retinitis. Am J Ophthalmol. 1995;120:219–226. doi: 10.1016/s0002-9394(14)72610-8. [DOI] [PubMed] [Google Scholar]
- 62.Short GA, Margolis TP, Kuppermann BD, et al. A polymerase chain reaction-based assay for diagnosing varicella-zoster virus retinitis in patients with acquired immunodeficiency syndrome. Am J Ophthalmol. 1997;123:157–164. doi: 10.1016/s0002-9394(14)71031-1. [DOI] [PubMed] [Google Scholar]
- 63.Mitchell SM, Fox JD, Tedder RS, et al. Vitreous fluid sampling and viral genome detection for the diagnosis of viral retinitis in patients with AIDS. J Med Virol. 1994;43:336–340. doi: 10.1002/jmv.1890430404. [DOI] [PubMed] [Google Scholar]
- 64.Hu H, Jabs DA, Forman MS, et al. Comparison of cytomegalovirus (CMV) UL97 gene sequences in the blood and vitreous of patients with acquired immunodeficiency syndrome and CMV retinitis. J Infect Dis. 2002;185:861–867. doi: 10.1086/339603. [DOI] [PubMed] [Google Scholar]
- 65.Jabs DA, Martin BK, Forman MS, et al. Mutations conferring ganciclovir resistance in a cohort of patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis. J Infect Dis. 2001;183:333–337. doi: 10.1086/317931. [DOI] [PubMed] [Google Scholar]
- 66.Liu W, Kuppermann BD, Martin DF, et al. Mutations in the cytomegalovirus UL97 gene associated with ganciclovir-resistant retinitis. J Infect Dis. 1998;177:1176–1181. doi: 10.1086/515293. [DOI] [PubMed] [Google Scholar]
- 67.Jabs DA, Martin BK, Ricks MO, et al. Detection of ganciclovir resistance in patients with AIDS and cytomegalovirus retinitis: correlation of genotypic methods with viral phenotype and clinical outcome. J Infect Dis. 2006;193:1728–1737. doi: 10.1086/504270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holland GN, Muccioli C, Silveira C, et al. Intraocular inflammatory reactions without focal necrotizing retinochoroiditis in patients with acquired systemic toxoplasmosis. Am J Ophthalmol. 1999;128:413–420. doi: 10.1016/s0002-9394(99)00300-1. [DOI] [PubMed] [Google Scholar]
- 69.Ronday MJ, Ongkosuwito JV, Rothova A, Kijlstra A. Intraocular anti-Toxoplasma gondii IgA antibody production in patients with ocular toxoplasmosis. Am J Ophthalmol. 1999;127:294–300. doi: 10.1016/s0002-9394(98)00337-7. [DOI] [PubMed] [Google Scholar]
- 70.Hazan A, Patel RM, Levinson D, et al. Atypical bilateral Toxoplasma retinochoroiditis in a bone marrow transplant patient with negative serum titers. J Ophthalmic Inflamm Infect. 2013;3:23. doi: 10.1186/1869-5760-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones CD, Okhravi N, Adamson P, et al. Comparison of PCR detection methods for B1, P30, and 18S rDNA genes of T. gondii in aqueous humor. Invest Ophthalmol Vis Sci. 2000;41:634–644. [PubMed] [Google Scholar]
- 72.Montoya JG, Parmley S, Liesenfeld O, et al. Use of the polymerase chain reaction for diagnosis of ocular toxoplasmosis. Ophthalmology. 1999;106:1554–1563. doi: 10.1016/S0161-6420(99)90453-0. [DOI] [PubMed] [Google Scholar]
- 73.Garweg JG, de Groot-Mijnes JD, Montoya JG. Diagnostic approach to ocular toxoplasmosis. Ocul Immunol Inflamm. 2011;19:255–261. doi: 10.3109/09273948.2011.595872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sugita S, Ogawa M, Inoue S, et al. Diagnosis of ocular toxoplasmosis by two polymerase chain reaction (PCR) examinations: qualitative multiplex and quantitative real-time. Jpn J Ophthalmol. 2011;55:495–501. doi: 10.1007/s10384-011-0065-8. [DOI] [PubMed] [Google Scholar]
- 75.Fardeau C, Romand S, Rao NA, et al. Diagnosis of toxoplasmic retinochoroiditis with atypical clinical features. Am J Ophthalmol. 2002;134:196–203. doi: 10.1016/s0002-9394(02)01500-3. [DOI] [PubMed] [Google Scholar]
- 76.Talabani H, Asseraf M, Yera H, et al. Contributions of immunoblotting, real-time PCR, and the Goldmann-Witmer coefficient to diagnosis of atypical toxoplasmic retinochoroiditis. J Clin Microbiol. 2009;47:2131–2135. doi: 10.1128/JCM.00128-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fekkar A, Bodaghi B, Touafek F, et al. Comparison of immunoblotting, calculation of the Goldmann-Witmer coefficient, and real-time PCR using aqueous humor samples for diagnosis of ocular toxoplasmosis. J Clin Microbiol. 2008;46:1965–1967. doi: 10.1128/JCM.01900-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okhravi N, Jones CD, Carroll N, et al. Use of PCR to diagnose Toxoplasma gondii chorioretinitis in eyes with severe vitritis. Clin Experiment Ophthalmol. 2005;33:184–187. doi: 10.1111/j.1442-9071.2005.00992.x. [DOI] [PubMed] [Google Scholar]
- 79.Grigg ME, Ganatra J, Boothroyd JC, Margolis TP. Unusual abundance of atypical strains associated with human ocular toxoplasmosis. J Infect Dis. 2001;184:633–639. doi: 10.1086/322800. [DOI] [PubMed] [Google Scholar]
- 80.Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis. 1995;172:1561–1566. doi: 10.1093/infdis/172.6.1561. [DOI] [PubMed] [Google Scholar]
- 81.Saeij JP, Boyle JP, Boothroyd JC. Differences among the three major strains of Toxoplasma gondii and their specific interactions with the infected host. Trends Parasitol. 2005;21:476–481. doi: 10.1016/j.pt.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 82.Switaj K, Master A, Borkowski PK, et al. Association of ocular toxoplasmosis with type I Toxoplasma gondii strains: direct genotyping from peripheral blood samples. J Clin Microbiol. 2006;44:4262–4264. doi: 10.1128/JCM.01786-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fekkar A, Ajzenberg D, Bodaghi B, et al. Direct genotyping of Toxoplasma gondii in ocular fluid samples from 20 patients with ocular toxoplasmosis: predominance of type II in France. J Clin Microbiol. 2011;49:1513–1517. doi: 10.1128/JCM.02196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Han DP, Wisniewski SR, Wilson LA, et al. Spectrum and susceptibilities of microbiologic isolates in the Endophthalmitis Vitrectomy Study. Am J Ophthalmol. 1996;122:1–17. doi: 10.1016/s0002-9394(14)71959-2. [DOI] [PubMed] [Google Scholar]
- 85.Okhravi N, Adamson P, Carroll N, et al. PCR-based evidence of bacterial involvement in eyes with suspected intraocular infection. Invest Ophthalmol Vis Sci. 2000;41:3474–3479. [PubMed] [Google Scholar]
- 86.Therese KL, Anand AR, Madhavan HN. Polymerase chain reaction in the diagnosis of bacterial endophthalmitis. Br J Ophthalmol. 1998;82:1078–1082. doi: 10.1136/bjo.82.9.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chiquet C, Cornut PL, Benito Y, et al. Eubacterial PCR for bacterial detection and identification in 100 acute postcataract surgery endophthalmitis. Invest Ophthalmol Vis Sci. 2008;49:1971–1978. doi: 10.1167/iovs.07-1377. [DOI] [PubMed] [Google Scholar]
- 88.Lohmann CP, Linde HJ, Reischl U. Improved detection of microorganisms by polymerase chain reaction in delayed endophthalmitis after cataract surgery. Ophthalmology. 2000;107:1047–1051. doi: 10.1016/s0161-6420(00)00083-x. discussion 1051–1042. [DOI] [PubMed] [Google Scholar]
- 89.Anand A, Madhavan H, Neelam V, Lily T. Use of polymerase chain reaction in the diagnosis of fungal endophthalmitis. Ophthalmology. 2001;108:326–330. doi: 10.1016/s0161-6420(00)00517-0. [DOI] [PubMed] [Google Scholar]
- 90.Gupta V, Arora S, Gupta A, et al. Management of presumed intraocular tuberculosis: possible role of the polymerase chain reaction. Acta Ophthalmol Scand. 1998;76:679–682. doi: 10.1034/j.1600-0420.1998.760609.x. [DOI] [PubMed] [Google Scholar]
- 91.Arora SK, Gupta V, Gupta A, et al. Diagnostic efficacy of polymerase chain reaction in granulomatous uveitis. Tuber Lung Dis. 1999;79:229–233. doi: 10.1054/tuld.1999.0210. [DOI] [PubMed] [Google Scholar]
- 92.Biswas J, Therese L, Madhavan HN. Use of polymerase chain reaction in detection of Mycobacterium tuberculosis complex DNA from vitreous sample of Eales’ disease. Br J Ophthalmol. 1999;83:994. doi: 10.1136/bjo.83.8.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yeh S, Sen HN, Colyer M, et al. Update on ocular tuberculosis. Curr Opin Ophthalmol. 2012;23:551–556. doi: 10.1097/ICU.0b013e328358ba01. [DOI] [PubMed] [Google Scholar]
- 94❖.Singh R, Toor P, Parchand S, et al. Quantitative polymerase chain reaction for Mycobacterium tuberculosis in so-called Eales’ disease. Ocul Immunol Inflamm. 2012;20:153–157. doi: 10.3109/09273948.2012.658134. The authors apply Mycobacterial PCR to 88 patients, of whom 28 were diagnosed with Eales’ disease, and find 57% positive for PCR, with copy numbers ranging from 104 to 106/mL. [DOI] [PubMed] [Google Scholar]
- 95.Thompson PP, Kowalski RP. A 13-year retrospective review of polymerase chain reaction testing for infectious agents from ocular samples. Ophthalmology. 2011;118:1449–1453. doi: 10.1016/j.ophtha.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 96❖.Sugita S, Ogawa M, Shimizu N, et al. Use of a Comprehensive Polymerase Chain Reaction System for Diagnosis of Ocular Infectious Diseases. Ophthalmology. 2013 doi: 10.1016/j.ophtha.2013.02.020. The authors employ a comprehensive PCR strategy essentially testing for all known common PCR-detectable pathogens from single samples, and find positive predictive and negative predictive values each in excess of 90%. [DOI] [PubMed] [Google Scholar]
- 97.Tripathy S, Jiang RH. Massively parallel sequencing technology in pathogenic microbes. Methods Mol Biol. 2012;835:271–294. doi: 10.1007/978-1-61779-501-5_17. [DOI] [PubMed] [Google Scholar]
- 98.Gui J, Patel IR. Recent advances in molecular technologies and their application in pathogen detection in foods with particular reference to yersinia. J Pathog. 2011;2011:310135. doi: 10.4061/2011/310135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muthappan V, Lee AY, Lamprecht TL, et al. Biome representational in silico karyotyping. Genome Res. 2011;21:626–633. doi: 10.1101/gr.115758.110. [DOI] [PMC free article] [PubMed] [Google Scholar]